Abstract
Significant growth phase-dependent differences were noted in the transcriptome of the hyperthermophilic bacterium Thermotoga maritima when it was cocultured with the hyperthermophilic archaeon Methanococcus jannaschii. For the mid-log-to-early-stationary-phase transition of a T. maritima monoculture, 24 genes (1.3% of the genome) were differentially expressed twofold or more. In contrast, methanogenic coculture gave rise to 292 genes differentially expressed in T. maritima at this level (15.5% of the genome) for the same growth phase transition. Interspecies H2 transfer resulted in three- to fivefold-higher T. maritima cell densities than in the monoculture, with concomitant formation of exopolysaccharide (EPS)-based cell aggregates. Differential expression of specific sigma factors and genes related to the ppGpp-dependent stringent response suggests involvement in the transition into stationary phase and aggregate formation. Cell aggregation was growth phase dependent, such that it was most prominent during mid-log phase and decayed as cells entered stationary phase. The reduction in cell aggregation was coincidental with down-regulation of genes encoding EPS-forming glycosyltranferases and up-regulation of genes encoding β-specific glycosyl hydrolases; the latter were presumably involved in hydrolysis of β-linked EPS to release cells from aggregates. Detachment of aggregates may facilitate colonization of new locations in natural environments where T. maritima coexists with other organisms. Taken together, these results demonstrate that syntrophic interactions can impact the transcriptome of heterotrophs in methanogenic coculture, and this factor should be considered in examining the microbial ecology in anaerobic environments.
Despite the fact that microorganisms interact significantly within their ecological niche, this factor is usually not taken into account in the context of microbial physiology. One key limitation in this regard is that methodologies for examining the influence of one microbial species on another are typically based on either identification (e.g., 16S rRNA phylogeny [46]) or enumeration (e.g., fluorescent in situ hybridization [12]). Additional insights into functional outcomes of interspecies interactions are difficult to obtain. Yet such information is needed to relate pure culture cellular physiology to the beneficial and antagonistic elements present in microbial ecosystems.
Syntrophic relationships between microorganisms with complementary growth physiologies are a key part of microbial ecosystems. One such example in certain anaerobic niches is the association between fermentative H2 producers and methanogenic H2 consumers, whereby the inhibitory H2 formed as the by-product of sugar or peptide metabolism serves as an energy source for the generation of methane (26). This syntrophy can be found in niches ranging from the mammalian digestive tract (24) to anaerobic digesters used for domestic waste treatment (12). Molecular hydrogen is also a key chemical species in hydrothermal environments (1) and likely supports this form of syntrophy; indeed, the pairing of hyperthermophilic fermentative anaerobes and methanogens in laboratory cocultures leads to higher growth rates and higher biomass yields of heterotrophs compared to monocultures (5). Furthermore, the growth rates of certain hyperthermophilic methanogens appear to be contingent upon the supply of available H2 that can be maximized through close spatial proximity with fermentative anaerobes (26).
The hyperthermophilic bacterium Thermotoga maritima grows heterotrophically by fermenting a variety of carbohydrates, producing H2 as an autoinhibitory by-product (16). In syntrophic coculture with a hydrogenotrophic methanogen, such as Methanococcus jannaschii (7), this inhibition is relieved and cell population density is enhanced through the interspecies transfer of hydrogen (26). In addition to the utilization of sugars as carbon and energy sources (8, 28), T. maritima also has been shown to produce exopolysaccharides (EPS) that serve as the basis for biofilms (35, 36). In fact, when grown to high cell densities through coculture with the hyperthermophilic methanogen M. jannaschii, T. maritima produces EPS that leads to formation of stable cellular aggregates, which presumably facilitate interspecies H2 transfer (26). Cellular aggregation was found to occur initially during mid-log phase in high-cell-density cocultures, apparently driven, at least in part, by quorum sensing and cyclic di-GMP regulation (18, 37). Calcofluor staining indicated that exopolysaccharide was produced in pure cultures but at a level much lower than that in the coculture, with aggregation noted only in the coculture (18). Carbohydrate analysis of the aggregates indicated that the exopolysaccharide consisted of a polymer of ribose, glucose, and mannose and was likely beta linked due to the positive calcofluor staining (18). Transcriptional profiling of exponential-phase T. maritima present in cell aggregates indicated an increase in uptake of extracellular sugars compared to that in the pure culture at the same stage of growth. However, sugar uptake by T. maritima in the exponential-phase coculture was not coupled to glycolysis and compatible solute formation but rather was directed toward the production of EPS (18).
The response of the T. maritima transciptome during the transition into, and existence in, stationary phase in both pure and syntrophic cultures has not been examined. In general, the transition from log to stationary phase and transitions within stationary phase follow different trajectories for microorganisms that depend on the causative agents that decelerate growth, such as growth substrate limitation (29), accumulation of inhibitory by-products (5), and imposition of certain forms of stress (43). The onset of stationary phase is known to be related to the action of specific regulators, such as sigma factors, that establish stationary-phase phenotypes involving changes in morphology, virulence, and survival strategy (29). Indeed, transcriptional analysis of microorganisms driven into stationary phase by nutrient depletion has highlighted the importance of global regulators, such as σH, RpoS, and guanosine-3′,5′-(bis)pyrophosphate (ppGpp), in limiting cellular processes and promoting scavenging and other survival activities (6, 43).
