Abstract
The determination of cell numbers or biomass in laboratory cultures or environmental samples is usually based on turbidity measurements, viable counts, biochemical determinations (e.g., protein and lipid measurements), microscopic counting, or recently, flow cytometric analysis. In the present study, we developed a novel procedure for the sensitive quantification of microbial cells in cultures and most-probable-number series. The assay combines fluorescent nucleic acid staining and subsequent fluorescence measurement in suspension. Six different fluorescent dyes (acridine orange, DAPI [4′,6′-diamidino-2-phenylindole], ethidium bromide, PicoGreen, and SYBR green I and II) were evaluated. SYBR green I was found to be the most sensitive dye and allowed the quantification of 50,000 to up to 1.5 × 108 Escherichia coli cells per ml sample. The rapid staining procedure was robust against interference from rRNA, sample fixation by the addition of glutaric dialdehyde, and reducing agents such as sodium dithionite, sodium sulfide, and ferrous sulfide. It worked well with phylogenetically distant bacterial and archaeal strains. Excellent agreement with optical density measurements of cell increases was achieved during growth experiments performed with aerobic and sulfate-reducing bacteria. The assay offers a time-saving, more sensitive alternative to epifluorescence microscopy analysis of most-probable-number dilution series. This method simplifies the quantification of microbial cells in pure cultures as well as enrichments and is particularly suited for low cell densities.
The quantification of microbial cells in pure cultures, enrichments, and environmental samples is a key measurement in microbiology, ecology, and biotechnology. Several methods are currently applied to obtain direct (light and epifluorescence microscopy and flow cytometry) or indirect (turbidimetry, nephelometry, and biochemical determinations [17]) measures of cell density or biomass. These methods vary considerably regarding their sensitivities and time requirements. While biomass determination by turbidimetric measurement is fast and easy to perform, and therefore still frequently used, its sensitivity is rather low, and it is susceptible to interference, for example, by precipitates or cell aggregate formation. Microscopic measures, in turn, are more sensitive, but particularly in the case of epifluorescence microscopy, are more time-consuming, require some experience, and may be influenced by biases between individuals. The application of flow cytometry is an emerging technique with a high sensitivity that simplifies the counting procedure, but it needs careful setup, and the equipment is not generally available (16, 18).
Cultivation-based estimates such as plate counts and most probable numbers (MPN) are widely used, since they offer the opportunity to target particular physiological groups (e.g., sulfate reducers) and can serve as a source for the isolation of pure cultures (12, 28, 30). However, some bacterial groups exhibit little growth, such as chemolithoautotrophic ammonia oxidizers (5) and oligocarbophilic marine bacteria (11), and thus are hardly analyzable by simple turbidimetric measurements. Therefore, following the growth of these organisms requires the use of fluorescence microscopy, flow cytometry, or chemical or activity measurements (1, 5, 12, 27).
During balanced growth of microbial cultures, every cell component is supposed to change at the same rate (23). Accordingly, the culture biomass and its particular constituents, such as proteins and nucleic acids, will occur at constant ratios, although for a single cell the biomass and nucleic acid content may vary significantly during the cell cycle (6, 23). Therefore, the increase in cellular constituents such as proteins or nucleic acids is supposed to serve as a reliable marker of biomass increase during balanced microbial growth. In recent years, the determination of cellular nucleic acid contents by fluorescent staining and flow cytometry has been successfully applied (10, 16). Hence, the determination of bulk nucleic acid content in microbial cultures is likely to provide a reliable and sensitive tool for the determination of microbial growth. However, little research has focused in this direction, although fluorescence measurements have already been applied for determinations of microbial cell numbers in samples from soils, sediments (34), and pelagic environments (14, 31).
The aim of the present study was to develop a sensitive method for the quantification of microbial cells in pure and enrichment cultures that is particularly suitable for low cell densities. A simple protocol combining nucleic acid staining of fixed or untreated cultures or enrichments and subsequent measurement of fluorescence emission was developed and applied to growth experiments and most-probable-number dilution series.
MATERIALS AND METHODS
Nucleic acid dyes and staining procedures.
Stock solutions (10 mg ml−1) of acridine orange (3,6-bis[dimethylamino]acridine hydrochloride) and DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) were freshly prepared from crystalline powder by dissolving the powder in double-distilled and sterilely filtered (0.2 μm) water and were stored at 4°C. Stock solutions of ethidium bromide (2,7-diamino-10-ethyl-9-phenyl-phenanthridinium bromide; 10 mg ml−1) were prepared in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) and subsequently sterilely filtered and stored at 4°C. Stock solutions of SYBR green I and II as well as of PicoGreen were purchased from Molecular Probes (Eugene, OR), subdivided into small aliquots upon receipt, and stored in 1.5-ml reaction tubes at −20°C. All other dyes were obtained from Sigma (Deisenhofen, Germany).
Working solutions of dyes were freshly prepared each day at five times the desired final assay concentrations in sterile plastic petri dishes. Working solutions of DAPI were prepared in sterilely filtered phosphate-buffered saline (PBS) (8.0 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter; pH 7.2). All other working solutions were prepared in sterilely filtered TE buffer concentrate (200 mM Tris-HCl, 50 mM sodium EDTA, pH 8.0). Unless otherwise stated, staining experiments were performed with 200 μl of sample plus 50 μl of dye working solution in black untreated 96-well microplates (Nunc 237108; VWR International, Darmstadt, Germany).
Comparison of nucleic acid dyes.
The applicability of nucleic acid dyes was tested with suspended Escherichia coli cells. Therefore, E. coli K-12 was cultured aerobically in liquid Luria-Bertani broth (17) at 37°C on a rotary shaker. Early-stationary-phase cultures were harvested by centrifugation for 20 min at 10,000 × g (Beckman J2-HS centrifuge with a JA20 rotor). Cells were washed, resuspended in sterilely filtered (0.2-μm) vitamin-free freshwater medium (see below) to a final cell density of approximately 2 × 109 cells per ml, and stored at 4°C. The cell suspension or cell-free medium was dispensed into black microplates, stained by the addition of dye working solution, incubated at room temperature in the dark, and subsequently analyzed by fluorescence measurement (see below). For dilution series of E. coli, the cell numbers were determined exactly by epifluorescence microscopy after SYBR green I staining and checked again after fluorescence measurements. For pH optimization of each dye, the respective dye working solution was adjusted to the desired pH by the addition of HCl or NaOH prior to use. Because of the small sample volume, the final pHs in the assays were checked with narrow-range pH paper (CS series; Whatman, Maidstone, United Kingdom), and they deviated no more than 0.1 pH units.
