Abstract
The tfdCIDIEIFI, and tfdDIICIIEIIFII gene modules of plasmid pJP4 of Ralstonia eutropha JMP134 encode complete sets of functional enzymes for the transformation of chlorocatechols into 3-oxoadipate, which are all expressed during growth on 2,4-dichlorophenoxyacetate (2,4-D). However, activity of tfdI-encoded enzymes was usually higher than that of tfdII-encoded enzymes, both in the wild-type strain grown on 2,4-D and in 3-chlorobenzoate-grown derivatives harboring only one tfd gene module. The tfdDII-encoded chloromuconate cycloisomerase exhibited special kinetic properties, with high activity against 3-chloromuconate and poor activity against 2-chloromuconate and unsubstituted muconate, thus explaining the different phenotypic behaviors of R. eutropha strains containing different tfd gene modules. The enzyme catalyzes the formation of an equilibrium between 2-chloromuconate and 5-chloro- and 2-chloromuconolactone and very inefficiently catalyzes dehalogenation to form trans-dienelactone as the major product, thus differing from all (chloro)muconate cycloisomerases described thus far.
Ralstonia eutropha JMP134, isolated by its capability to mineralize 2,4-dichlorophenoxyacetate (2,4-D) (7), is one of the most intensively studied chloroaromatic-degrading organisms. 2,4-D, 4-chloro-2-methylphenoxyacetate (MCPA) (37), 3-chlorobenzoate (3CB) (16), 2,4,6-trichlorophenol (4), and 4-fluorobenzoate (43) as well as various other aromatic substrates (11, 35, 37, 42) are growth substrates for this strain. The genes necessary for the metabolism of 2,4-D and MCPA are localized on a 22-kb DNA fragment of plasmid pJP4 (24). The first genes encoding enzymes involved in 2,4-D degradation, identified more than 10 years ago (8, 47), include the tfdA gene, encoding a 2,4-D/α-ketoglutarate dioxygenase (15), the tfdB gene, encoding a 2,4-dichlorophenol hydroxylase (13), and the chlorocatechol tfdCDEF gene cluster. Two identical regulatory genes, tfdR and tfdS, whose products belong to the LysR family of transcriptional regulators and which are positioned as inverted repeats, have been localized (18, 27), with tfdS being positioned divergently from tfdA (Fig. 1). A third putative regulator gene, tfdT, localized directly upstream of tfdC, is interrupted by insertional sequence ISJP4, which renders it inactive (25). Therefore, it has been proposed that the tfdR and tfdS genes are the master regulators, which control the activity of tfd genes (25). Downstream and positioned divergently from tfdR, an open reading frame with significant similarities to tfdD was observed (27), and a whole new set of tfd genes between tfdR and tfdT (i.e., tfdDIICIIEIIFII, tfdBII, and tfdK) has recently been localized. More recently, both tfdCDEF-B and tfdDIICIIEIIFII-BII, as well as tfdA and tfdK, have been shown to be expressed upon exposure to 2,4-D (24). However, the contributions of the different tfd-encoded enzymes to the complete metabolism of pathway intermediates have not yet been determined, nor has it been shown that all genes encode functional enzymes. Pérez-Pantoja et al. (34), by evaluating the function of the tfdCDEF (tfdCIDIEIFI; module I) and tfdDIICIIEIIFII (module II) genes in R. eutropha JMP222 for growth on 3CB, have shown that both modules encode functional enzymes for chlorocatechol metabolism. Similarly, Laemmli et al. (22) reported that all genes carried by tfd module II code for functional enzymes. However, a low level of dienelactone (4-carboxymethylenebut-2-ene-4-olide) hydrolase activity and an even lower activity against 2-chloromuconate were observed in strains containing module II genes, whereas a very poor maleylacetate reductase activity was found in strains containing only module I (34). Therefore, despite its high similarity to genes encoding functional maleylacetate reductases (19), tfdFI has been said to encode a poor or nonfunctional enzyme. Moreover, attempts to purify maleylacetate reductase from R. eutropha JMP134 yielded a protein with an N-terminal sequence matching that of TfdFII (45), indicating a critical role of TfdFII for growth of strain JMP134 on 2,4-D. In addition to TfdFI and TfdFII, yet another maleylacetate reductase was evident in strain JMP134 by the induction of such an activity during growth of pJP4-cured derivative JMP222 on 4-fluorobenzoate (43). The possibility that the low maleylacetate reductase activity observed in strain JMP222 containing module I is due to recruitment of the chromosomally encoded gene cannot, therefore, be excluded. A chromosomally encoded dienelactone hydrolase, which converts trans-dienelactone much faster than cis-dienelactone, is also present in strain JMP222 (43) and probably is responsible for the poor activity observed in strain JMP222 containing module II with cis-dienelactone (34). Thus, the contributions of the different tfd gene modules to chloroaromatic degradation in strain JMP134 are not yet clear. Interestingly, interruption of tfdCI, -DI, or -EI by transposon mutagenesis resulted in mutants no longer capable of growing on 2,4-D (8), suggesting that the products of these genes play a major role in the metabolism of this substrate. In contrast, interruption of tfdFI resulted in only a slight retardation of growth. The critical importance of the tfdDI-encoded chloromuconate cycloisomerase is supported by the fact that diverse attempts to purify chloromuconate cycloisomerase from strain JMP134 resulted in purification of only TfdDI (17, 21). In the present investigation, the importance of the different tfd-encoded enzymes, chlorocatechol 1,2-dioxygenase, chloromuconate cycloisomerase, dienelactone hydrolase, and maleylacetate reductase, for growth of R. eutropha on chloroaromatics was characterized biochemically.
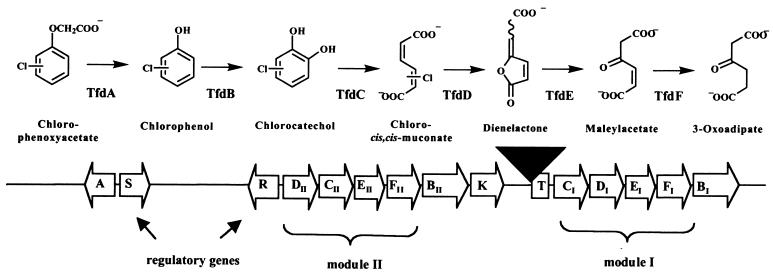
FIG. 1.
Overview of genes and enzymes involved in the degradation of chloroaromatics by R. eutropha JMP134. The enzymes catalyzing the indicated catabolic steps are TfdA, 2,4-D/α-ketoglutarate dioxygenase, TfdB, chlorophenol hydroxylase, TfdC, chlorocatechol dioxygenase, TfdD, chloromuconate cycloisomerase, TfdE, dienelactone hydrolase, and TfdF, maleylacetate reductase. The tfd genes in R. eutropha are localized on plasmid pJP4. Arrows, orientations of the tfd genes described thus far; black triangle, insertion element ISJP4.
MATERIALS AND METHODS
Bacterial strains.
The 2,4-D-degrading organism R. eutropha JMP134 was isolated by Don and Pemberton (7). R. eutropha JMP222 is a derivative of strain JMP134, cured of plasmid pJP4. R. eutropha JMP222(pBBR1M-I) and R. eutropha JMP222(pBBR1M-II) are derivatives of strain JMP222 containing either module I (pBBR1M-I) or module II chlorocatechol genes as described recently (34).
Culture conditions and preparation of cell extracts.
Growth in liquid culture was performed in the mineral salts medium described by Dorn et al. (9) containing 50 mM phosphate buffer (pH 7.4). The medium was supplemented with a carbon source, usually at 2.5 or 5 mM. Cells were grown in fluted Erlenmeyer flasks, incubated at 30°C on a rotary shaker at 150 rpm. Growth was monitored spectrophotometrically. Harvested cells were resuspended in Tris-HCl buffer (100 mM, pH 7.5) supplemented with 2 mM MnCl2 and disrupted with a French press (Aminco, Silver Spring, Md.). Cell debris was removed by centrifugation at 100,000 × g for 1 h at 4°C.