The influence of interspecies interactions is typically not considered when examining growth phase transitions but clearly can have a significant impact on microbial physiology. To examine this issue, a cDNA microarray-based functional genomics approach was used to determine the influence of interspecies H2 transfer on growth phase-related transcriptional phenotypes of T. maritima in coculture with M. jannaschii. It is clear from the results of this study that markedly different physiological and transcriptional states arise from interspecies interactions that are variable both between and within growth phases. While the focus of this work is on hyperthermophilic microorganisms, the findings point out the importance of ecological relationships in examining microbial physiology in anaerobic environments.
MATERIALS AND METHODS
Growth of microorganisms.
T. maritima strain MSB8 and M. jannaschii strain DSM2661 were grown at 80°C on medium (BSMII) containing: 40 g/liter sea salts, 5 g/liter tryptone, 3 g/liter yeast extract, 3 g/liter maltose, 3 g/liter piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES), 0.5 mg/liter NaSeO3 · 5H2O, 0.5 mg/liter NiCl2 · 6H2O, 0.25 g/liter NH4Cl, 10 mg/liter Fe(NH4)2(SO4)2 · 6H2O, 1.25 g/liter sodium acetate anhydrate, 1 ml/liter 1% Resazurin, 10 ml/liter Wolfe's trace elements, and 10 ml/liter Wolfe's trace vitamins solution (all from Sigma Aldrich; components of Wolfe's vitamin and mineral solutions can be found in ATCC medium no. 1343). The medium was adjusted to pH 7.0, autoclaved for sterility, and stored at 4°C until use. For H2 inhibition experiments, 400 ml of medium was dispensed into a 1-liter glass jar sealed with a rubber septum (Corning, Corning, NY) and heated to 80°C. The medium was then sparged through a syringe needle with the appropriate gas (100% N2 or 80% H2-20% CO2) for 1 minute and reduced with 4 ml of a 10% sodium sulfide-10% l-cysteine HCl solution (pH 8.0) (Sigma-Aldrich). A 1% inoculum of log-phase cells was used. Culture bottles were agitated at 100 rpm in an oil bath at 80°C. To study the effects of hydrogen removal, three cases were examined in 400-ml batch culture. The first case was a reference culture grown with an enclosed headspace initially filled with nitrogen. The second case tested the effect of N2 sparging on T. maritima monocultures by having two syringe needles inserted through the bottle septum, the first of which was supplied with 250 ml per minute of nitrogen and the second of which was used as a vent. For the coculture of T. maritima growth with M. jannaschii, both species were inoculated simultaneously into a bottle with an initial atmosphere of 80% H2-20% CO2 (2 bars). Duplicate cultures for all conditions were harvested at mid-log phase and pooled with equal amounts of RNA.
For growth phase experiments, T. maritima was grown with and without M. jannaschii at 80°C in a 16-liter Microgen sterilizable-in-place fermentor (New Brunswick Scientific). Ten liters of nonsterile BSMII medium adjusted to pH 7.0 was charged to the fermentor, which was sterilized in place by heating to 100°C for 20 min. The fermentor was then cooled to 80°C and stirred at 500 rpm with three Rushton-style impellors for agitation. Temperature was controlled by utilizing the sterilization mode on the fermentor, which heats with steam through an internal heat exchanger. Once at 80°C, sparging was started using N2 fed at 0.25 liter/min to maintain anaerobic conditions. The medium was reduced by the addition of 60 ml of 10% sodium sulfide through a septum, and 200 ml of inoculum was then added via a growth flask fitted with a dip tube. Anaerobic inoculum (200 ml of either T. maritima or T. maritima/M. jannaschii coculture) was forced into the fermentor via positive pressure with nitrogen gas through an addition port on the fermentor. Sparging was not utilized after inoculation in the coculture experiment in order to prevent removal of hydrogen essential for methanogen growth. Samples from both the pure culture and the coculture were taken at mid-log, early stationary, and late stationary phases, after completing duplicate trial runs under each growth condition to determine accurate growth curves.
Fluorescence microscopy.
Cells were enumerated by fixing culture samples with glutaraldehyde (Sigma Aldrich; 0.25% final concentration) for 5 min and then staining the cells by adding small aliquots (∼10 μl) to 5 ml of 0.05% acridine orange (Sigma) and filtering the cells onto 0.2-μm polycarbonate filter membranes (Osmonics), with the goal of having 100 to 300 cells in the field of view. Ten replicate counts were completed using an eyepiece grid (Nikon). Error analysis yielded a 15% error associated with counting 100 to 300 cells per field of view. To capture images, an epifluorescence microscope (Nikon) with appropriate filter sets (Southern Micro Instruments, Marietta, GA) that was fitted with a digital camera (Spot system; Diagnostic Instruments Inc.) was utilized.
Gas chromatography.
Headspace gases were analyzed using a Gow-Mac 400 series gas chromatogram outfitted with a thermal conductivity detector and an 8-ft by 1/8-in. HayeSep Q 80/100 mesh column (Gow-Mac). Standard mixed gases were used for calibration (National Welders), with a bridge current of 150 mA; oven and detector temperatures of 90°C and 105°C, respectively; and helium as the carrier gas at a flow rate of 30 ml/min. Cultures used for headspace analysis were sparged with helium in place of nitrogen during initial reduction.
Microarray protocols.