Nucleic acid quantification.
For DNA quantification, a λ phage DNA standard (100 μg ml−1; Molecular Probes, Eugene, OR) was diluted 100-fold in sterilely filtered TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8). Aliquots were dispensed into black microplates and brought up with TE buffer to a final volume of 200 μl. Staining was performed by the addition of 50 μl SYBR green I working solution (2,000-fold dilution of the stock solution). After 5 minutes of incubation in the dark, the samples were analyzed on a fluorescence microplate reader (see below).
The quantification of rRNAs in the presence of DNA was performed with a λ phage DNA standard and E. coli rRNAs (16S and 23S rRNAs at 4 mg ml−1; Roche, Mannheim, Germany). Briefly, RNase-free TE buffer was prepared by incubation in the presence of 0.1% diethyl pyrocarbonate at 37°C overnight and subsequent autoclaving. The DNA standard (200 ng ml−1) and rRNA standard (0 to 1,600 ng ml−1) were prepared with RNase-free TE buffer. One-hundred-microliter aliquots of DNA standard were mixed with 100 μl diluted rRNA standard and 50 μl SYBR green I working solution (2,000-fold dilution of the stock solution in RNase-free concentrated TE buffer) in sterile and RNase-free microplates (Costar 3596; Corning Inc., NY). Fluorescence measurements were performed after 5 minutes of incubation in the dark.
Cultivation of pure cultures.
Pure cultures were grown in artificial seawater, brackish water, or freshwater. Artificial seawater contained (in g liter−1) NaCl (24.3), MgCl2 · 6H2O (10.0), CaCl2 · 2H2O (1.5), KCl (0.66), and Na2SO4 (4.0). One milliliter each of stock solutions of KBr (0.84 M), H3BO3 (0.4 M), SrCl2 (0.15 M), NH4Cl (0.4 M), KH2PO4 (0.04 M), and NaF (0.07 M) was added per liter of medium. Brackish water contained (in g liter−1) NaCl (5.5), MgCl2 · 6H2O (2.3), CaCl2 · 2H2O (0.34), KCl (0.15), and Na2SO4 (0.91). A 0.23-ml volume each of stock solutions of KBr (0.84 M), H3BO3 (0.4 M), SrCl2 (0.15 M), NH4Cl (0.4 M), KH2PO4 (0.04 M), and NaF (0.07 M) was added per liter of medium. Freshwater medium contained (in g liter−1) NaCl (1.0), NH4Cl (0.3), MgSO4 · 7H2O (0.025), CaCl2 · 2H2O (0.1), MgCl2 · 6H2O (0.4), and KH2PO4 (0.6). For cultures of methanogens, a slightly modified freshwater medium was used, which contained (in g liter−1) KH2PO4 (0.1), NH4Cl (0.1), NaCl (0.25), KCl (0.1), MgCl2 · 6H2O (0.31), CaCl2 · 2H2O (0.1), and resazurine (0.5 mg liter−1). All oxic media were buffered by the addition of HEPES (2.38 g liter−1) and by adjusting the pH to 7.2 with 1 M NaOH. Anoxic media were cooled under a N2/CO2 atmosphere (80/20 [vol/vol]) after being autoclaved and were supplemented with 30 ml of a sodium bicarbonate solution (1 M). The pH was adjusted to pH 7.2 with sterile 1 M HCl or NaOH, if necessary. All media received 10 ml of a sterilely filtered vitamin solution (4), 1 ml trace element solution SL10 (36) (for methanogens, SL9 was used [32]), and 0.2 ml of selenite tungstate solution (35) per liter. Anoxic media were reduced by adding separately 1.2 ml of 1 M Na2S and 0.6 ml 1 M FeSO4 per liter of medium (FeS-reduced medium) or by the addition of a few crystals of sodium dithionite to medium containing resazurine as a redox indicator (dithionite-reduced medium). The pure cultures used in the present study and the respective cultivation conditions are given in Table 1.
TABLE 1.
Bacterial and archaeal strains used for the present study
| Domain | Phylum | Strain | Cultivation conditions and substrate
|
Fluorescence/OD ratiod
|
|||
|---|---|---|---|---|---|---|---|
| Medium (oxygen status) | Temp (°C) | Substrate (concn) | Fresh cells | Fixed cells | |||
| Bacteria | Alphaproteobacteria | Ruegeria algicola ATCC 51440T | Marine (oxic) | 20 | Glucose (10 mM) | 8.06 ± 3.52 | 5.09 ± 1.55 |
| Gammaproteobacteria | Shewanella baltica DSM 9439T | Brackish (oxic) | 20 | Monomer mixa | 3.40 ± 1.34 | 3.39 ± 1.20 | |
| Oceanospirillum sp. strain GM1b | Brackish (oxic) | 20 | Acetate (100 μM) | NDc | 3.43 ± 1.15 | ||
| Deltaproteobacteria | Desulfovibrio acrylicus DSM 10141T | Marine (anoxic/FeS) | 30 | Lactate (20 mM) | 4.29 ± 0.40 | NAc | |
| Epsilonproteobacteria | Arcobacter sp. strain Na105b | Marine (oxic) | 20 | Monomer mixa | 3.02 ± 1.35 | 1.49 ± 0.41 | |
| “Firmicutes” | Desulfosporosinus orientis DSM 765T | Freshwater (anoxic/dithionite) | 28 | Lactate (10 mM) | 5.23 ± 1.20 | 8.16 ± 2.14 | |
| “Bacteroidetes” | Muricauda ruestringensis DSM 13258T | Marine (oxic) | 20 | Mannose (10 mM) | 6.93 ± 0.82 | 11.6 ± 3.79 | |
| Archaea | Euryarchaeota | Methanospirillum hungatei DSM 864T | Freshwater (anoxic/sulfide) | 30 | Acetate (10 mM), H2 | 8.31 | 9.66 |
| Methanosarcina barkeri DSM 800T | Freshwater (anoxic/sulfide) | 30 | Acetate (10 mM), H2 | 7.94 | 9.22 | ||
According to the work of Süß et al. (30).
Sass and Martens-Habbena (unpublished).
NA, was not fixed because of a possible reaction of aldehydes with sulfide. ND, not done.
Data are means ± standard deviations.
Growth experiments.