Enzyme assays.
Muconolactone isomerase (EC 5.3.3.4) was assayed as described by Prucha et al. (40) in 50 mM sodium phosphate (pH 7.5) with 0.1 mM (4S,5S/4R,5R)-5-chloro-3-methylmuconolactone as the substrate. Accumulation of 3-methyldienelactone was analyzed spectrophotometrically at 270 nm (ɛ3-methyl-trans-dienelactone = 15,200 M−1 cm−1). Catechol 1,2-dioxygenase (EC 1.13.11.1), chlorocatechol dioxygenase, muconate cycloisomerase (EC 5.5.1.1), chloromuconate cycloisomerase (EC 5.5.1.7), and dienelactone hydrolase (EC 3.1.1.45) were measured as previously described (10, 43, 44) with catechol, 3-chlorocatechol, muconate, 3-chloromuconate, or cis-dienelactone as the substrate. Substrate concentrations in the enzymatic tests were usually 0.05 mM, except for catechols and substituted catechols, which were added at initial concentrations of 0.2 mM. 3-Chloromuconate, as the substrate for chloromuconate cycloisomerase, was prepared in situ from 4-chlorocatechol by using TfdCII that was partially purified by anion-exchange chromatography and that was free of any interfering enzyme activity. Activity of partially purified chloromuconate cycloisomerases was determined in the presence of an excess of dienelactone hydrolase. Generally, TfdEI, partially purified by hydrophobic interaction chromatography and free of any interfering enzyme activity, was used. Maleylacetate reductase (EC 1.3.1.32) was measured as described by Seibert et al. (45) by using 0.05 mM maleylacetate freshly prepared by alkaline hydrolysis of cis-dienelactone or through transformation by TfdEI partially purified by hydrophobic-interaction chromatography. For determination of the kinetic properties of TfdDII, the formation of cis-dienelactone was recorded at 305 nm. A reaction coefficient of 5,300 M−1 cm−1 was calculated after complete transformation of 3-chloromuconate (20 to 50 μM) into cis-dienelactone.
Specific activities are expressed as micromoles of substrate converted or product formed per minute per gram of protein at 25°C. Protein concentrations were determined by the Bradford procedure (2).
Chromatographic separation of enzyme activities.
Cells were harvested during late-exponential growth, and the cell extract (usually containing between 8 and 20 mg of protein per ml) was applied directly to a MonoQ HR5/5 column (Amersham Pharmacia Biotech) or mixed with an equal volume of 2 M (NH4)2SO4 and, after centrifugation, applied to a phenyl-Superose HR5/5 column (Amersham Biosciences, Freiburg, Germany). At least three independent experiments were performed for each type of growth condition to verify the reproducibility of the method. Elution of the respective activities was highly reproducible and varied between independent experiments by only ±1 fraction, which corresponds to differences of 0.01 M NaCl or 0.02 M (NH4)2SO4. Proteins were eluted from the MonoQ HR5/5 column by a linear gradient of NaCl (0 to 0.5 M) in Tris-HCl (50 mM, pH 7.5, supplemented with 2 mM MnCl2) in a total volume of 25 ml or by a stepwise gradient of 0 to 0.14 M over 2 ml, 0.14 to 0.29 M over 17 ml, and 0.25 to 0.5 M over 6 ml. Proteins were eluted from the phenyl-Superose HR5/5 column by a linear gradient of (NH4)2SO4 (1 to 0 M) in Tris-HCl (50 mM, pH 7.5, supplemented with 2 mM MnCl2) over 25 ml or by a stepwise gradient of 1 to 0.8 M over 2 ml, 0.8 to 0.5 M over 8 ml, 0.5 to 0.3 M over 2 ml, 0.3 to 0.2 M over 6 ml, and 0.2 to 0 M over 10 ml. For chromatographic separation of maleylacetate reductases, proteins were eluted from the MonoQ HR5/5 column by the gradients described above, with Tris-HCl buffer exchanged for phosphate buffer. The flow rate was always 1 ml/min, and the fraction volume was 0.5 ml. Partial purification of TfdDII was achieved by successive separation using hydrophobic interaction and gel filtration. The two fractions from the phenyl-Superose HR5/5 chromatography containing the most TfdDII activity were pooled, concentrated by ultrafiltration to a final volume of 0.5 ml, applied to a Superose 12 column (HR10/30) (Amersham Biosciences), and eluted with 50 mM Tris-HCl, pH 7.5, at a flow rate of 0.3 ml/min (fraction volume, 0.5 ml). Similarly, fractions from MonoQ HR5/5 chromatography containing TfdFI activity were pooled, concentrated, and subjected to gel filtration.
Electrophoretic methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a Bio-Rad Miniprotein II essentially as described by Laemmli (23). The acrylamide concentrations for the concentrating and separating gels were 5 and 10% (wt/vol), respectively. The proteins were stained by Coomassie brilliant blue R250. Molecular mass standards (Bio-Rad, Munich, Germany) were lysozyme (14.4 kDa), trypsin inhibitor (21.5 kDa), carbonic anhydrase (31.0 kDa), albumin (45.0 kDa), bovine serum albumin (66.2 kDa), and phosphorylase b (97.4 kDa). For quantification of TfdDII in partially purified fractions, gels were stained with the fluorescent dye Sypro Ruby (Molecular Probes Inc., MoBiTec GmbH, Göttingen, Germany). Gels were scanned with a Fujifilm LAS-1000 charge-coupled device camera. The relative amount of the TfdDII protein band was determined with the AIDA, version 2.1, software package (Raytest Isotopenmessgeräte GmbH, Straubenhardt, Germany).
For two-dimensional gel electrophoresis, a 250-μl aliquot of a TfdFI-containing fraction obtained by anion-exchange chromatography was precipitated with 10% trichloroacetic acid, and then the precipitate was washed with ice-cold acetone. The protein was dissolved in 300 μl of reswelling solution containing 7.4 M urea, 2 M thiourea, 4% CHAPS (3-[{3-cholamidopropyl}dimethylammonio]-1-propanesulfonate), 20 mM Tris base, 30 mM dithiothreitol (DTT), one-half tablet of protease inhibitor (MiniComplete; Roche Applied Biosciences, Mannheim, Germany), 5% IPG buffer, pH 4 to 7 (Amersham Biosciences) and applied to isoelectric focusing (IEF) ReadyStrips (Bio-Rad). After 100 kV · h focusing time in the first dimension (IEF), the gel strip was equilibrated for the second dimension (SDS-PAGE) twice for 15 min by using 5 ml of equilibration solution consisting of 6 M urea, 30% glycerol, 2% SDS, 0.06 mM bromophenol blue, 50 mM Tris base, pH 8.8, 65 mM DTT, and 260 mM iodoacetamide. The strip was applied then to an SDS-10 to 15% PAGE gel (1.5 mm thick) and developed overnight in an IsoDALT chamber (Amersham Biosciences).
N-terminal amino acid sequencing.
Proteins were electroblotted onto a polyvinylidene difluoride membrane, and the membrane was stained with Coomassie brilliant blue R250. N-terminal amino acid sequencing was performed with an Applied Biosystems model 494A Procise HT sequencer.
Analytical methods.
High-performance liquid chromatography (HPLC) of low-molecular-weight compounds was performed with a Lichrospher SC 100 RP8 reverse-phase column (125 by 4.6 mm; Bishoff, Leonberg, Germany). Methanol-H2O containing 0.1% (vol/vol) H2PO4 was used as the eluant at a flow rate of 1 ml/min. The column effluent was monitored simultaneously at 210, 260, and 270 nm by a diode array detector (Shimadzu). Typical retention volumes using 25% (vol/vol) methanol were 7.7 (2-chloro-cis,cis-muconate), 2.5 (2-chloromuconolactone), 1.1 (5-chloromuconolactone), 4.1 (cis-dienelactone), and 1.8 ml (trans-dienelactone); those using 40% (vol/vol) methanol were 2.1 (2-chloro-cis,cis-muconate) and 1.4 ml (cis-dienelactone); those using 50% (vol/vol) methanol were 4.4 (3CB) and 0.7 ml (2-chloro-cis,cis-muconate). Kinetic measurements were recorded on an UV 2100 spectrophotometer (Shimadzu Corporation, Kyoto, Japan).