A T. maritima microarray was synthesized from PCR products as discussed previously (8), representing over 99% of the open reading frames in the sequenced genome (28). For each RNA extraction, 400-ml samples were harvested, split into four 100-ml samples, and quickly cooled to 0°C by immersion in ice water. The harvested cells were then immediately centrifuged at 13,000 × g (25 min at 4°C). The supernatant was discarded, and the RNA was extracted from cell pellets, from which cDNA was synthesized, labeled through indirect incorporation, and hybridized to the microarray by the methods described previously (38). The experimental designs for the gene expression studies were completed following recommended guidelines (20) by utilizing independent loops for both the hydrogen removal study (three-condition loop) and the comparison of growth phases (six-condition loop). Microarray slides were scanned using the appropriate laser power to balance the Cy3 and Cy5 signals with a Perkin-Elmer ExpressLite scanner (Perkin-Elmer). Raw, uncorrected quantitation of spots was completed using ScanArray (Perkin-Elmer), and data analysis was completed as discussed previously (8), utilizing a mixed-model analysis-of-variance-based analysis to generate both least-squared mean expression data and Bonferroni-corrected (47) fold change data. Unless otherwise noted, gene annotation is from the COG database at NCBI (44).
The cross-hybridization of M. jannaschii labeled cDNA with the T. maritima microarray was expected to be minimal, since M. jannaschii comprised a small fraction of the coculture and the genomes for these two organisms show less than 80% identity at the nucleotide level (32, 48). To verify this, RNA from M. jannaschii was labeled using the protocols explained above and tested for cross-hybridization by hybridizing the labeled samples to the T. maritima microarray. No cross-hybridization of the M. jannaschii RNA was detected on the scanned slides at full laser power.
Independent confirmation with real-time PCR.
A biological repeat of both the pure and coculture growth curve experiments was completed, with selected genes analyzed for expression in both replicate experiments. The following genes and primers were utilized: TM0767 (5′-CGCATTGCCACAAGAAAGTA-3′ and 5′-CCCGTGTTCACATAGGGAAT-3′), TM1451 (5′-AGGCTTGTTGTCAGTATAGCCA-3′ and 5′-CTACCGCCTTCAGAAGTCCTAT-3′), TM1598 (5′-GATGTCGTTCAGGATGTGTTTT-3′ and 5′-GACATTCACGGCTATCCTGTAG-3′),TM1662 (5′-TTGCGTACTCCACTACAGGAAC-3′ and 5′-ACGATGAGATCGACCCTTTTAT-3′), and TM1834 (5′-AGGAGCAGATTCTGAAATAGGC-3′ and 5′-GGACGGATTTTCTTTGATGAAC-3′). cDNA for each growth condition and replicate was synthesized using Superscript III (Invitrogen). Real-time PCRs were carried out with iQ SYBR Green Supermix (Bio-Rad) with 20-μl reaction mixtures containing both primers at a concentration of 250 nM, using the iCycler iQ real-time PCR detection system (Bio-Rad). Template cDNA was added to the test reaction mixtures at 2.4 ng per reaction, and a standard curve was completed for each gene represented using cDNA added in fourfold serial dilutions in the range from 0.15 ng to 38.4 ng of cDNA. The PCR efficiency and cycle thresholds were calculated using the iCycler iQ software (Bio-Rad), and the fold changes were calculated by the method of Pffafl (33). A melt curve analysis was completed and confirmed that homogeneous PCR products were generated with no nonspecific amplification. Calculated fold changes from real-time PCR for both biological repeats were in agreement with the microarray results (Table 1).
TABLE 1.
Confirmation of microarray-based transcriptional results for the transition from mid-log to early stationary phase with real-time PCR complemented with an independent biological repeat
| Gene | Annotation | Fold changea
|
|||||
|---|---|---|---|---|---|---|---|
| Coculture
|
Pure culture
|
||||||
| Biological repeat 1, real-time PCR | Biological repeat 2, real-time PCR | Biological repeat 1, array results | Biological repeat 1, real-time PCR | Biological repeat 2, real-time PCR | Biological repeat 1, array results | ||
| TM0767 | Maltosyltransferase | −1.8 | −2.6 | −3.3 | −1.1 | −1.0 | NCb |
| TM1451 | RNA polymerase, sigma A subunit | 3.3 | 2.2 | 2.0 | 1.6 | 1.1 | NC |
| TM1598 | RNA polymerase, sigma E subunit | 6.6 | 4.0 | 5.0 | 1.3 | 1.8 | NC |
| TM1662 | SurE stationary-phase survival protein | 1.0 | −1.3 | NC | −1.2 | 1.1 | NC |
| TM1834 | Alpha-glucosidase | −1.3 | −1.9 | NC | −1.1 | −1.1 | NC |
All microarray-deduced fold changes are significant based on the Bonferroni correction. Real-time PCR data analysis was conducted as previously described (33).
NC, no change.
RESULTS AND DISCUSSION
The objective of this study was to examine transcriptional and physiological changes with respect to growth phase and population density in T. maritima as a consequence of syntrophic coculture with M. jannaschii. A whole genome cDNA microarray was used to interrogate samples of T. maritima obtained at different stages of growth in both pure culture and coculture. The fact that no significant cross-hybridization of the T. maritima microarray with M. jannaschii was noted facilitated the functional genomics approach described here.
Transition from log to stationary phase in T. maritima pure culture and coculture.
Cell densities (Fig. 1B), epifluorescence micrographs (Fig. 1A), and volcano plots (Fig. 1C) are shown for T. maritima pure culture and coculture with M. jannaschii as a function of growth phase, comparing mid-log phase, early stationary phase, and late stationary phase. T. maritima was indeed inhibited in growth by the presence of hydrogen and could not be grown with a hydrogen-pressurized headspace. While the growth rates of the pure culture and coculture were similar (Fig. 1B), the corresponding maximum cell densities were appreciably different; the coculture peaked at more than 109 cells/ml, compared to 2 × 108 to 3 × 108 cells/ml for the monoculture. The lower cell density of the T. maritima pure culture relative to the coculture was apparently more the result of H2 inhibition than nutrient limitation and could be the reason for the more abrupt transition to stationary phase in the pure culture. Gas chromatogram analysis of the headspace of stationary-phase cultures indicated that no methane is produced by T. maritima, while M. jannaschii cultures averaged 5.1% methane in their headspace, compared to 11.1% methane in M. jannaschii/T. maritima cocultures, suggesting that the coculture relationship enhances the growth of the methanogen.