Growth experiments were performed with selected bacterial strains (Table 2). All oxic growth experiments were performed in 250-ml Erlenmeyer flasks on a rotary shaker at 90 rpm. Anoxic growth experiments were performed in 100-ml serum bottles or 100-ml screw-cap bottles. Subsamples (2 ml) were taken aseptically from the growth vessels, and the optical density at 436 nm (OD436) was immediately determined on a spectrophotometer (Shimadzu RF-1501) relative to distilled water. Samples were diluted with growth medium if the optical density exceeded 0.3. Samples were then stored in 2-ml reaction tubes on ice until fluorescence analysis. Parallel samples were fixed by the addition of glutaric dialdehyde (2% final concentration).
TABLE 2.
Comparison of growth rates of selected strains
| Phylum | Strain | Growth rate μ (d−1)a
|
||
|---|---|---|---|---|
| OD436 | FH | FX | ||
| Alphaproteobacteria | Ruegeria algicolaT | 0.15 | 0.17 | 0.17 |
| Gammaproteobacteria | Shewanella balticaT | 0.18 | 0.22 | 0.18 |
| Oceanospirillum sp. strain GM1 | 0.09 | n.d. | 0.09 | |
| Deltaproteobacteria | Desulfovibrio acrylicusT | 0.16 | 0.16 | n.d. |
| Epsilonproteobacteria | Arcobacter sp. strain NA105 | 0.13 | 0.18 | 0.16 |
| “Firmicutes” | Desulfosporosinus orientisT | 0.13 | 0.16 | 0.15 |
| “Bacteroidetes” | Muricauda ruestringensisT | 0.05 | 0.05 | 0.05 |
Growth rates were calculated from optical density (436 nm) and fluorescence data obtained upon SYBR green I staining of fresh (FH) and glutaric dialdehyde-fixed cells (FX).
Fluorescence analyses of all culture samples were performed in duplicate by the addition of 50 μl SYBR green I working solution (1,000-fold dilution of stock), incubation in the dark for at least 2 hours, and subsequent analysis on a microplate reader (see below).
Environmental samples and total cell counts (TCC).
Water was collected at Buzzards Bay (Woods Hole, MA) in July 2003, approximately 1 km offshore. About 20 liters of water from a 50-cm water depth was used to fill a clean plastic container that had been thoroughly flushed five times with deionized water and once with sample water before being filled. Immediately after return to the laboratory, a 50-ml subsample was transferred aseptically into a 50-ml Falcon tube, fixed with 3 ml 37% formaldehyde, and stored at 4°C in the dark until further processing.
Total cell counts were performed according to the protocol of Noble and Fuhrman (25). Briefly, 1 ml fixed sample was mixed with 100 μl staining solution (SYBR green I; 400-fold dilution of the stock solution) and incubated for 10 min in the dark. A 100-μl aliquot of this mixture was filtered through a black polycarbonate filter (0.2-μm pore size; Millipore, Eschborn, Germany), rinsed with particle-free (0.2-μm sterilely filtered) PBS, air dried, and fixed on a microscopic slide by the addition of 10 μl mounting solution (50% glycerol, 50% PBS, 0.1% p-phenylenediamine [Sigma]). Samples were counted using a Zeiss Axioscope microscope equipped with a 100-W mercury vapor lamp and filter set 09 (BP450-490, FT510, and LP515). At least 20 fields and 400 cells were counted.
Most-probable-number dilution series.
Most-probable-number dilution series were inoculated with the Buzzards Bay sample immediately after return to the laboratory, using three different media based on oxic artificial seawater (described above) with reduced vitamin contents (0.02 ml liter−1 instead of 2 ml liter−1). Medium A was supplemented with 70 mg liter−1 Bacto peptone, 14 mg liter−1 Bacto yeast extract (both from BD Biosciences, San Diego, CA), 1.4 mg liter−1 ferric citrate, and 1 ml liter−1 substrate mix (glucose, lactose, cellobiose, fructose, glucosamine, sodium salts of acetate, malate, succinate, tartrate, and pyruvate, and amino acids [cysteine, methionine, leucine, proline, glycine, alanine, and phenylalanine] [1 mM each], as well as 0.05% chitin). Medium B received only 1 ml liter−1 substrate mix. Medium C received no organic substrates at all.
MPN series were prepared in 2-ml 96-well polypropylene deep-well plates. First, 800 μl of medium was dispensed into each well of the plates, and then 200-μl samples were added as the inoculum to seven wells of the first row. One row of wells served as a control without inoculum. Subsequently, the contents of the first row of wells were diluted fivefold into consecutive wells, creating 12 dilutions. The MPN series were incubated in total for 141 days at 15°C. After 3, 17, and 141 days, 200-μl subsamples from the wells were transferred to black microplates. Fifty microliters of SYBR green I working solution was added to each well. Fluorescence was measured after 2 h of incubation in the dark. Fluorescence measurements of MPN samples after 3 and 17 days of incubation were conducted in a Tecan SPECTRAFluor Plus microplate reader (Tecan GmbH, Gröding, Austria) (excitation, 485 nm; emission, 540 nm). After 141 days, fluorescence measurements were done as described below. The mean fluorescence emission of uninoculated controls was 45.7 relative fluorescence units (RFU), and the standard deviation was 6.1. Growth was scored as positive if the fluorescence emission in the inoculated wells was at least 80 RFU. This value exceeds the average of the controls by at least five times the standard deviation. The rationale for this was that it would detect significant but low growth, as can be expected for assays with little or no substrate addition.
Bias-corrected most probable numbers with approximate Cornish and Fisher confidence limits were calculated using the MPN calculator software described by Klee (22).
Fluorescence measurements.
Fluorescence intensities were determined in a microplate reader (Fluostar Optima; BMG Labtechnologies, Offenburg, Germany) at the following excitation/emission wavelengths: 485/520 nm (acridine orange), 360/460 nm (DAPI), 540/590 nm (ethidium bromide), and 485/520 nm (SYBR green dyes and PicoGreen). All measurements were carried out in three reading cycles, with integration of 20 flashes, a 0.5-s delay between plate movement and reading, and 10 s of shaking before each cycle. During comparisons of the different dyes, the gain of the photomultiplier was adjusted for each dye separately so that the fluorescence intensity (FI) of the highest E. coli cell density tested (ca. 2 × 109 cells per ml) equaled approximately 60,000 RFU. All subsequent measurements of SYBR green I-stained samples were carried out with a constant detector gain of 1,300 arbitrary units. Unless otherwise stated, stained samples were incubated overnight before fluorescence measurement (see Results).
