Abstract
The capacity of ribosomal modification to improve antibiotic production by Streptomyces spp. has already been demonstrated. Here we show that introduction of mutations that produce streptomycin resistance (str) also enhances α-amylase (and protease) production by a strain of Bacillus subtilis as estimated by measuring the enzyme activity. The str mutations are point mutations within rpsL, the gene encoding the ribosomal protein S12. In vivo as well as in vitro poly(U)-directed cell-free translation systems showed that among the various rpsL mutations K56R (which corresponds to position 42 in E. coli) was particularly effective at enhancing α-amylase production. Cells harboring the K56R mutant ribosome exhibited enhanced translational activity during the stationary phase of cell growth. In addition, the K56R mutant ribosome exhibited increased 70S complex stability in the presence of low Mg2+ concentrations. We therefore conclude that the observed increase in protein synthesis activity by the K56R mutant ribosome reflects increased stability of the 70S complex and is responsible for the increase in α-amylase production seen in the affected strain.
Of the various starch-hydrolyzing enzymes, the α-amylases (1,4-α-d-glucan glucanohydrolase; EC 3.2.1.1) are of particular importance, as they are responsible for the solubilization of starch. As such, these enzymes are currently among the most widely utilized in biotechnology. Although α-amylases can be derived from plants and animals, it is the enzymes from microbial sources (typically Bacillus spp.) that are generally used to meet the expanding industrial demands. In addition to well-established applications in starch saccharification and in the textile, food, brewing, and distilling industries, bacterial α-amylases are now also used in areas of clinical, medicinal, and analytical chemistry (for a review, see reference 23). Contributing to the appeal of bacterial α-amylases is their high degree of optimization (e.g., highly thermostable or alkali tolerant), conferred by means of protein engineering or discovered through investigation of novel microorganisms (7; also reviewed in references 2 and 19).
α-Amylase is an end-type enzyme that hydrolyzes α-1,4-glucosidic linkages from starch and various other types of oligosaccharides, and several of the enzymes from Bacillus subtilis have been cloned, sequenced, and subjected to three-dimensional structural analysis (5, 18). The production of extracellular α-amylase by B. subtilis is known to be controlled by several genes, including amyR and pap, and to vary with changes in the environmental conditions as well as with changes in the structure and function of the cell envelope (1). Mutagenesis induced by chemicals such as N-methyl-N′-nitro-N-nitrosoguanidine or by UV radiation has been employed to obtain hyperproducing strains, in which α-amylase synthesis can often be doubled or even tripled (32; for a review, see reference 23). Current developments in gene engineering also make possible improvements in amylase production through molecular breeding (13).
We previously showed that a certain streptomycin resistance-producing mutation (str) in rpsL, which encodes the ribosomal protein S12, also gives rise to antibiotic production in Streptomyces lividans and Streptomyces coelicolor (8, 26). Later, we used other bacterial genera to demonstrate that introducing a specific str mutation together with a gentamicin resistance-producing mutation (gen) gives rise to a marked increase in antibiotic production and to shed light on the mechanism by which antibiotic is produced in S. coelicolor (10). It was also demonstrated that by introducing various combinations of drug resistance-producing mutations, we could increase the production of an antibiotic in a stepwise manner (11).