FIG. 1.
Growth of T. maritima in pure culture and in coculture with M. jannaschii. (A) Epifluorescence micrographs of pure culture and coculture corresponding to the growth curves are shown from left to right. After inoculation, the pure culture grew without aggregating to a density of above 108 cells/ml before entering a prolonged stationary phase, during which time the T. maritima cell morphology changed from rods to cocci. The coculture began to aggregate once cell densities reached approximately 5 × 108 cells/ml until entering stationary phase (∼109 cells/ml). In stationary phase, cells detached from aggregates, and by 24 h they displayed coccoid morphology similar to what is seen in the pure culture during late stationary phase. (B). Growth curves for T. maritima grown in pure culture and in coculture with M. jannaschii. Error bars indicate standard deviations. (C) Volcano plots comparing expression profiles of pure T. maritima culture (top) and coculture of T. maritima with M. jannaschii (bottom) during the transition of growth phases from mid-log to early stationary phase. The x axis is the log2 fold change from mid-log to early stationary phase, and the y axis is the −log10 P value for the calculated fold change.
Genes encoding starvation response proteins, such as SurE, an acid phosphatase that acts to scavenge phosphate under conditions of phosphorus limitation (51), and known carbohydrate utilization pathways in T. maritima (8) did not respond in the transition from mid-log to early stationary phase in the pure culture. In addition, regulatory proteins, including sigma factors, were not differentially expressed during this transition. A likely cause for the onset of stationary phase in the pure culture was the buildup of inhibitory H2, bottlenecking core metabolic processes in T. maritima.
Indeed, contrasts in differential expression between a quiescent pure culture, a pure culture sparged with inert gas to remove inhibitory H2, and a coculture with M. jannaschii supported this hypothesis. ABC transporters for amino acids, peptides, and sugars were up-regulated in both the sparged culture and coculture cases relative to the quiescent culture, suggesting that hydrogen inhibition was alleviated to some extent by H2 removal (see Table S1 in the supplemental material). In the quiescent culture, compared to the other two cases, up-regulation of core metabolism genes was noted, consistent with reduced metabolic efficiency attributed to H2 inhibition and a shift to a lower redox state. Furthermore, heterotrophic thermophiles have been observed to form lactate and ethanol as by-products in the absence of elemental sulfur (45). Ethanol accumulation in T. maritima could be responsible for the observed up-regulation of σE, which is known to be responsive to thermal stress in T. maritima (34). In Escherichia coli mutants resistant to elevated ethanol concentrations, the overexpression of chaperones was found to be necessary for survival (13). Here, chaperones and protein repair-related open reading frames were expressed at the highest levels in the quiescent pure culture.
Not only was population density a distinguishing factor between pure culture and coculture, but the corresponding transcriptomes were substantially different (Fig. 1C). Transcriptional response analysis showed that 24 genes changed twofold or more in the pure culture (1.3% of the T. maritima genome) between mid-log and early stationary phases (Table 2). Of note was the down-regulation of molecular chaperones, such as GroES (eightfold) and GroEL (ninefold), presumably a reflection of decreased protein synthesis in stationary phase. In contrast to the case for the pure culture, 292 genes responded twofold or more (15.5% of the T. maritima genome) for the same phase transition in the coculture (see Table S2 in the supplemental material). Cell aggregation was extensive during mid-log phase (when cell densities reached 5 × 108 cells/ml). In mesophilic microorganisms, aggregate formation leading to biofilms can be triggered by stresses, such as starvation (10), or by a cell density-dependent response, such as quorum sensing (11). Indeed, previous work showed that population density-triggered EPS formation occurred in T. maritima cocultures during exponential phase (18). Here, however, aggregation was significantly reduced as the coculture entered stationary phase and was nonexistent in late stationary phase (Fig. 1B). The reasons for this may be related to observations that some mesophilic bacteria deliberately detach during certain growth phases to escape from the biofilm matrix (3, 19, 39). The mechanism for detachment is usually enzyme based, with specific activity towards elements comprising the biofilm (3, 19). Furthermore, microorganisms clustered together stand a greater chance of colonizing new environments than do individual cells; this is especially important in situations where syntrophic relationships exist so that deliberate detachment is advantageous compared to gradual biofilm erosion (39). A similar strategy may have been operational in the T. maritima coculture. While transporters and nutrient-scavenging genes are often more highly expressed in stationary phase due to nutrient limitation (15), this does not appear to be the case in the coculture. ABC transporters and carbohydrate-active enzymes up-regulated during the stationary-phase transition were from a broad distribution of carbohydrate-active enzymes (Table 3). Despite the fact that the growth medium was maltose based, maltose-directed ABC transporters (TM1836 andTM1839) and related glycosidases (TM1835) were down-regulated, while genes encoding enzymes for the degradation of β-glycans (e.g., TM0024, TM0070, TM1231, TM1524, and TM1752) were up-regulated during the transition to stationary phase in the coculture (Table 3). This suggests that the β-linked saccharide components of EPS produced during exponential growth may have been degraded, released, and transported back into the cell to be reused. It also appears that the onset of stationary phase for T. maritima in the coculture was not driven by nutrient limitation. For example, no differential expression was noted for the gene encoding a stationary-phase survival protein, SurE (TM1662), and as mentioned above, the maltose-specific transport and degradation genes were down-regulated in the coculture. In addition, the rich growth medium used here had relatively high carbon and nitrogen contents compared to the quantity of biomass produced. Neither cell aggregate formation nor differential expression of any carbohydrate-active enzymes and transporters was noted in the pure culture during the transition into stationary phase.