However, the time dependence of SYBR green I fluorescence emission was analyzed by transferring the microplates to the reader directly after the addition of the SYBR green I working solution. Fluorescence data were acquired over 50 to 200 reading cycles, with integration of 10 flashes. Data points represent the averages of two parallels.
No data manipulation was applied to the raw data, except for subtraction of a blank when noted.
RESULTS
Comparison of nucleic acid dyes.
For staining of E. coli cells, the optimal final dye concentrations were found to be 2 μg ml−1 (ethidium bromide and DAPI), 1:5,000 to 1:10,000 (SYBR green I and II), and 1:400 (PicoGreen). Acridine orange turned out to be unsuitable, since it displayed very high background fluorescence even at very low concentrations (Fig. 1) and did not allow discrimination between a cell-free control and 109 E. coli cells ml−1. While DAPI worked best in PBS (pH 7.2), the best signal-to-noise ratios for all other dyes were found at pH 8.0 in TE buffer (data not shown). In particular, SYBR green I and II, PicoGreen, and ethidium bromide were found to be pH sensitive. Therefore, working solutions of these dyes (5× final concentration) were prepared at elevated buffer concentrations (200 mM Tris, 50 mM EDTA, pH 8.0) to ensure stable pH conditions even during the analysis of very acid or alkaline samples.
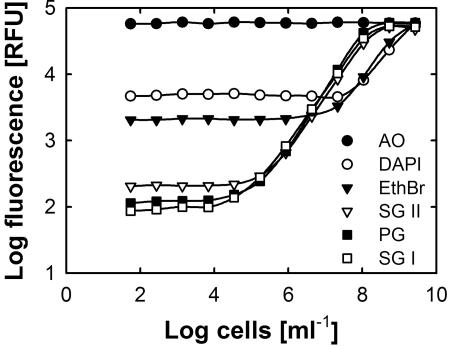
FIG. 1.
Comparison of six nucleic acid dyes for the detection of bacterial cells in a microplate assay. Each data point represents the mean of four (PicoGreen) or eight replicates. Standard deviations were omitted for clarity. AO, acridine orange; EthBr, ethidium bromide; PG, PicoGreen; SG II, SYBR green II; SG I, SYBR green I.
The fluorescence yields and background fluorescence differed remarkably among the different dyes. With SYBR green I (1:10,000) or PicoGreen (1:400) in a sample volume of 200 μl, even as few as 10,000 E. coli cells per well (equivalent to 50,000 cells ml−1) were unambiguously detected (Fig. 1), while SYBR green II (1:10,000) allowed the detection of 1.7 × 105 E. coli cells ml−1. Ethidium bromide and DAPI were far less sensitive and required cell densities of at least 2 × 107 and 108 cells ml−1, respectively (Fig. 1). However, further dilution to final concentrations of 1:20,000 (SYBR green dyes) and 1:800 (PicoGreen) did not result in the expected decrease in background fluorescence (data not shown). For further experiments, SYBR green I was chosen, because it is less prone to interference with various chemical compounds than SYBR green II or PicoGreen (20, 37).
Characteristics of quantitative SYBR green I cell staining.
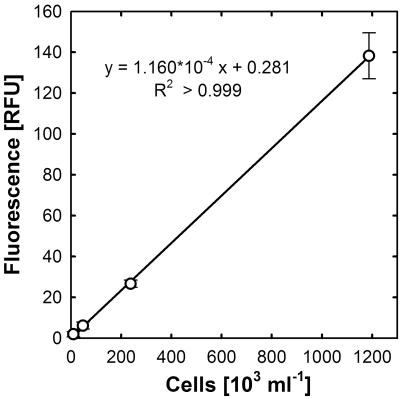
Linear and log-transformed calibration curves obtained with washed E. coli cells are shown in Fig. 2 and 3. A double logarithmic plot of fluorescence intensities versus cell densities showed a linear correlation (R2 > 0.999) over 4 orders of magnitude, from 50,000 to 2 × 108 cells ml−1, following the equation FI = (cells ml−1)0.945 × 10−3.631, indicating that over a broad range of cell numbers the calibration curve follows a power law function. However, at low cell densities, when scattering and quenching of emitted fluorescence can be neglected, a direct linear correlation between cell numbers and fluorescence intensity was observed.
FIG. 2.
Quantification of washed E. coli cells at low cell densities by SYBR green I staining. Error bars represent standard deviations for eight replicates.
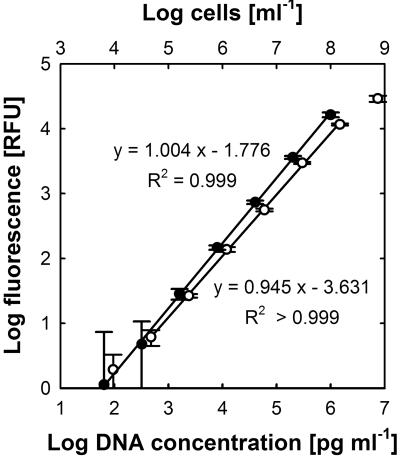
FIG. 3.
Quantification of washed E. coli cells (open circles) and lambda phage DNA (closed circles) in a microplate assay. Samples were serially diluted, and fluorescence intensities were measured after staining with SYBR green I. Linear regression was calculated from log-transformed data. Error bars represent standard deviations for eight replicates.
SYBR green I quantification of DNA.
Quantification experiments with purified double-stranded DNA were performed to confirm the results of the cell staining experiments described above but also to investigate potential interference by RNA and fixation agents. The increase in fluorescence was linearly correlated (R2 > 0.999) with increases in the DNA concentration from 102 to 106 pg ml−1 DNA, following the equation (Fig. 3) FI = [DNA]1.004 × 10−1.776, for DNA concentrations in pg ml−1.
In contrast to the case for SYBR green I-stained cells, for purified DNA the slope of the regression curve was nearly 1.0 over almost the whole range of DNA concentrations tested. However, the quantification of very low DNA concentrations was limited by the intrinsic background fluorescence, and at DNA concentrations above 106 pg ml−1, the amount of SYBR green I in the assays became limiting. This could be circumvented by the use of a dye working solution with a higher SYBR green I concentration. For the experiments presented here, a 1:10,000 dilution of the dye was chosen, since it worked well with the desired range of cell counts per ml. Higher cell numbers were avoided, since fluorescence emission would be negatively influenced by scattering and quenching.