Transformation of 2-chloromuconate by chloromuconate cycloisomerase.
Transformation of 2-chloromuconate was usually performed in 50 mM Tris-HCl, pH 8. The reaction mixtures contained 500 μM 2-chloromuconate and 20 to 100 mU (as determined with 50 μM 3-chloromuconate as the substrate) of chloromuconate cycloisomerase per ml. Substrate transformation was monitored by HPLC.
Chemicals.
Chemicals were purchased from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany; Baker Chemikalien, Griesheim, Germany; and Merck AG, Darmstadt, Germany. 3-Chlorocatechol was obtained from Promochem, Wesel, Germany. 2-Chloro-cis,cis-muconate and (4R,5R/4S,5S)-5-chloro-3-methylmuconolactone were prepared as described by Pieper et al. (36, 38). (4R,5S)-5-Chloromuconolactone and (4S)-2-chloromuconolactone were prepared as described by Prucha et al. (40). cis-Dienelactone was a generous gift from Stefan Kaschabek and Walter Reineke (Bergische Universität-Gesamthochschule, Wuppertal, Germany).
RESULTS
Growth of R. eutropha JMP222 derivatives containing pBBR1M-I or pBBR1M-II on 3CB and expression of Tfd gene products.
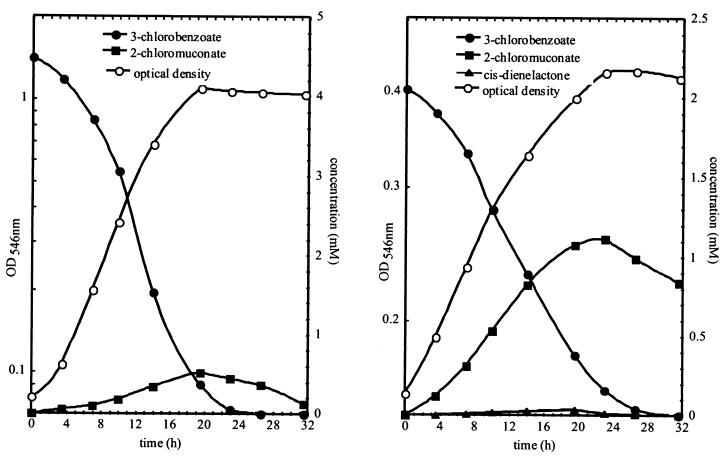
Differences in the growth of R. eutropha JMP222 derivatives harboring tfd module I or II on 3CB have been reported previously (34). Accordingly, R. eutropha JMP222(pBBR1M-I) exhibited significantly higher growth rates (0.19/h) and higher growth yields (0.22 units of optical density at 546 nm (OD546)/mM 3CB) during growth on 3CB than did R. eutropha JMP222(pBBR1M-II) (0.06/h and 0.13 OD546 units/mM 3CB), as indicated by the turbidity of the culture obtained after complete transformation of the 3CB growth substrate (Fig. 2). The observed differences in growth yield seem to be due to the significantly different amounts of intermediates produced during growth of the two strains. Thus, whereas R. eutropha JMP222(pBBR1M-I) accumulated 15% ± 5% of added 3CB as 2-chloromuconate, more than 50% of added 3CB appeared as 2-chloromuconate in the culture supernatant during growth of R. eutropha JMP222(pBBR1M-II).
FIG. 2.
Growth of R. eutropha JMP222(pBBR1M-I) (left) and R. eutropha JMP222(pBBR1M-II) (right) on 3CB. Substrate depletion and product formation were monitored by HPLC.
Whereas 2-chloromuconate was the only intermediate excreted during the growth of R. eutropha JMP222(pBBR1M-I), R. eutropha JMP222(pBBR1M-II) also excreted minute amounts of cis-dienelactone, corresponding to 2% of the substrate transformed. The accumulation of large amounts of 2-chloromuconate in strain JMP222 expressing module II, compared to the amounts accumulated in strain JMP222 expressing module I, can be explained by the recently reported very low activity of TfdDII chloromuconate cycloisomerase with this substrate (34). Accumulation of cis-dienelactone can be assumed to have been due to the observed low activity of TfdEII dienelactone hydrolase in R. eutropha JMP222(pBBR1M-II) (Table 1).
TABLE 1.
Specific activities of catechol and chlorocatechol catabolic enzymes of 2,4-D- or 3-CB-grown cells of R. eutropha JMP134, JMP222(pBBR1M-I), and JMP222(pBBR1M-II)
| Enzyme activity | Assay substrate | Sp act (U/g of protein)a for indicated strain/substrate
|
|||
|---|---|---|---|---|---|
| JMP134(pJP4)/ 5 mM 2,4-D | JMP134(pJP4)/ 5 mM 3CB | JMP222(pBBR1M-I)/ 5 mM 3CB | JMP222(pBBR1M-II)/ 2 mM 3CB | ||
| (Chloro) catechol 1,2-dioxygenase | Catechol | 430 ± 90 | 840 ± 120 | 550 ± 90 | 540 ± 90 |
| 3-Chlorocatechol | 420 ± 90 | 380 ± 80 | 540 ± 90 | 290 ± 70 | |
| 4-Chlorocatechol | 410 ± 80 | 370 ± 80 | 520 ± 90 | 260 ± 50 | |
| (Chloro) muconate cycloisomerase | Muconate | <5 | 120 ± 20 | <5 | 90 ± 30 |
| 2-Chloromuconate | 25 ± 5 | 15 ± 5 | 35 ± 10 | <5 | |
| 3-Chloromuconate | 270 ± 40 | 180 ± 40 | 360 ± 40 | 140 ± 50 | |
| Dienelactone hydrolase | cis-Dienelactone | 1,050 ± 120 | 980 ± 110 | 1,200 ± 400 | 55 ± 15 |
| Maleylacetate reductase | Maleylacetate | 750 ± 170 | 510 ± 150 | 470 ± 150 | 1,520 ± 300 |
Values are averages of at least three independent experiments.
Maleylacetate, produced by the activity of dienelactone hydrolase on cis- or trans-dienelactone, was never observed in the culture supernatants, indicating that maleylacetate reductase did not constitute a pathway bottleneck in R. eutropha JMP222(pBBR1M-II) or in R. eutropha JMP222(pBBR1M-I), despite the fact that only poor maleylacetate reductase activities have been recently reported to be induced during growth of strain JMP222 expressing tfd module I (34). This discrepancy can be explained by the fact that the former enzyme activity measurements were performed with maleylacetate produced in situ by cell extracts. However, by using maleylacetate freshly prepared from cis-dienelactone by alkaline hydrolysis or purified dienelactone hydrolase and by measuring the enzymatic activity immediately after the preparation of a cell extract containing at least 5 mg of protein per ml, it was possible to reproducibly detect significant maleylacetate reductase activities in R. eutropha JMP222(pBBR1M-I) (Table 1). All other enzyme activities were as previously reported (Table 1) (34).
Levels of Tfd activity during growth of R. eutropha JMP134 on chloroaromatics.
Due to the fact that all four enzymes involved in the chlorocatechol degradation of tfd modules I and II were obviously active and based on detection of the respective mRNAs, which showed that both gene modules were expressed during growth on 2,4-D (22, 24), the importance of each gene module for the degradation of chloroaromatics in wild-type strain R. eutropha JMP134(pJP4) was investigated. Chromatographic methods were applied to partially purify and separate the respective isoenzymes from cell extracts without significant loss of activity. Strains R. eutropha JMP222(pBBR1M-I) and R. eutropha JMP222(pBBR1M-II) were used for the establishment of an optimized purification scheme.