TABLE 2.
Differentially expressed genes in T. maritima during the transition from mid-log to early stationary phase in pure culture growth
| Gene | Annotation | Fold changea |
|---|---|---|
| TM_rnpB | RNAse P RNA | 3.0 |
| TM0317 | Cation transport ATPase | 2.2 |
| TM0373 | Molecular chaperone | −5.5 |
| TM0374 | Small heat shock chaperone | −3.4 |
| TM0456 | Ribosomal protein L10 | −2.0 |
| TM0504 | Predicted signaling peptide (15) | 2.2 |
| TM0505 | Cochaperonin GroES | −7.9 |
| TM0506 | Chaperonin GroEL | −8.8 |
| TM0571 | Trypsin-like serine proteases | 2.3 |
| TM0729 | ppGpp synthetases | 2.5 |
| TM0807 | Peroxiredoxin | −3.5 |
| TM0849 | DnaJ-class molecular chaperone | −2.3 |
| TM1286 | Methionine synthase II | −3.1 |
| TM1369 | Possible ABC-type transport system | 2.5 |
| TM1370 | Possible ABC-type transport system | 2.5 |
| TM1371 | Selenocysteine lyase | 2.8 |
| TM1372 | NifU homolog (Fe-S clusters) | 2.0 |
| TM1375 | Spermidine-binding periplasmic protein | 2.0 |
| TM1434 | Conserved hypothetical protein | 2.0 |
| TM1435 | Predicted coenzyme A-binding protein | 2.0 |
| TM1533 | Ferredoxin-like protein | 2.2 |
| TM1536 | Predicted membrane protein | 4.0 |
| TM1867 | Malate/lactate dehydrogenases | 2.2 |
| TM1874 | Cold shock proteins | 2.4 |
All fold changes are significant based on the Bonferroni correction.
TABLE 3.
Carbohydrate-active enzymes and ABC transporters in T. maritima that were differentially regulated during the transition from mid-log to early stationary phase in coculture with M. jannaschiia
| Gene(s) | Annotation | Fold change |
|---|---|---|
| TM0024 | Laminarinase | 3.0 |
| TM0030-31 | β-Glucan ABC transporter subunits | 2.1, 3.4 |
| TM0070 | Xylanase | 2.2 |
| TM0071 | Xylan ABC transporter subunit | 7.2 |
| TM0114 | Putative monosaccharide ABC transporter subunit | 2.5 |
| TM0123 | Metal ABC transporter subunit | 2.4 |
| TM0300-302 | Unknown ABC transporter subunits | 3.3, 2.1, 2.0 |
| TM0310 | Galactosidase | 2.2 |
| TM0418 | Maltose ABC transporter subunit | 2.2 |
| TM0430-432 | Putative pectin ABC transporter subunits | 3.1, 2.5, 4.4 |
| TM0433 | Pectate lyase | 2.4 |
| TM0533 | Unknown ABC transporter | −2.1 |
| TM0624 | F1 glycosyltransferase | 2.4 |
| TM0627 | F1 glycosyltransferase | −2.0 |
| TM0633 | Predicted GH73 | 2.6 |
| TM0752 | Glucuronidase | 2.3 |
| TM0767 | Maltosyltransferase | −3.3 |
| TM0958 | Ribose ABC transporter subunit | 2.7 |
| TM1064 | Rhamnose ABC transporter subunit | 2.0 |
| TM1068 | Glururonidase | 1.9 |
| TM1199 | Lactose ABC transporter subunit | 2.0 |
| TM1223 | β-Glucan ABC transporter subunit | 2.3 |
| TM1227 | Mannase | 1.9 |
| TM1231 | Mannosidase | 2.9 |
| TM1232-35 | Unknown ABC transporter subunits | 3.4, 2.1, 2.7, 5.3 |
| TM1524 | Endoglucanase | 2.1 |
| TM1525 | Endoglucanase | 1.8 |
| TM1650 | Amylase | 2.1 |
| TM1746-49 | Mannan ABC transporter subunits | 2.1, 2.9, 2.4, 2.1 |
| TM1751 | Endoglucanase | 2.0 |
| TM1752 | Endomannase | 2.0 |
| TM1834 | Glucosidase | NCb |
| TM1835 | Cyclomaltodextrinase | −1.9 |
| TM1836 | Maltose ABC transporter subunit | −2.3 |
| TM1839 | Maltose ABC transporter subunit | −2.8 |
| TM1840 | Amylase | 1.9 |
| TM1848 | Cellobiose phosphorylase | 2.1 |
| TM1851 | Mannosidase | 3.0 |
| TM1853-55 | Putative sugar ABC transporter subunits | 2.9, 5.7, 22.6 |
Comparisons between phase-dependent phenotypes in pure culture and coculture.