From the plots of fluorescence versus DNA concentration or cell number (lower parts of the curves), it can be estimated that 1 RFU corresponds to approximately 9,250 cells ml−1 or 55.5 pg DNA ml−1 (Fig. 2 and 3). From these data, a DNA content of 6.0 × 10−3 pg per cell can be inferred. Considering a genome size of 4.75 × 106 bp for E. coli K-12 (3) and an average molar weight of 618 g for nucleotides (6), it can be calculated that the E. coli K-12 cells used in the present study contained, on average, 1.23 genomes per cell.
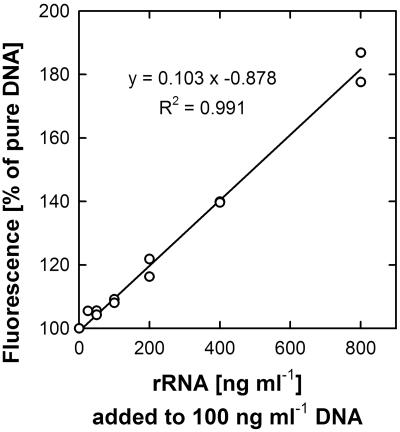
The addition of rRNA to DNA-containing assays led to a noticeable increase in fluorescence (Fig. 4). As expected, the increase in fluorescence correlated linearly with the amount of rRNA added. However, RNA yielded much less fluorescence than DNA. Even at the highest rRNA-to-DNA ratio tested in this study (eightfold), the increase in fluorescence compared to that with DNA alone was only by a factor of 0.8.
FIG. 4.
Interference of rRNA with SYBR green I staining of double-stranded DNA. Increasing amounts of E. coli rRNA were added to 100 ng ml−1 lambda phage DNA and stained with SYBR green I. The fluorescence was measured and is displayed as a relative increase compared to that of pure DNA (defined as 100%). Linear regression was calculated from two parallel experiments.
Influence of sample fixation on quantification of dissolved DNA.
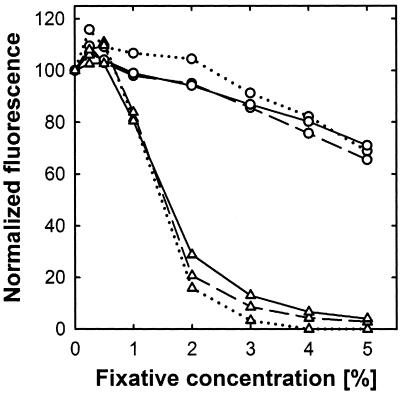
At concentrations commonly used for sample fixation, ethanol strongly inhibited fluorescence upon SYBR green I staining. After staining of glutaric dialdehyde- or formaldehyde-fixed samples, a fixative concentration-dependent decrease in fluorescence emission was also observed but was much weaker than that caused by ethanol. This effect seemed to be independent of the actual DNA concentration (Fig. 5). Up to 2% glutaric dialdehyde did have a minor effect on fluorescence. However, at higher concentrations, the fluorescence decrease seemed to be linearly correlated with the glutaric dialdehyde concentration. With 5% fixative, an approximately 30% reduction in fluorescence was observed (Fig. 5). Overall, formaldehyde had a significantly stronger effect than glutaric dialdehyde. While 0.25 or 0.5% (vol/vol) formaldehyde had no effect on fluorescence, concentrations of 1% and 2% already resulted in 20% and 80% reductions in fluorescence, respectively. However, it was critical to measure the fluorescence of formaldehyde- or glutaric dialdehyde-fixed DNA samples within 10 minutes of dye addition, since a longer incubation regularly caused a further decrease in the fluorescence signal. After several hours of incubation, glutaric dialdehyde-fixed DNA samples stained with SYBR green I developed a visible yellowish color.
FIG. 5.
Effects of different glutaric dialdehyde (circles) and formaldehyde (triangles) concentrations on fluorescence emission of SYBR green I-stained DNA. Different DNA concentrations (dotted lines, 10 ng ml−1; dashed lines, 100 ng ml−1; solid lines, 1,000 ng ml−1) were incubated with fixative for 1 hour prior to staining. The resulting fluorescence was recorded and is displayed as a relative decrease compared to that of pure DNA (defined as 100%). Each data point represents the mean of three replicates. Standard deviations were negligible and were omitted for clarity.
SYBR green I staining of viable and glutaric dialdehyde-fixed cells.
Fresh as well as glutaric dialdehyde-fixed cultures could be stained with SYBR green I, with similar fluorescence intensities, but they had different response times. Fresh (stained directly after sampling or stored on ice for some hours) cells needed several hours to reach stable fluorescence signals after the addition of dye, and it was advisable to incubate samples at least overnight or for approximately 20 h. Fixed samples attained stable signals after a minimum of 4 hours of incubation, while fluorescence could already be measured with sufficient accuracy after 2 hours. In contrast to the case with fresh samples, fluorescence in fixed samples should be measured within a maximum of 4 to 5 hours after the addition of dye, since glutaric dialdehyde-fixed cell suspensions developed a visible yellowish color after continued incubation, accompanied by an increasingly fluorescent background. However, in contrast to the case with glutaric dialdehyde-treated DNA samples, fluorescence levels in fixed cell suspensions did not decrease and remained constant for at least 6 hours. Controls without the addition of DNA or cells, however, also turned slightly yellowish with time, indicating an interaction of dye, buffer, and glutaric dialdehyde.
Growth experiments.
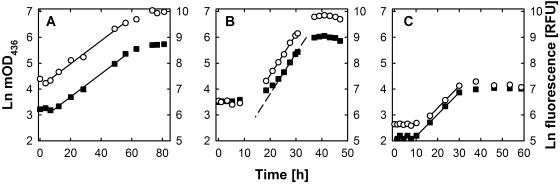
Growth experiments were performed with several bacterial strains representing different physiological types, including aerobes and sulfate reducers. As an example, growth curves of aerobic bacteria growing in rich (panel A) and substrate-poor (panel C) media as well as of sulfate-reducing bacteria in ferrous sulfide-reduced medium (panel B) are shown in Fig. 6. Growth was monitored by both optical density and fluorescence measurements after SYBR green I staining. The latter was performed for comparison with glutaric dialdehyde-fixed cells and cells that were stored on ice between sampling and staining (fresh cells) but yielded no significant differences. Generally, both approaches (optical density and fluorescence determinations) showed similar time courses (Fig. 6) and consequently resulted in almost the same growth rates (Table 2). Minor deviations were observed only during the late lag and stationary phases. During late lag phase, some cultures, such as Oceanospirillum sp. strain GM1, already exhibited increased fluorescence while the turbidity remained stagnant, whereas in some stationary-phase cultures (Fig. 6C) the fluorescence started to decline earlier than the optical density.