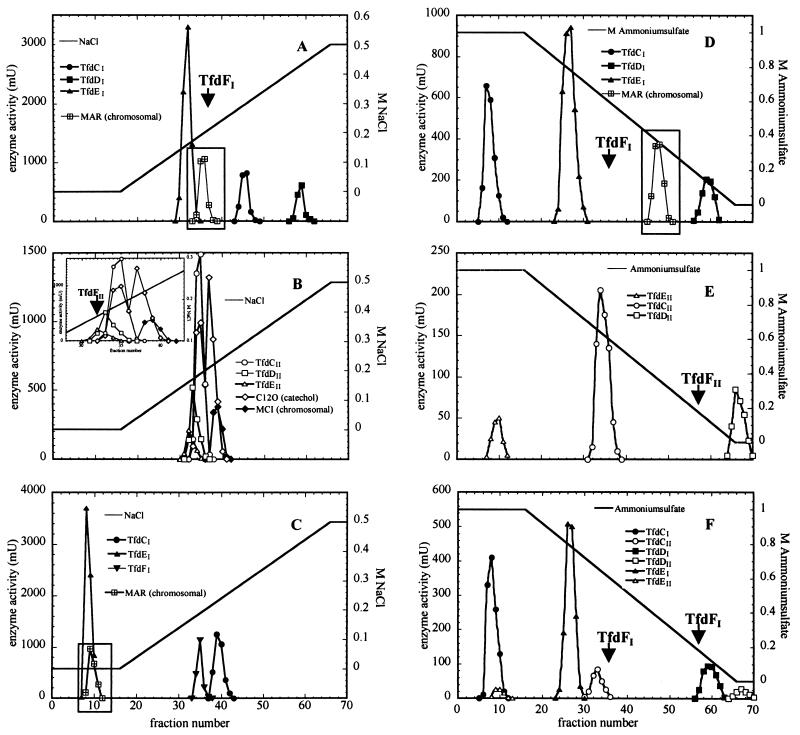
Anion-exchange chromatography of cell extracts of 3CB-grown R. eutropha JMP222(pBBR1M-I) or JMP(pBBR1M-II) resulted in high recoveries of TfdCI (60% ± 10% of applied activity), TfdCII (80% ± 10%), TfdDI (70% ± 10%), TfdDII (60% ± 10%), TfdEI (90% ± 10%), and TfdEII (90% ± 10%) activity (Fig. 3A and B), although only low activities of maleylacetate reductase (activity of TfdFI eluting at 0.21 ± 0.02 M NaCl was usually lower than 5% of the applied activity, and activity of TfdFII eluting at 0.16 ± 0.01 M NaCl was usually lower than 10%) could be recovered. However, TfdCI (eluting at 0.29 M NaCl) showed a distinctly different retention behavior than did TfdCII (eluting at 0.19 M NaCl). TfdDI and TfdDII also exhibited different properties (eluting at 0.43 and 0.17 M NaCl, respectively). TfdEI and TfdEII could not be separated by such a chromatographic method (eluting at 0.16 M NaCl). Optimization of the applied NaCl gradient did not result in a significant enhancement of separation.
FIG. 3.
Separation of proteins from cell extracts of R. eutropha JMP134, JMP222, and derivatives by means of a MonoQ HR5/5 anion-exchange (A to C) or a phenyl-Superose HR5/5 hydrophobic-interaction column (D to F). Cell extracts were either directly applied (A to C) or applied after addition of ammonium sulfate (final concentration of 1 M) (D to E), and proteins were eluted with a linear gradient of NaCl (0 to 0.5 M) or (NH4)2SO4 (1 to 0 M). The eluted fractions (0.5 ml) were analyzed for activities against the respective assay substrates (usually applied at a concentration of 50 μM, except for catechol and 3-chlorocatechol, which were added at a concentration of 200 μM). Enzymes eluting with a yield of >40% are shown. Arrows, enzymes eluting with a low yield (>20%; TfDFI and TfdFII). Elution was performed in Tris-HCl buffer, except for panel C, where phosphate buffer was used. (A) Separation of a cell extract of 3CB-grown cells of JMP222(pBBR1M-I) (6.1 mg of protein). (Inset) Activity of chromosomally encoded maleylacetate reductase after separation of a cell extract of 4-fluorobenzoate (4FB)-grown cells of JMP222 (7.9 mg of protein). (B) Separation of a cell extract of 3CB-grown cells of JMP222(pBBR1M-II) (11.6 mg of protein). (Inset) Detailed view of the activities observed in fractions 30 to 41. (C) Separation of a cell extract of 3CB-grown cells of JMP222(pBBR1M-I) (5.6 mg of protein). (Inset) Activity of chromosomally encoded maleylacetate reductase after separation of a cell extract of 4FB-grown cells of JMP222 (6.5 mg of protein). Muconate cycloisomerase activities could not be recovered after elution with phosphate buffer, as phosphate completely inhibited those enzymes. (D) Separation of a cell extract of 3CB-grown cells of JMP222(pBBR1M-I) (4.7 mg of protein). (Inset) Activity of chromosomally encoded maleylacetate reductase after separation of a cell extract of 4FB-grown cells of JMP222 (8.1 mg of protein). (E) Separation of a cell extract of 3CB-grown cells of JMP222(pBBR1M-II) (4.5 mg of protein). (F) Separation of a cell extract of 2,4-D-grown cells of JMP222(pBBR1M-I) (3.6 mg of protein).
Resolution of soluble enzymes by anion-exchange chromatography clearly indicated that two peaks of activity against catechol (Fig. 3B) were recovered from R. eutropha JMP222(pBBR1M-II). Thus, the high level of catechol 1,2-dioxygenase activity with catechol, compared to that with 3-chlorocatechol in 3CB-grown cells of strain JMP222(pBBR1M-II) (Table 1), is due to the induction of the chromosomally encoded catechol 1,2-dioxygenase (eluting at 0.22 M NaCl) in addition to TfdCII. Whereas TfdCII exhibited similar activities against both catechol and 3-chlorocatechol, transformation of 3-chlorocatechol by fractions containing catechol 1,2-dioxygenase was negligible. A chromosomally encoded muconate cycloisomerase (exhibiting significant activity only with cis,cis-muconate and eluting at 0.22 M NaCl) was also induced under these conditions (Fig. 3B). A similar induction of enzymes of the 3-oxoadipate pathway has previously been observed in 3CB-grown cells of R. eutropha JMP134 (36), whereas those enzymes were not induced during growth of this strain on 2,4-D (21). However, neither catechol 1,2-dioxygenase nor muconate cycloisomerase was induced during growth of strain JMP222(pBBR1M-I) with 3CB. The observed dienelactone hydrolase activity in strain JMP222(pBBR1M-II) was not identical to that of chromosomally encoded dienelactone hydrolase of strain JMP222 observed during growth on 4-fluorobenzoate (43), as the enzyme induced in strain JMP222(pBBR1M-II) exhibited similar activities against both cis- and trans-dienelactone (activity with trans-dienelactone was 60% of that with cis-dienelactone at substrate concentrations of 50 to 100 μM). The chromosomally encoded enzyme, in contrast, has been shown to exhibit activity only against trans-dienelactone (43).
Hydrophobic interaction chromatography was observed to be superior to anion-exchange chromatography, allowing separation of the different chlorocatechol dioxygenases and chloromuconate cycloisomerases, as well as the dienelactone hydrolases (Fig. 3D and E). However, the yields (50% ± 10% for TfdDI and TfdDII; 60% ± 10% for TfdEI, TfdEII, and TfdCII; and 70% ± 10% for TfdCI) were usually slightly lower than those obtained by anion-exchange chromatography, and hydrophobic-interaction chromatography also failed to give highly active preparations of maleylacetate reductase. Although a reasonable maleylacetate reductase activity, corresponding to less than 20% of the applied activity (eluting at 0.16 M [NH4]2SO4), could be observed after hydrophobic-interaction chromatography of a cell extract of 3CB-grown strain JMP222(pBBR1M-II), only negligible activity (corresponding to less than 5% of the applied activity and eluting at 0.60 ± 0.04 M [NH4]2SO4) could be recovered from a cell extract of strain JMP222(pBBR1M-I) grown on 3CB.