In addition to tracking differential expression for points along the growth curves for the pure culture and coculture, transcriptional contrasts between the pure culture and coculture for the equivalent points on their growth phase trajectories were examined. Thirty-five genes were differentially expressed twofold or more between the mid-log phase of the pure culture and the mid-log phase of the coculture, including 24 genes that were up-regulated in the pure culture, mostly related to central metabolism and chaperones (see Table S3 in the supplemental material). The 11 genes that were up-regulated in the coculture included those for an α-glucuronidase (41) (TM0434, +2.0-fold in coculture), an α-glucosidase (4) (TM1834, +4.6-fold in coculture), and several hypothetical proteins. Forty-four genes were differentially expressed twofold or more between late-stationary-phase pure culture and late-stationary-phase coculture, mainly central metabolism genes (see Table S3 in the supplemental material). Late-stationary-phase cells in both the pure culture and coculture were comparable in appearance, characterized by a shift in T. maritima from the rod-like morphology noted in exponential growth to a coccoid-like morphology in stationary phase, with no evidence of dividing cells. To investigate the viability of T. maritima and T. maritima/M. jannaschii coculture in extended stationary phase, both were inoculated and allowed to grow at 80°C into stationary phase. Samples were drawn from each culture every 12 h and inoculated as a 1% inoculum into fresh, sterile medium. Even after 120 h at 80°C, both cultures contained viable cells, which grew quickly (lag phase of less than 3 h) to cell densities characteristic of pure culture and cocultures.
The most significant differences between the pure culture and coculture were associated with early stationary phase, where 240 genes were differentially expressed twofold or more (see Table S3 in the supplemental material). These consisted of 127 genes expressed higher in the pure culture, of which over half were central metabolism-related genes, and 113 genes detected at higher levels in the coculture. Genes up-regulated in the stationary-phase coculture included those for a large number of ABC transporter components (30 genes in all) and carbohydrate hydrolases, including an α-glucosidase (TM1834, +3.8-fold), a pectate lyase (TM0433, +3.5-fold), an α-mannosidase (TM1231, +3.0-fold), two alpha-glucuronidases (TM0055, +2.1-fold; TM1068, +2.6-fold), two endoglucanases (TM0305, +2.0-fold; TM1751, +2.1-fold), a laminarinase (TM0024, 2.1-fold), an α-galactosidase (TM0310, +2.1-fold), and an endoxylanase (TM0061, +2.2-fold). The up-regulation of the carbohydrate hydrolases and transporters in the coculture likely reflects the processing of EPS associated with cell aggregates as discussed above.
Differential expression of regulatory proteins.
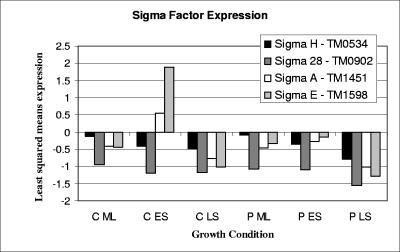
Regulatory mechanisms in T. maritima that potentially contribute to the transition between growth phases and aggregation, maturation, and detachment of cell aggregates include sigma factors, ppGpp, and cyclic di-GMP. Equally important to note are stationary-phase regulatory proteins that apparently do not exist in the T. maritima genome but are present and necessary for survival in model mesophilic microorganisms. Examples of such regulatory proteins not encoded in the T. maritima genome include starvation response regulators FadR (14) and Lrp (52), global regulators UspA (23) and RpoS (21), the programmed cell death system MazEF (2), DNA protection protein Dps (27), and the SOS polymerase system, which facilitates beneficial mutations (49). The roles of the few regulatory systems that do exist in T. maritima are likely to be particularly important for growth phase transitions and survival. Expression profiles (based on least-squares mean) of four identifiable sigma factors in the T. maritima genome (σH [TM0534], σ28 [TM0902], σA [TM1451], and σE [TM1598] [28]) were examined (Fig. 2). σH responds strongly during stationary phase in Bacillus subtilis, controlling the formation of spores and inducing the organism into stationary phase (6). Here, the σH homolog (TM0534) was not differentially expressed during or between exponential and stationary phases for either pure culture or coculture (Fig. 2). Many genes under control of σH in B. subtilis are involved in adaptation to nutrient deficiency (6). Since T. maritima was grown here in a nutrient-rich medium, the fact that σH was not differentially expressed is not surprising. In E. coli, RpoS (σS) is required for stationary-phase survival, along with a required lipoprotein (NlpD), an l-isoaspartate methyltransferase (Pcm), and a stationary-phase survival protein (SurE) (51). An RpoS ortholog has yet to be identified in T. maritima, and therefore a different sigma factor or regulatory protein likely regulates stationary-phase physiology. Putative T. maritima homologs do exist for NlpD (TM0409) and for Pcm (TM0704) and SurE (TM1662) (51). It is interesting to note that all three genes were not differentially expressed in the growth phase contrasts or in the sparged culture and coculture comparisons.
FIG. 2.
Relative expression as shown by the least-squares mean expression of putative sigma factors in T. maritima under conditions of hydrogen removal (C, coculture) compared to that of hydrogen accumulation (P, quiescent pure culture) for mid-log (ML), early-stationary (ES), and late-stationary (LS) phases transitions.
Both σA and σE were up-regulated during the transition from mid-log to early stationary phase in the coculture (Fig. 2). σE in mesophilic bacteria is often involved in cellular stress responses, such as heat shock (50), protein aggregation (50), and oxidative stress (29). σA in mesophiles is known to regulate housekeeping functions and RNA genes transcribed during active growth (6). In previous studies with T. maritima, it was noted that σA was up-regulated in biofilm cells compared to planktonic cells (35) and under conditions of heat stress (34). In this study, σA and σE both responded to H2 inhibition in the quiescent pure culture during exponential growth and were up-regulated compared to conditions of hydrogen removal (σA, +4.0-fold versus coculture and +4.3-fold versus sparged; σE, +2.1-fold versus coculture and +2.1-fold versus sparged). Up-regulation of σA and σE likely reflects a lowered metabolic efficiency and increased levels of stress in H2-inhibited T. maritima, which in the absence of sulfur may generate lactate and ethanol as by-products (45), leading to intracellular protein denaturation.