FIG. 6.
Growth curves of Muricauda ruestringensis (A), Desulfovibrio acrylicus (B), and Oceanospirillum sp. strain GM1 (C) obtained by determining the optical density at 436 nm (filled squares) and by fluorescence measurement after SYBR green I staining (circles). The resulting growth rates of the strains in panels A to C and the other strains tested are given in Table 2.
The ratio of fluorescence to optical density was calculated separately for each strain. For all cultures (Table 1), this ratio remained almost constant, even during the different growth phases. However, pronounced differences were found between the different strains, ranging from 1.5 × 104 (Arcobacter sp. strain NA105) to 11.6 × 104 (Muricauda ruestringensisT) RFU OD−1, and in some cases, between fresh and glutaric dialdehyde-fixed samples of a single strain (Table 1).
Most-probable-number dilution series.
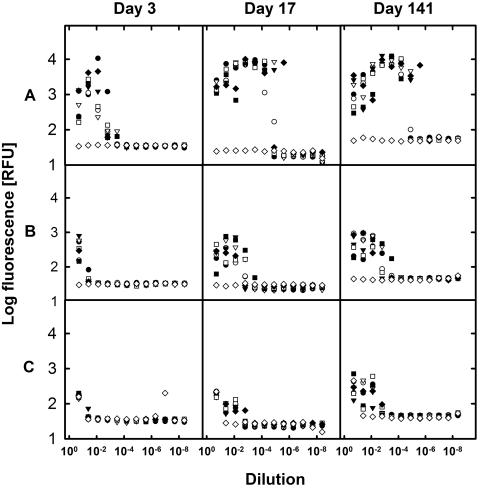
MPN series were monitored for growth after 3, 17, and 141 days (Fig. 7). Among the different series, pronounced differences were observed that could be easily linked to nutrient concentrations. Medium A, with the highest nutrient additions, stimulated the fastest growth, already reaching maximum cell numbers, as indicated by fluorescence, after 17 days. Growth on medium B commenced later and resulted in lower cell numbers than that on medium A. Surprisingly strong growth was obtained even without substrate addition (medium C). According to the DNA calibration curves (Fig. 3), the maximum fluorescence levels of 10,000, 1,000, and 700 RFU achieved with the different media correspond to DNA concentrations of approximately 500, 50, and 38 ng ml−1, respectively. Assuming a DNA content of 5 × 10−3 pg DNA per cell (6), this would represent 108, 107, and 7.5 × 106 cells ml−1, respectively. With a total cell count of 9.8 × 105 cells ml−1 in the original sample, an inoculum size of approximately 200,000 cells in the first dilution can be estimated, which would amount to a fluorescence of about 20 RFU. The values observed after 3 days even with the substrate-free series were clearly higher, demonstrating cell reproduction.
FIG. 7.
Analysis of most-probable-number dilution series inoculated with water from Buzzards Bay. MPN series were set up in 96-well plates using three different media with different substrate additions. To 800 μl medium in the first dilution, 200 μl of sample was added and consecutively diluted into the following 11 wells. Subsamples were taken after 3, 17, and 141 days of incubation and were analyzed by SYBR green I staining and fluorescence determination. Different symbols represent seven parallel dilutions, and open diamonds represent uninoculated controls (by accident, the least diluted control of series C was inoculated but not further diluted). Medium A (top) contained peptone, yeast extract, and a substrate mix, while medium B (middle) received only the substrate mix and medium C (bottom) received no organic substrates.
For evaluation of the fluorescence data, single wells of the MPN plates were checked by microscopy as well. Cells were never found in wells shown to be negative by SYBR green I staining. Because of their high sensitivity, fluorescence measurements revealed positive scores in cases when direct microscopic observation remained questionable and would require filtering of a larger volume of sample. The MPN count obtained with medium A was 42,000 cells ml−1 (4.3% of the TCC), while those with medium B and C reached 610 cells ml−1 (0.06% of the TCC) and 153 cells ml−1 (0.016% of the TCC), respectively.
DISCUSSION
A novel assay for the detection and quantification of microbial cells in aqueous suspension was developed. The described assay is simple, sensitive, and fast to perform, allowing a reliable quantification of cells in pure culture experiments as well as estimations of microbial biomass in enrichment procedures or MPN series. In combination with molecular phylogenetic analysis of successful enrichments, the selection of promising enrichments for subsequent isolation procedures is made easier. In addition, this assay can facilitate physiological investigations of strains which do not exhibit high growth yields, e.g., oligocarbophilic or strictly lithotrophic microorganisms.
Comparison of nucleic acid dyes.
For the detection of microbial cells in suspension, fluorescent nucleic acid dyes were chosen because they stain any type of microbial cell (14, 25, 26, 34), irrespective of phylogeny or physiological capacities. However, the different dyes were found to vary remarkably in terms of sensitivity and the range of cell numbers they can detect. Cyanine dyes (PicoGreen and SYBR green) that are commonly employed for the quantification of dissolved DNA (20, 33, 37) were the most sensitive and allowed reliable quantification of microbial cells. Consistent with this, PicoGreen has previously been used for the determination of microbial biomass in environmental samples (14, 31). However, it is sensitive to interference from a broad range of organic compounds and salt concentrations present in seawater (20). Similarly, cations can impair the SYBR green I fluorescence yield. However, this effect can be minimized by maintaining a clear molar dye surplus. Zipper et al. (37) estimated an approximate concentration of 10 mM in the bulk stock solution of SYBR green I and discovered that at molar-dye-to-base-pair ratios higher than 10:1 (mol:mol nucleotide), this effect was negligible. Thus, with a 1:10,000 dilution of the SYBR green I stock, as used in the present study, DNA concentrations of up to 100 nM base pairs, corresponding to 1.2 × 107 E. coli cells per ml, can be expected to be determined free of interference. However, using freshwater media, even up to 1.5 × 108 E. coli cells per ml could reliably be determined (Fig. 3). For marine media or higher cell densities, a narrower working range is expected, but for these applications dye working solutions with a higher SYBR green I concentration can be prepared.