Neither TfdCI nor TfdEII interacted with the column material under the applied conditions (i.e., an initial ammonium sulfate concentration of 1 M; Fig. 3D and E). Further increases in the initial ammonium sulfate concentration resulted in precipitation of dienelactone hydrolase activities. However, no catechol 1,2-dioxygenase activity against 3-chlorocatechol was observed in the initially eluting fractions during separation of the JMP222(pBBR1M-II)-derived extract. Furthermore, no activity against cis-dienelactone could be found in the respective fractions after separation of a JMP222(pBBR1M-I)-derived extract, indicating that the activities assigned to TfdCI or TfdEII were not due to chromatographic artifacts.
R. eutropha JMP134 was grown on 2,4-D, and, subsequently, the soluble proteins were separated by means of hydrophobic-interaction chromatography (Fig. 3F). The activities of tfdI-encoded enzymes TfdCI, TfdDI, and TfdEI were approximately 40 to 70% of those induced during growth of strain JMP222(pBBR1M-I) on 3CB, and those of tfdII-encoded enzymes TfdCII, TfdDII, and TfdEII were approximately 25 to 50% of those induced during growth of strain JMP222(pBBR1M-II) on 3CB. As the yields of the respective isoenzymes after chromatographic separation were always similar, the comparison of total activities in the respective fractions indicates the actual importance of the respective enzymes for the degradation process. As shown in Fig. 3F, in 2,4-D-grown cells of R. eutropha JMP134, the TfdII-derived enzyme activities comprised 20% ± 5% of the observed total activity against 3-chlorocatechol and 3-chloromuconate but only approximately 5% of the observed activity against cis-dienelactone.
Characterization of maleylacetate reductases induced during growth of R. eutropha JMP134 and derivatives on chloroaromatics.
R. eutropha JMP222 is capable of growth on 4-fluorobenzoate, and induction of a chromosomally encoded maleylacetate reductase under these conditions has been reported (43). Therefore, the possibility that the maleylacetate reductase activities observed during growth of strain JMP222(pBBR1M-I) or JMP222(pBBR1M-II) on 3CB (Table 1) are, at least partially, due to recruitment of this chromosomally encoded maleylacetate reductase cannot be excluded. Partial purification of this enzyme activity, which was usually induced at levels of 300 ± 50 U/g of protein in 4-fluorobenzoate-grown cells of strain JMP222, showed that this enzyme, in contrast to the activities induced in strains JMP222(pBBR1M-I) and JMP222(pBBR1M-II), exhibits high stability. Approximately 50% of the applied activity could be recovered by hydrophobic-interaction chromatography; the activity eluted at 0.38 M ammonium sulfate (Fig. 3D, inset). As no significant activity was observed in the respective fractions after separation of JMP222(pBBR1M-I)- or JMP222(pBBR1M-II)-derived cell extracts, it can be concluded that the chromosomally encoded maleylacetate reductase activity was not recruited in these strains for growth on 3CB. Similarly, as no significant activity was observed in the respective fractions after separation of a cell extract of 2,4-D-grown cells of strain JMP134 (Fig. 3F), it seems that the chromosomally encoded maleylacetate reductase does not play a major role in the degradation of this substrate in the wild-type strain.
Approximately 90% of the applied maleylacetate reductase activity of 4-fluorobenzoate-grown cells of R. eutropha JMP222 was recovered by anion-exchange chromatography; the activity eluted at 0.2 M NaCl (Fig. 3A, inset), a condition similar to those under which the activities of JMP222(pBBR1M-I) and JMP222(pBBR1M-II) eluted. However, the yields of the latter maleylacetate preparations were <5 and 10% ± 5%, respectively, again indicating that the chromosomally encoded maleylacetate reductase plays no significant role during 3CB degradation by strain JMP222(pBBR1M-I) or JMP222(pBBR1M-II).
As neither hydrophobic-interaction chromatography nor anion-exchange chromatography resulted in sufficient resolution of the three maleylacetate reductase activities or in reasonable yields of TfdFII or, particularly, TfdFI, anion-exchange chromatography was applied and Tris-HCl buffer in the eluant was replaced by phosphate buffer. This simple change had a marked effect on the retention behavior of the chromosomally encoded maleylacetate reductase, as well as on the TfdEI enzyme, as their interactions with the column material were nearly abolished (Fig. 3C). Moreover, the use of phosphate buffer dramatically increased the yield of TfdFI. In a typical separation run, after application of a cell extract of strain JMP222(pBBR1M-I) containing 15 mg of protein, the two most active fractions contained, together, 7,050 mU of TfdFI activity and 0.48 mg of protein (corresponding to a specific activity of 14,700 U/g of protein) in a complete volume of 1 ml. Further purification by gel filtration resulted in a significant loss of activity. Less than 10% of the applied activity was recovered, with specific activities of up to only 800 U/g of protein. To further demonstrate that the maleylacetate reductase induced in 3CB-grown strain JMP222(pBBR1M-I) is identical to TfdFI, an aliquot of a highly active fraction obtained by anion-exchange chromatography, containing 240 μg of protein, was subjected to two-dimensional gel electrophoresis and blotted onto a polyvinylidene difluoride membrane and a major protein spot corresponding to the estimated molecular mass of the TfdFI subunit (37.9 kDa) was analyzed by N-terminal sequencing. The amino-terminal sequence was identified as MKKFTLDYLSPR; this sequence was identical to the sequence deduced from the tfdFI gene. Therefore, it can be assumed that the maleylacetate reductase activity present in 3CB-grown cells of strain JMP222(pBBR1M-I) is, in fact, due to expression of tfdFI.
Thus, the use of anion-exchange chromatography with phosphate buffer has enabled the characterization of the expression of TfdFI, as well as of the different dienelactone hydrolases and catechol 1,2-dioxygenases, during growth of R. eutropha JMP134 and JMP222 derivatives.
2,4-D-grown cells of strain JMP134 induce significant levels of TfdFI, comprising roughly half of the total maleylacetate reductase activity. Calculations of the levels of TfdCI versus TfdCII and of TfdEI versus TfdEII in those cells confirmed the results obtained above by hydrophobic interaction chromatography, with 20% of the total chlorocatechol 1,2-dioxygenase activity due to TfdCII and 5% of the total dienelactone hydrolase activity due to TfdEII.
Characterization of chloromuconate cycloisomerase TfdDII.
Compared to the previously described chloromuconate cycloisomerases from gram-negative microorganisms (21, 51) the chloromuconate cycloisomerase activity encoded by tfdDII exhibited very poor activity against 2-chloromuconate, possibly resembling chloromuconate cycloisomerase of Rhodococcus opacus 1CP (46). Therefore, this enzyme activity was further characterized. TfdDII was partially purified from 3CB-grown cells of strain JMP222(pBBRM1-II) by anion-exchange chromatography and gel filtration. A total of 2.1 U (calculated for a substrate concentration of 50 μM 3-chloromuconate) of chloromuconate cycloisomerase (specific activity of 185 U/g) was applied to a MonoQ column. The three most active fractions eluting from the column were pooled (1.8 U with a specific activity of 650 U/g) and subjected to gel filtration. A total of 0.8 U was recovered in four fractions, with specific activities as high as 3,800 U/g. Kinetics experiments were performed by quantification of cis-dienelactone formation at 305 nm, and a Km value of 190 ± 15 μM was calculated. For quantification of TfdDII in active fractions, aliquots were separated by PAGE (40 to 300 ng of protein per fraction) and stained with Sypro Ruby. A major band with a molecular mass of 41 ± 1 kDa, which contained 30 to 60% of the total protein in the respective fractions, was observed. The N-terminal sequence of this protein (MLTEKAIADSPN) was identical to that expected for TfdDII. Thus, maximal transformation rates calculated during kinetics experiments could be related to the amount of TfdDII present. Taking into account the predicted subunit molecular mass of 36.9 kDa, a kcat value of 1,950 ± 150 min−1 was calculated.