TM0902, which belongs to the COG 1191 group of σ28-related regulators, was down-regulated in both sparged culture and coculture compared to the quiescent culture (−2.5-fold for coculture and −2.4-fold for sparged pure culture). Members of this COG have been shown to induce the expression of flagellar proteins in E. coli (22). For example, the σ28-related sigma factor identified in the genome of another hyperthermophilic bacterium, Aquifex aeolicus, and closely related to TM0902 (31% identity and 60% similarity over 90% of the protein), was shown to restore the motility of an E. coli σ28 mutant (40). Higher expression of TM0902 in the pure culture perhaps indicates an attempt by T. maritima cells to move from stressful conditions. Hydrogen levels have been shown to modulate flagellum expression in M. jannaschii (25), and it could be hypothesized that a similar system may exist in T. maritima. Several chemotaxis genes were down-regulated in both the sparged culture and coculture, with a greater number down-regulated in the coculture (listed here are fold changes in coculture and sparged culture, respectively, where NC indicates less than twofold: TM0429, −3.1 and −2.8; TM0701, −2.8 and NC; TM0702, −4.5 and −2.6; TM0718, −2.2 and NC; TM0904, NC and −2.0; TM0918, −2.2 and NC; TM1428, −2.0 and NC; predicted flagellar protein TM0219, −3.1 and NC; and predicted flagellar protein TM0908, 2.1 and NC).
Secondary messengers.
Secondary messengers, such as guanosine-3′,5′-(bis)pyrophosphate and cyclic di-GMP, play a role in regulating gene expression in microorganisms by influencing the efficiency of binding of sigma factors to RNA polymerase (29-31) and modulating enzyme activity (17), respectively. ppGpp, a hormone-like nucleotide which mediates the preferential binding of sigma factors in response to the nutritional quality of the extracellular environment, is produced by two different classes of enzymes that are utilized to detect amino acid starvation (RelA), and carbon starvation or stress (SpoT) (29). T. maritima contains putative homologs to RelA (TM0729) and its accessory gene product GppA (TM0195), while a homolog to SpoT has yet to be identified. In T. maritima, TM0195 was up-regulated nearly twofold during the transition from mid-log to early stationary phase in the coculture, while TM0729 was up-regulated during the same transition in both the pure culture and coculture (+2.4-fold for coculture and +2.5-fold for pure culture), suggesting an enhanced ability to detect amino acid starvation in both cultures in stationary phase.
Cyclic di-GMP is apparently involved in controlling biofilm formation processes in T. maritima (18, 37) as well as in mesophilic bacteria, such as Acetobacter xylinum (42). Here, differential expression of cyclic di-GMP-related genes was not observed during the transition to stationary phase in either the pure culture or coculture. However, differential regulation of this system was seen during batch growth comparing the coculture to the hydrogen-accumulating condition. In particular, a putative cyclic di-GMP cyclase (TM1788) was up-regulated 4.2-fold in the coculture compared to the quiescent pure culture, while the putative cyclic di-GMP phosphodiesterase (TM1184) was down-regulated 2.6-fold under the same comparison. In the comparison between the sparged and quiescent cultures, TM1184 was down-regulated 3.4-fold. Because GGDEF domain proteins are known to respond to environmental factors for the regulation of cyclic di-GMP (17), it may be that the differential expression of these genes relates to the nutritional quality of the extracellular environment more than to growth rate or population density.
Summary.
In this study, it was shown that, when grown at high cell densities through syntrophic coculture with M. jannaschii, T. maritima cycles through aggregate formation, maturation, and detachment likely mediated by the action of carbohydrate-active enzymes and transporters. While it is likely the transition to stationary phase in the T. maritima coculture falls under the control of specific sigma factors and ppGpp regulation, the aggregation life cycle may also be affected by quorum sensing. It has been previously shown the aggregation process appears to be regulated, at least in part, by TM0504, a small peptide that induces the transcription of EPS-forming glycosyltransferases upon its accumulation in the culture supernatant (18). In this study, the expression of this gene changed very little during the transition from mid-log phase to early stationary phase in the coculture. However, it was already expressed within the top 7% of all genes in mid-log phase, suggesting that the maximum dynamic range of this sensing system may have been reached early in coculture growth.
The importance of interspecies interaction in examining microbial physiology is underlined by the results of this study. T. maritima grown in pure batch culture was driven into stationary phase at a relatively low cell density by H2 inhibition. The cell aggregation cycle and the effects of population density were seen for T. maritima only when it was grown in high-density coculture with M. jannaschii. Overall, this work illustrates the point that ecological interactions are an essential element to be considered in studying microbial physiology and that functional genomics approaches can be used to complement classical microbiological methods for this purpose.