In addition, SYBR green I has been reported to exhibit some inherent limitations that are commonly found with cyanine dyes, as its binding behavior to double-stranded DNA depends on the GC content, fragment size, and conformation of DNA (33, 37). These effects were not apparent, however, in the present study and seemed not to impair the applicability of SYBR green I for sensitive biomass determinations. A much more important effect was the sensitivity of SYBR green I stock solution to multiple freeze-thaw cycles, which was avoided by subdivision into small aliquots upon receipt (20, 24).
Possible constraints by fluorescent compounds.
The use of nucleic acid stains for the measurement of fluorescence in aqueous cell suspensions is possible only if concentrations of interfering compounds emitting autofluorescence upon excitation are negligible. This effect can easily be recognized by fluorescence measurement prior to nucleic acid staining. Examples of fluorescent compounds are aromatics that may be present in considerable concentrations in culture media (e.g., amino acids or vitamins) but also compounds present in the inoculum, such as humic acids (34, 38). The latter, however, would probably be diluted out during the MPN procedure but still might interfere if only low cell growth occurs or even result in false-positive results for the low dilutions. Interference by autofluorescence of the medium, however, can be circumvented by separating cells from the medium by filtration prior to analysis. No loss of accuracy was found if samples were filtered onto black polycarbonate membranes and subjected to fluorescence measurements (data not shown).
Interference with ribosome content and sample fixation.
rRNA is the most abundant nucleic acid in bacterial cells and in E. coli can surpass the DNA content by a factor of 11.5 (6). It is therefore likely to interfere with the quantification of microbial cells by nucleic acid staining. However, the fluorescence yield of rRNA was only 10% that of DNA (Fig. 4), confirming the results obtained for single-stranded DNA by Zipper et al. (37) and indicating that even in extremely fast-growing E. coli cells, rRNA might only double the fluorescence yield. Nevertheless, for growth rate determinations the fluorescence of rRNA can be neglected, since during exponential growth rRNA contents are supposed not to change dramatically (6, 9). A slight influence might be seen only during the lag phase, when ribosome contents are built up, and during stationary phase, when cells reduce their cellular rRNA contents, like the case for Oceanospirillum sp. strain GM1 in Fig. 6. However, this effect is too weak to impede even the determination of cell numbers from fluorescence data. The present approach was particularly intended for the quantification of environmental isolates or chemolithoautotrophs that are slow growing and reach only low cell densities. These organisms are generally characterized by lower rRNA-to-DNA ratios, and therefore RNA interference should be of minor importance.
In many cases, an immediate analysis of cultures after sampling is not possible, and therefore sample fixation is required. Of the typical preservation protocols involving formaldehyde, glutaric dialdehyde, or ethanol, glutaric dialdehyde was most recommendable and could be used at final concentrations of up to 2%. Formaldehyde can be used as an alternative to glutaric dialdehyde, for example, for fluorescence in situ hybridization or flow cytometry (10), but must not be present at final concentrations exceeding 0.5%. Ethanol was found to be completely incompatible with SYBR green I staining. Since ethanol leads to a quantitative secession of SYBR green I from the DNA (20), it can be expected that it in turn also inhibits binding of the dye to DNA.
Alternatively, storing samples on ice until processing, such as during growth experiments, may be considered, since it avoids side reactions, but it requires several hours of incubation (for example, overnight) to achieve stable fluorescence signals. An advantage of this method, however, is that samples can serve subsequently for other purposes, e.g., nucleic acid extraction.
Biomass determination during growth experiments.
Classical growth experiments typically rely on measurements of optical density, or in some cases, on microscopic counting. Counting is time-consuming and often associated with large standard deviations. Optical density, however, was found to be in good agreement with culture biomass for single bacterial types (9, 23), but there might be variations depending on the growth phase (6). Optical densities obtained for different organisms can hardly be compared, since cell size and shape vary between different species and strongly influence the optical density (19). This may be reflected by the variation in the FI/OD ratios found among the different strains in the present study (Table 1). DNA concentrations in prokaryotic cultures are directly dependent on cell numbers and are related to the growth phase (6). Each cell contains at least one and normally not more than two genomes, but genome sizes may vary from approximately 1 to 12 Mbp per cell among different prokaryotic taxa (15). However, the majority of bacterial and archaeal species seem to contain in the range of 1.5 to 5 Mbp (15). Similarly, little variation in DNA contents per cell (approximately 2.5 fg per cell) was found for aquatic prokaryotes and appeared to be independent of the phylogenetic position of the single cells (10, 29). Therefore, it seems that apart from exceptions with very large genomes, DNA fluorescence upon SYBR green I staining allows a better comparison of different strains than optical density.
Measuring the optical density is also hampered by cells forming chains, filaments, clumps, or aggregates or by the presence of particles (e.g., FeS in cultures of sulfate-reducing bacteria). It is also reliable over only 1 to 2 orders of magnitude (OD, ∼0.01 to 0.3). At lower densities, no reliable signal is obtained, while at higher densities the OD deviates from linearity and cultures have to be diluted. Similarly, SYBR green I fluorescence may be scattered and quenched at very high cell densities. However, it offers a lower detection limit (by 1 to 2 orders of magnitude) and reliable signals over 4 to 5 orders of magnitude (Fig. 1 and 3). It therefore allows growth detection even in substrate-limited cultures (Fig. 6C) that are hardly or not analyzable by simple optical density determination.
Particles seem to affect fluorescence determinations less than they affect optical density measurements. During growth experiments with the sulfate-reducing bacterium Desulfovibrio acrylicus, OD measurements resulted in a rather sigmoid growth curve (Fig. 6B). A reliable determination of the growth rate was possible only several hours after exponential growth commenced, as could be seen by comparison with the plot of fluorescence data. Similarly, it is to be expected that fluorescence delivers more reliable data for filamentous cultures, clumping cultures, etc. OD measurements rely on the relative surface area of the cell and a homogenous distribution of cells in the sample. Fluorescence determination, in contrast, depends on the DNA (and RNA) content of the cells, and since normally the whole sample is scanned and integrated, the allocation of the cells within the sample is of minor importance.
Most-probable-number enrichments.