By the photometric test (depletion in the presence of an excess of dienelactone hydrolase as measured at λ = 260 nm), TfdDII exhibited significant activity only against 3-chloromuconate and poor activity against 2-chloro-substituted muconate, as well as against unsubstituted muconate. At a substrate concentration of 0.1 mM, the activity with muconate was only 0.4% ± 0.1% of that with 3-chloromuconate and the activity with 2-chloromuconate was 0.8% ± 0.2%.
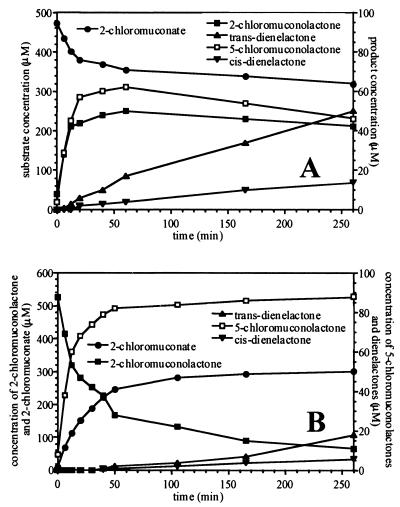
This substrate spectrum significantly resembles that reported for chloromuconate cycloisomerase of Rhodococcus opacus (46). This cycloisomerase differs from those of previously described gram-negative organisms in that it lacks the ability to convert 2-chloromuconolactone and to form trans-dienelactone from 2-chloromuconate. Thus, the kinetic properties of the TfdDII chloromuconate cycloisomerase were assessed by using the partially purified enzyme fractions described above. These fractions were free of any muconate cycloisomerase (capable of transforming 2-chloromuconolactone), muconolactone isomerase (capable of dehalogenating intermediate 5-chloromuconolactone), or dienelactone hydrolase activities. As shown in Fig. 4, the predominant products formed from 2-chloromuconate were identified as 2- and 5-chloromuconolactone. The ratio between 2-chloromuconate, 5-chloromuconolactone, and 2-chloromuconolactone, after a short incubation period, was about 20:4:3, as previously reported for the equilibrium between those compounds after addition of muconate cycloisomerases (50). Further incubation resulted in the formation of mainly trans-dienelactone and minor amounts of cis-dienelactone. A control incubation using partially purified muconate cycloisomerase resulted in the formation of a similar equilibrium between 2-chloromuconate and 5- and 2-chloromuconolactone. However, the rate of trans-dienelactone formation was negligible in those assays (less than 2 μM h−1 in the presence of 100 mU of muconate cycloisomerase per ml, compared to 12 μM h−1 in the presence of 100 mU of TfdII per ml).
FIG. 4.
HPLC analyses of the conversion of 2-chloro-cis,cis-muconate (A) and 2-chloromuconolactone (B) by TfdDII chloromuconate cycloisomerase of R. eutropha JMP222. Reaction mixtures (0.2 ml) contained 50 mM Tris-HCl (pH 8) supplemented with 1 mM MnCl2, 0.5 mM 2-chloro-cis,cis-muconate (A) or 0.5 mM 2-chloromuconolactone (B) and 20 (as determined with 3-chloromuconate as the substrate) (A) or 10 mU (B) of TfdDII.
Incubation of TfdDII with 2-chloromuconolactone resulted in formation of the equilibrium between 2-chloromuconate and 2- and 5-chloromuconolactone described above, demonstrating that TfdDII, in contrast to the Rhodococcus enzyme, can transform 2-chloromuconolactone. Considerable levels of trans-dienelactone formation were visible only after significant accumulation of 5-chloromuconolactone.
DISCUSSION
Chloroaromatic degradation by R. eutropha JMP134 has been the subject of investigations for decades. The recently discovered tfdDII, tfdCII, tfdEII, tfdFII, and tfdBII genes have elucidated a new layer of complexity for chloroaromatic degradation by this strain. It had been shown that tfdII genes are transcribed in R. eutropha JMP134 upon exposure to 2,4-D (22, 24). Moreover, R. eutropha JMP222 strains containing tfdII genes were capable of growing on 3CB (34), and expression in Escherichia coli showed that tfdII genes encode functional enzymes (22). We have demonstrated here that, besides tfdFII (45), at least tfdDII, tfdCII, and tfdEII are translated into functional enzymes in R. eutropha JMP134. We have also shown that tfdFI, which was previously thought to be nonfunctional or poorly functional (34, 45), is transcribed and translated into a functional enzyme. Thus, tfdI, as well as tfdII, gene modules encode a complete set of functional and active enzymes for the transformation of chlorocatechols into 3-oxoadipate. However, whereas significant activities of TfdDII, TfdCII, and TfdFII were obvious during the growth of strain JMP134 on 2,4-D and also of strain JMP222 containing tfdII genes, activities of TfdEII were negligible compared with those of TfdEI. The basis for this poor activity remains to be elucidated. A second obvious difference between tfdI- and tfdII-encoded proteins lies in the TfdD proteins. On the first view, both TfdDI (21, 51) and TfdDII are characterized by their poor activity with 2-chloromuconate and muconate, compared to that with 3-chloromuconate and thus differ only slightly in substrate specificity. However, whereas the relative TfdDI activity with 2-chloromuconate at substrate concentrations between 10 and 100 μM is 6 to 9% of that with 3-chloromuconate (51), the relative activity of TfdDII is less than 1%. This poor activity with 2-chloromuconate is reflected in the higher accumulation of this intermediate and lower growth yield of strain JMP222 containing tfdII genes, compared to those of strain JMP222 containing tfdI genes, when the strain is grown on 3CB.
However, the most striking difference between TfdDI and TfdDII is the poor capability of TfdDII to dechlorinate during cycloisomerization of 2-chloromuconate. In contrast to what was found for other gram-negative bacterium chloromuconate cycloisomerases, where only small amounts of 2- and 5-chloromuconolactone accumulated during 2-chloromuconate turnover (52), dehalogenation to form trans-dienelactone by TfdDII is slow compared to cycloisomerization. Thus, TfdDII seems to be specifically dedicated to the transformation of 3-chloro-substituted muconates, a feature recently described for chloromuconate cycloisomerase of Rhodococcus opacus 1CP (46). However, whereas the Rhodococcus enzyme neither transforms 2-chloromuconolactone nor dehalogenates 2-chloromuconate or 5-chloromuconolactone during cycloisomerization, TfdDII exhibits both capabilities. TfdDII thus appears to be only distantly related, by sequence homology (12) as well as by kinetic properties, to all chloromuconate cycloisomerases described to date (Fig. 5). With respect to biochemical properties, TfdDII is intermediate to muconate and chloromuconate cycloisomerases of gram-negative bacteria, making TfdDII an interesting subject for evolutionary study. Regarding 3-chloromuconate turnover, no differences between TfdDII and other chloromuconate cycloisomerases described so far (e.g., a kcat/Km value of 10 μM−1min−1 compared to 7 to 38 μM−1min−1 for the Pseudomonas sp. strain P51- and Pseudomonas sp. strain B13-derived and TfdDI chloromuconate cycloisomerases) were evident.
FIG. 5.