Supplementary Material
Acknowledgments
This work was supported in part through grants from the NASA Exobiology Program, the NSF Biotechnology Program, and the DOE Energy Biosciences Program.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, M. W. W. 1990. The metabolism of hydrogen by extremely thermophilic, sulfur-dependent archaebacteria. FEMS Microbiol. Rev. 75:219-238. [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 4.Bibel, M., C. Brettl, U. Gosslar, G. Kriegshauser, and W. Liebl. 1998. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol. Lett. 158:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Bonch-Osmolovskaya, E. A., and K. O. Stetter. 1991. Interspecies hydrogen transfer in cocultures of thermophilic Archaea. Syst. Appl. Microbiol. 14:205-208. [Google Scholar]
- 6.Britton, R., P. Eichenberger, J. Gonzales-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary phase sigma factor (sigma H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 8.Chhabra, S., K. R. Shockley, S. Conners, K. Scott, R. Wolfinger, and R. Kelly. 2003. Carbohydrate induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 9.Conners, S. B., C. I. Montero, D. A. Comfort, K. R. Shockley, M. R. Johnson, S. Chhabra, and R. M. Kelly. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 187:7267-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137-140. [DOI] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.de los Reyes, F. L., D. B. Oerther, M. F. de los Reyes, M. Hernandez, and L. Raskin. 1998. Characterization of filamentous foaming in activated sludge systems using oligonucleotide hybridization probes and antibody probes. Water Sci. Technol. 37:485-493. [Google Scholar]
- 13.Echave, P., M. Esparza-Cerón, E. Cabiscol, J. Tamarit, J. Ros, J. Membrillo-Hernández, and E. C. C. Lin. 2002. DnaK dependence of mutant ethanol oxidoreductases evolved for aerobic function and protective role of the chaperone against protein oxidative damage in Escherichia coli. Proc Natl. Acad. Sci. USA 99:4626-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farewell, A., A. Diez, C. C. Di Russo, and T. Nystrom. 1996. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J. Bacteriol. 178:6443-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208-213. [DOI] [PubMed] [Google Scholar]
- 16.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp-nov represents a new genus of unique extremely thermophilic eubacteria growing up to 90 degrees C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 17.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messanger involved in modulating cell surface structures in bacteria. Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, M. R., C. I. Montero, S. B. Conners, K. R. Shockley, S. L. Bridger, and R. M. Kelly. 2004. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 55:664-674. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr, M. K., and G. A. Churchill. 2001. Experimental design for gene expression microarrays. Biostatistics 2:183-201. [DOI] [PubMed] [Google Scholar]
- 21.King, T., and T. Ferenci. 2005. Divergent roles of RpoS in Escherichia coli under aerobic and anaerobic conditions. FEMS Microbiol. Lett. 244:323-327. [DOI] [PubMed] [Google Scholar]
- 22.Kundu, T. K., S. Kusano, and A. Ishihama. 1997. Promoter selectivity of Escherichia coli RNA polymerase sigma F holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 179:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 24.Morvan, B., F. Bonnemoy, G. Fonty, and P. Gouet. 1996. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr. Microbiol. 32:129-133. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel p(H2) control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97:11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muralidharan, V., K. D. Rinker, I. S. Hirsh, E. J. Bouwer, and R. M. Kelly. 1997. Hydrogen transfer between methanogens and fermentative heterotrophs in hyperthermophilic cocultures. Biotechnol. Bioeng. 56:268-278. [DOI] [PubMed] [Google Scholar]
- 27.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, L. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 29.Nystrom, T. 2002. Aging in bacteria. Curr. Opin. Microbiol. 5:596-601. [DOI] [PubMed] [Google Scholar]
- 30.Nystrom, T. 2003. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 48:17-23. [DOI] [PubMed] [Google Scholar]
- 31.Nystrom, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, J., L. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pysz, M., D. Ward, K. Shockley, C. Montero, S. Conners, M. Johnson, and R. Kelly. 2004. Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8:209-217. [DOI] [PubMed] [Google Scholar]
- 35.Pysz, M. A., S. C. Conners, C. I. Montero, K. R. Shockley, M. R. Johnson, D. E. Ward, and R. M. Kelly. 2004. Transcriptional analysis of biofilm formation in the anaerobic hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinker, K. D., and R. M. Kelly. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 69:537-547. [DOI] [PubMed] [Google Scholar]
- 37.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shockley, K. R., D. E. Ward, S. Chhabra, S. Conners, C. Montero, and R. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studholme, D. J., and M. Buck. 2000. The alternative sigma factor 28 of the extreme thermophile Aquifex aeolicus restores motility to an Escherichia coli fliA mutant. FEMS Microbiol. Lett. 191:103-107. [DOI] [PubMed] [Google Scholar]
- 41.Suresh, C., A. A. Rus'd, M. Kitaoka, and K. Hayashi. 2002. Evidence that the putative alpha-glucosidase of Thermotoga maritima MSB8 is a pNP alpha-d-glucuronopyranoside hydrolyzing alpha-glucuronidase. FEBS Lett. 517:159-162. [DOI] [PubMed] [Google Scholar]
- 42.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatusov, R., D. Natale, I. Garkavtsev, T. Tatusova, U. Shankavaram, B. Rao, B. Kiryutin, M. Galperin, N. Fedorova, and E. Koonin. 1997. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Niel, E. W., P. A. Claassen, and A. J. M. Stams. 2003. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 81:255-262. [DOI] [PubMed] [Google Scholar]
- 46.Woese, C. R. 1998. The universal ancestor. Proc. Natl. Acad. Sci. USA 95:6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfinger, R., G. Gibson, E. Wolfinger, L. Bennet, H. Hamadeh, P. Bushel, C. Afshari, and R. Paules. 2001. Assesing gene significance from cDNA microarray expression data via mixed models. J. Comp. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 48.Wu, L. Y., D. K. Thompson, G. S. Li, R. A. Hurt, J. M. Tiedje, and J. Z. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon, S., M. Han, S. Lee, K. Jeong, and J. Yoo. 2003. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 81:753-767. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, R. G., T. Skarina, J. E. Katz, S. Beasley, A. Khachatryan, S. Vyas, C. H. Arrowsmith, S. Clarke, A. Edwards, A. Joachimiak, and A. Savchenko. 2001. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure 9:1095-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.