Similar to dilution-to-extinction methods (11, 13), which are generally applied to aquatic habitats, during MPN analysis samples are serially diluted. Therefore, MPN series offer the opportunity to enrich potentially abundant cells from the highest positive dilutions (12, 21, 28, 30). For analyses of MPN series, dilution-to-extinction cultures, or dilution culturing assays (2), generally a large number of samples needs to be screened for growth. This is often achieved by visual inspection of color changes related to growth, turbidity measurement, or microscopic observation. During recent years, the use of low-substrate media helped to improve cultivation by yielding higher viable counts and more diversity (8, 13, 27, 30). However, the addition of small amounts of substrate allows only low cell densities to develop and makes growth detection more difficult. Microscopy is reliable, but screening of a single MPN series can take considerable time (several hours). Furthermore, an approximate detection limit of only 5 × 106 cells ml−1 has to be considered. Detection could be improved by filtration through membranes, but this cannot be applied to novel approaches with reduced culture volumes (13, 27, 30). The high sensitivity of the fluorescence assay, however, allowed growth detection in MPN series, even with low or no substrate additions allowing cell growth that was hardly detectable by light microscopy.
The fluorescence microplate assay is particularly advantageous if deep well plates are used for preparing MPN series (30), since multichannel pipettes can be used for the transfer of cultures samples to fluorescence assay plates. Therefore, the method presented in this study is faster, as it requires about 30 min of work for analysis of a 96-well plate plus the incubation of the dye, and is comparable in terms of sensitivity to flow cytometry (13) or microgrowth assays (7). Since it requires only small sample volumes, the remaining sample volume can be used for subculturing or chemical or molecular analysis.
Acknowledgments
We thank Alfred Spormann and the Microbial Diversity Summer Course, Woods Hole, 2003, for inspiration and help during the initial work of this project. Alice Child and Kendra Williams are acknowledged for providing access to a fluorescence plate reader (Tecan). Torsten Brinkhoff, Yvonne Hilker, and Uwe Maschmann are gratefully acknowledged for providing type strains.
W.M.-H. gratefully acknowledges financial support by the Lower Saxony Graduate Program.
REFERENCES
- 1.Aakra, Å., J. B. Utåker, I. F. Nes, and L. R. Bakken. 1999. An evaluated improvement of the extinction dilution method for isolation of ammonia-oxidizing bacteria. J. Microbiol. Methods 39:23-31. [DOI] [PubMed] [Google Scholar]
- 2.Ammerman, J. W., J. A. Fuhrman, Å. Hagström, and F. Azam. 1984. Bacterioplankton growth in seawater. I. Growth kinetics and cellular characteristics in seawater culture. Mar. Ecol. Prog. Ser. 18:9-31. [Google Scholar]
- 3.Bachmann, B. J. 1990. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 54:130-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollmann, A., M.-J. Bär-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer, H., and P. P. Dennis. 1996. Modulations of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss II, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 7.Brewster, J. D. 2003. A simple micro-growth assay for enumerating bacteria. J. Microbiol. Methods 53:77-86. [DOI] [PubMed] [Google Scholar]
- 8.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the Central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunschede, H., T. L. Dove, and H. Bremer. 1977. Establishment of exponential growth after a nutritional shift-up in Escherichia coli B/r: accumulation of deoxyribonucleic acid, ribonucleic acid, and protein. J. Bacteriol. 129:1020-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Button, D. K., and B. R. Robertson. 2001. Determination of DNA content of aquatic bacteria by flow cytometry. Appl. Environ. Microbiol. 67:1636-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture—theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotner, J. B., M. L. Ogdahl, and B. A. Biddanda. 2001. Double-stranded DNA measurement in lakes with the fluorescent stain PicoGreen and the application to bacterial bioassays. Aquat. Microb. Ecol. 25:65-74. [Google Scholar]
- 15.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 16.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Å. Hagström. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerhard, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.). 1994. Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
- 18.Gruden, C., S. Skerlos, and P. Adriaens. 2004. Flow cytometry for microbial sensing in environmental sustainability applications: current status and future prospects. FEMS Microbiol. Ecol. 49:37-49. [DOI] [PubMed] [Google Scholar]
- 19.Günther, H. H., and F. Bergter. 1971. Determination of dry mass of cell suspensions by extinction measurements. Z. Allg. Mikrobiol. 11:191-198. [DOI] [PubMed] [Google Scholar]
- 20.Haugland, R. P. 1996. Handbook of fluorescent probes and research chemicals, 6th ed. Molecular Probes, Eugene, Oreg.
- 21.Jaspers, E., K. Nauhaus, H. Cypionka, and J. Overmann. 2001. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton. FEMS Microbiol. Ecol. 36:153-164. [DOI] [PubMed] [Google Scholar]
- 22.Klee, A. J. 1993. A computer program for the determination of most probable number and its confidence limits. J. Microbiol. Methods 18:91-98. [Google Scholar]
- 23.Koch, A. L. 1994. Growth measurement, p. 248-277. In P. Gerhard, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
- 24.Lunau, M., A. Lemke, K. Walther, W. Martens-Habbena, and M. Simon. 2005. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ. Microbiol. 7:961-968. [DOI] [PubMed] [Google Scholar]
- 25.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 26.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 27.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 28.Sass, H., H. Cypionka, and H. D. Babenzien. 1997. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol. Ecol. 22:245-255. [Google Scholar]
- 29.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 30.Süß, J., B. Engelen, H. Cypionka, and H. Sass. 2004. Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol. 51:109-121. [DOI] [PubMed] [Google Scholar]
- 31.Tranvik, L. J. 1997. Rapid fluorometric assay of bacterial density in lake water and seawater. Limnol. Oceanogr. 42:1629-1634. [Google Scholar]
- 32.Tschech, A., and N. Pfennig. 1984. Growth-yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163-167. [Google Scholar]
- 33.Vitzthum, F., G. Geiger, H. Bisswanger, H. Brunner, and J. Bernhagen. 1999. A quantitative fluorescence-based microplate assay for the determination of double-stranded DNA using SYBR green I and a standard ultraviolet transilluminator gel imaging system. Anal. Biochem. 276:59-64. [DOI] [PubMed] [Google Scholar]
- 34.Weinbauer, M. G., C. Beckmann, and M. G. Höfle. 1998. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl. Environ. Microbiol. 64:5000-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3372. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 36.Widdel, F., G.-W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 37.Zipper, H., H. Brunner, J. Bernhagen, and F. Vitzthum. 2004. Investigations on DNA intercalation and surface binding by SYBR green I, its structure determination and methodological implications. Nucleic Acids Res. 32:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipper, H., C. Buta, K. Lämmle, H. Brunner, J. Bernhagen, and F. Vitzthum. 2003. Mechanisms underlying the impact of humic acids on DNA quantification by SYBR green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res. 31:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]