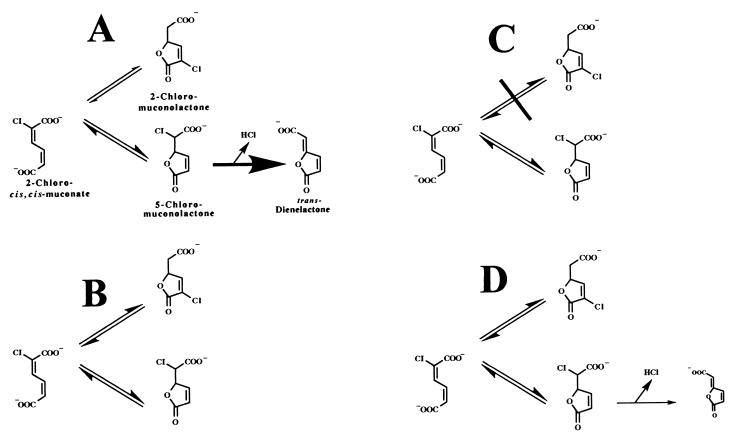
Reactions involved in 2-chloro-cis,cis-muconate conversion by muconate and chloromuconate cycloisomerases. (A) Reactions catalyzed by chloromuconate cycloisomerases such as TfdDI. These enzymes are supposed to preferentially catalyze a 3,6-cycloisomerization, followed by a fast dehalogenation to form trans-dienelactone. (B) Reactions catalyzed by muconate cycloisomerases such as the chromosomally encoded enzyme of R. eutropha JMP134. These enzymes catalyze the formation of an equilibrium between 2-chloro-cis,cis-muconate and 2- and 5-chloromuconolactone. (C) Reactions catalyzed by muconate and chloromuconate cycloisomerase of Rhodococcus opacus 1CP. These enzymes catalyze exclusively a 3,6-cycloisomerization and are not capable of transforming 2-chloromuconolactone. (D) Reactions catalyzed by chloromuconate cycloisomerase TfdDII. This enzyme catalyzes the formation of an equilibrium between 2-chloro-cis,cis-muconate and 2- and 5-chloromuconolactone and is capable of a slow dehalogenation to form trans-dienelactone. Minor amounts of cis-dienelactone were also observed as products.
It has been speculated recently (22) that the main reason for maintenance the tfdII gene cluster, other than as a mechanism for supplying auxiliary functions, such as the facilitated uptake of 2,4-D mediated by TfdK (26), is the supply of a functional transcriptional activator (TfdR) (25). Furthermore it has been assumed that TfdFII is the major maleylacetate reductase complementing a nonfunctional or poorly functional TfdFI gene product. However, we can demonstrate now that TfdFI is, in fact, functional and induced at a high level during growth on 2,4-D.
However, besides these observations, the tfdII genes definitely increase the dosage of chlorocatechol genes and of active gene products in strain JMP134 when the strain is grown on chloroaromatics. As has been recently shown, a duplication of the tfd gene modules results in an increased growth rate on 3CB (5). It can be proposed that the number of tfd genes present in the wild-type strain is limiting for growth on 3CB. As TfdDII, TfdCII, and TfdFII enzymes constitute more than 20% of the total activity during growth on chloroaromatics, it is evident that maintenance of the tfdII genes would be advantageous for the strain. The fact that an elevated number of chlorocatechol genes is necessary to achieve growth on chloroaromatics has been reported in various cases. The transfer of single copies of chlorocatechol genes (tcbR and tcbCDEF) originating from Pseudomonas sp. strain P51 (48) into Pseudomonas putida KT2442 did not result in the expected 3CB-degrading derivatives; multiple chromosomal copies (at least two) were needed to achieve this phenotype (20). Similarly, the transfer of single copies of tfdI or tfdII genes into the chromosome of R. eutropha JMP222 did not result in a 3CB-degrading phenotype, in contrast to the situation where those genes were introduced on a medium-copy-number vector (34). P. putida F1 transconjugants containing two copies of the Pseudomonas sp. strain B13-derived clc element were unable to grow on chlorobenzene, and characterization of chlorobenzene-degrading transconjugants revealed that three to eight copies of the clc element were required for growth, with a larger number of clc elements being associated with increasingly vigorous growth (41). Moreover, the currently available genetic evidence suggests that gene amplification plays an important role in the adaptation of bacteria to chloroaromatic degradation in contaminated environments (31, 32, 49).
High levels of catechol 1,2-dioxygenase and muconate cycloisomerase were observed in 3CB-grown cells of strain JMP222 harboring tfdII genes and of strain JMP134 but not in 3CB-grown cells of strain JMP222 harboring tfdI genes or 2,4-D-grown cells of strain JMP134 (Table 1). Expression of catechol and chlorocatechol operons usually requires LysR-type transcriptional activators and inducer muconate (catechol operons) or 2-chloromuconate (chlorocatechol operons) (28). In strain JMP134, the tfdR gene product is responsible for expression of the tfd operons (25) and it is activated by chloromuconates (14). It has been proposed that, even in the absence of tfdR, low-level expression of tfdCDEF occurs, implying a cross-activation by chromosomally encoded regulatory elements (14). In P. putida it has been shown that CatR, the regulator of the catBC operon (for catechol degradation), interacts with the clcABD promoter region and, likewise, clcR, the regulator of the clcABD operon (for chlorocatechol degradation), was shown to interact with the catBC promoter region. CatR could even complement a ClcR− mutant P. putida strain harboring the clcABD operon for growth on 3CB (29, 33). Moreover, it was recently demonstrated that the LysR-type regulator of the cbn operon of R. eutropha NH9 is activated by both muconate and 2-chloromuconate (30), implying the presence of significant cross talk among the homologous transcriptional activators. Consequently, it seems likely that the accumulated 2-chloromuconate may drive expression of the catechol pathway genes by acting on a putative catR-like element in the chromosomes of strains JMP134 and JMP222. However, induction was observed only in a subset of growth conditions or genetic backgrounds, specifically those that resulted in accumulation of a high level of 2-chloromuconate during growth. Thus, evidently, highly elevated levels of intracellular 2-chloromuconate are necessary to achieve expression of catechol catabolic genes. This agrees with observations of Cosper et al. (6), who proposed that versions of Acinetobacter sp. strain ADP1 containing variants of the muconate cycloisomerase with reduced catalytic properties have increased intracellular levels of muconate available, which are responsible for the activation of LysR-type regulator CatM, which is, in turn, capable of activating the expression of genes encoding enzymes for benzoate degradation.
On one hand, the activation of the catechol pathway genes in R. eutropha can be regarded as a burden for the strain. However, an induction of this type can be proposed to have advantageous effects, as well, specifically for strains such as JMP222(pBBR1M-II). Catechol 1,2-dioxygenase and muconate cycloisomerase have some activity on chlorinated substrate analogues (21, 37) and thus to a certain extent can be responsible for transformation of pathway intermediates. 2-Chloro- and 3-chloromuconate will, thereby, be transformed into chloromuconolactones (50) and protoanemonin (1), respectively. Protoanemonin accumulation, when this compound is produced at low rates, can be prevented by dienelactone hydrolase (3), and muconolactone isomerase can support the dehalogenation of any 5-chloromuconolactone (39) formed during cycloisomerization of 2-chloromuconate by muconate cycloisomerase or TfdDII. Thus, it can be assumed that part of 3CB in strain JMP222(pBBR1M-II) is mineralized by a complex metabolic interplay between chromosome- and plasmid-encoded enzymes.
Acknowledgments
The work was supported by EC grant EVK1-CT-1999-00023. D.P.-P., D.H.P., and B.G. acknowledge support by the collaborative grant BMBF-IB/CONICYT-Chile and FONDECYT 8990004.
We thank Josef Wissing for his support in protein quantification experiments, Rita Getzlaff for N-terminal amino acid sequencing, and Eva Medina and Edward Moore for critical reading of the manuscript.
Footnotes
For a commentary on this article, see page 4049 in this issue.
REFERENCES
- 1.Blasco, R., R.-M. Wittich, M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1995. From xenobiotic to antibiotic. Formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J. Biol. Chem. 270:29229-29235. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brückmann, M., R. Blasco, K. N. Timmis, and D. H. Pieper. 1998. Detoxification of protoanemonin by dienelactone hydrolase. J. Bacteriol. 180:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. [DOI] [PubMed] [Google Scholar]
- 5.Clément, P., D. H. Pieper, and B. González. 2000. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134 (pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 6.Cosper, N., L. Collier, T. Clark, R. Scott, and E. Neidle. 2000. Mutations in catB, the gene encoding muconate cycloisomerase, activate transcription of the distal ben genes and contribute to a complex regulatory circuit in Acinetobacter sp. strain ADP1. J. Bacteriol. 182:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don, R. H., A. J. Weightman, H.-J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 10.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem. J. 174:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ecker, S., T. Widmann, H. Lenke, O. Dickel, P. Fischer, C. Bruhn, and H.-J. Knackmuss. 1992. Catabolism of 2,6-dinitrophenol by Alcaligenes eutrophus JMP 134 and JMP 222. Arch. Microbiol. 158:149-154. [Google Scholar]
- 12.Eulberg, D., E. M. Kourbatova, L. A. Golovleva, and M. Schlömann. 1998. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J. Bacteriol. 180:1082-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farhana, L., and P. B. New. 1997. The 2,4-dichlorophenol hydroxylase of Alcaligenes eutrophus JMP134 is a homotetramer. Can. J. Microbiol. 43:202-205. [DOI] [PubMed] [Google Scholar]
- 14.Filer, K., and A. R. Harker. 1997. Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl. Environ. Microbiol. 63:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukumori, F., and R. P. Hausinger. 1993. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosal, D., I.-S. You, D. K. Chatterjee, and A. M. Chakrabarty. 1985. Genes specifying degradation of 3-chlorobenzoic acid in plasmids pAC27 and pJP4. Proc. Natl. Acad. Sci. USA 82:1638-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer, A., T. Hildenbrand, H. Hoier, K.-L. Ngai, M. Schlömann, and J. J. Stezowski. 1993. Crystallization and preliminary X-ray data of chloromuconate cycloisomerase from Alcaligenes eutrophus JMP134 (pJP4). J. Mol. Biol. 232:305-307. [DOI] [PubMed] [Google Scholar]
- 18.Harker, A. R., R. H. Olsen, and R. J. Seidler. 1989. Phenoxyacetic acid degradation by the 2,4-dichlorophenoxyacetic acid (TFD) pathway of plasmid pJP4: mapping and characterization of the TFD regulatory gene, tfdR. J. Bacteriol. 171:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasberg, T., D. L. Daubaras, A. M. Chakrabarty, D. Kinzelt, and W. Reineke. 1995. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J. Bacteriol. 177:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemba, M., B. Jakobs, R. M. Wittich, and D. H. Pieper. 2000. Chromosomal integration of the tcb chlorocatechol degradation pathway genes as a means of expanding the growth substrate range of bacteria to include haloaromatics. J. Bacteriol. 182:3255-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhm, A. E., M. Schlömann, H.-J. Knackmuss, and D. H. Pieper. 1990. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem. J. 266:877-883. [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, C. M., J. H. J. Leveau, A. J. B. Zehnder, and, J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182:4162-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Leveau, J. H. J., F. Konig, H. Fuchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 25.Leveau, J. H. J., and J. R. van der Meer. 1996. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 178:6824-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leveau, J. H. J., A. J. B. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrubutham, U., and A. R. Harker. 1994. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alcaligenes eutrophus JMP134. J. Bacteriol. 176:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFall, S. M., S. A. Chugani, and A. M. Chakrabarty. 1998. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene 223:257-267. [DOI] [PubMed] [Google Scholar]
- 29.McFall, S. M., T. J. Klem, N. Fujita, A. Ishihama, and A. M. Chakrabarty. 1997. DNase I footprinting, DNA bending and in vitro transcription analyses of ClcR and CatR interactions with the clcABD promoter: evidence of a conserved transcriptional activation mechanism. Mol. Microbiol. 24:965-976. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Chakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa, N., and K. Miyashita. 1999. The chlorocatechol-catabolic transposon Tn5707 of Alcaligenes eutrophus NH9, carrying a gene cluster highly homologous to that in the 1,2,4-trichlorobenzene-degrading bacterium Pseudomonas sp. strain P51, confers the ability to grow on 3-chlorobenzoate. Appl. Environ. Microbiol. 65:724-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa, N., and K. Miyashita. 1995. Recombination of a 3-chlorobenzoate catabolic plasmid from Alcaligenes eutrophus NH9 mediated by direct repeat elements. Appl. Environ. Microbiol. 61:3788-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsek, M. R., S. M. McFall, D. L. Shinabarger, and A. M. Chakrabarty. 1994. Interactions of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc. Natl. Acad. Sci. USA 91:12393-12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Pantoja, D., L. Guzmán, M. Manzano, D. H. Pieper, and B. González. 2001. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieper, D. H., K.-H. Engesser, R. H. Don, K. N. Timmis, and H.-J. Knackmuss. 1985. Modified ortho-cleavage pathway in Alcaligenes eutrophus JMP134 for the degradation of 4-methylcatechol. FEMS Microbiol. Lett. 29:63-67. [Google Scholar]
- 36.Pieper, D. H., H.-J. Knackmuss, and K. N. Timmis. 1993. Accumulation of 2-chloromuconate during metabolism of 3-chlorobenzoate by Alcaligenes eutrophus JMP134. Appl. Microbiol. Biotechnol. 39:563-567. [Google Scholar]
- 37.Pieper, D. H., W. Reineke, K.-H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP 134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 38.Pieper, D. H., K. Stadler-Fritzsche, K.-H. Engesser, and H.-J. Knackmuss. 1993. Metabolism of 2-chloro-4-methylphenoxyacetate by Alcaligenes eutrophus JMP 134. Arch. Microbiol. 160:169-178. [DOI] [PubMed] [Google Scholar]
- 39.Prucha, M., A. Peterseim, K. N. Timmis, and D. H. Pieper. 1996. Muconolactone isomerase of the 3-oxoadipate pathway catalyzes dechlorination of 5-chloro-substituted muconolactones. Eur. J. Biochem. 237:350-356. [DOI] [PubMed] [Google Scholar]
- 40.Prucha, M., V. Wray, and D. H. Pieper. 1996. Metabolism of 5-chlorosubstituted muconolactones. Eur. J. Biochem. 237:355-366. [DOI] [PubMed] [Google Scholar]
- 41.Ravatn, R., S. Studer, D. Springael, A. J. B. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schenzle, A., H. Lenke, P. Fischer, P. A. Williams, and H.-J. Knackmuss. 1997. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP 134. Appl. Environ. Microbiol. 63:1421-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seibert, V., K. Stadler-Fritzsche, and M. Schlömann. 1993. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 175:6745-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solyanikova, I. P., O. V. Maltseva, M. D. Vollmer, L. A. Golovleva, and M. Schlömann. 1995. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J. Bacteriol. 177:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Meer, J. R., R. I. L. Eggen, A. J. B. Zehnder, and W. M. de Vos. 1991. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J. Bacteriol. 173:2425-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vollmer, M. D., P. Fischer, H.-J. Knackmuss, and M. Schlömann. 1994. Inability of muconate cycloisomerases to cause dehalogenation during conversion of 2-chloro-cis,cis-muconate. J. Bacteriol. 176:4366-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollmer, M. D., U. Schell, V. Seibert, S. Lakner, and M. Schlömann. 1999. Substrate specificities of the chloromuconate cycloisomerases from Pseudomonas sp. B13, Ralstonia eutropha JMP134 and Pseudomonas sp. P51. Appl. Microbiol. Biotechnol. 51:598-605. [DOI] [PubMed] [Google Scholar]
- 52.Vollmer, M. D., and M. Schlömann. 1995. Conversion of 2-chloro-cis,cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J. Bacteriol. 177:2938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]