The finding that certain rpsL mutations induce dramatic activation of antibiotic production prompted us to hypothesize that bacterial gene expression may be altered dramatically by modifying ribosomal proteins or rRNA. Thus, our ultimate aim has been to develop “ribosome engineering” (20) as a rational approach to taking full advantage of bacterial capabilities. Because certain extracellular enzymes (such as α-amylase and protease) are known to be produced during the late growth phase, it seemed plausible that synthesis of these enzymes might be enhanced by introducing certain streptomycin resistance-producing (rpsL) mutations. The aim of the present study, therefore, was to assess the efficacy with which ribosome engineering could be used to enhance enzyme synthesis in B. subtilis by examining its effect on α-amylase (and protease) production.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacillus subtilis rpsL mutants (WL1, WL2, WL3, WL4, WL5, and WL9) were all derived from strain 168 (10, 14), which is a standard (Marburg) strain frequently used for studying sporulation. Mutants WL6 and WL15, which contain a K56H and a K101E substitution, respectively, were constructed by site-directed mutagenesis using the plasmid pKF19k-rpsL as a template (14). The oligonucleotides 5′-AGTTCGGTTTGTGCGGTGTCATTG-3′ (for K56H) and 5′-GGTAAGTCTTCTACACGTCCG-3′ (for K101E), which include the mutation sites (underlined), were used to generate rpsL6 and rpsL15. The mutant WL10 (G105W) was prepared using PCR random mutagenesis on the basis of this mutant's ability to resist streptomycin, as described previously (14). Detection of the rpsL mutations was accomplished using PCR, the products of which were directly sequenced using a sequence analyzer (14).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotype | Amino acid exchange in ribosome protein S12 | Source and/or reference |
|---|---|---|---|
| Bacillus subtilis strains | |||
| 168 | trpC2 | Laboratory stock | |
| KO272 | trpC2 str-10 (rpsL5) Smr | K56Q | Spontaneous rpsL mutant; 10 |
| WL1 | trpC2 rpsL1 Smr | K56R | 14 |
| WL2 | trpC2 rpsL2 Smr | K56N | 14 |
| WL3 | trpC2 rpsL3 Smr | K56T | 14 |
| WL4 | trpC2 rpsL4 Smr | K56I | 14 |
| WL5 | trpC2 rpsL5 Smr | K56Q | KO272→168a |
| WL6 | trpC2 rpsL6 Smr | K56H | pKF19k-rpsL6→168 |
| WL9 | trpC2 rpsL9 Smr | P104S | 14 |
| WL10 | trpC2 rpsL10 Smr | G105W | PCR product→168 |
| WL15 | trpC2 rpsL15 Smr | K101E | pKF19k-rpsL15→168 |
| Plasmids | |||
| pKF19k-rpsL | Template for site-directed mutagenesis | 14 | |
| pKF19k-rpsL6 | pKF19k-rpsL containing the rpsL6 (K56H) mutation | This study | |
| pKF19k-rpsL15 | pKF19k-rpsL containing the rpsL15 (K101E) mutation | This study |
The strain to the right of the arrow was transformed with the chromosomal DNA, PCR product, or plasmid to the left of the arrow.
Media and growth conditions.
NG medium, which was originally developed for antibiotic production by B. subtilis (10), was used for α-amylase production. It contained (per liter) 10 g of nutrient broth (Difco), 10 g of glucose, 2 g of NaCl, 5 mg of CuSO4 · 5H2O, 7.5 mg of FeSO4 · 7H2O, 3.6 mg of MnSO4 · 5H2O, 15 mg of CaCl2 · 2H2O, 9 mg of ZnSO4 · 7H2O, and 50 mg of tryptophan, as required (adjusted to pH 7.2 with NaOH). Strains were initially grown for 12 h in NG medium (10 ml in 100-ml flasks) at 37°C. Thereafter, aliquots (0.1 ml) of the culture broth were inoculated into 10 ml of NG medium in 100-ml flasks and cultured for the indicated times on a rotary shaker (200 rpm) at 45°C instead of 37°C (due to higher productivity of α-amylase at 45°C). Protease production was carried out the same as α-amylase production, except that cells were grown in PM medium, which contained (per liter) 5 g of peptone (Difco), 5 g of meat extract (Difco), 5 g of NaCl, 1 g of glucose, and 50 mg of tryptophan, as required (adjusted to pH 7.2 with NaOH).
Assay for α-amylase and protease.
α-Amylase activity was assayed using the method of Fuwa (6). A sample (0.2 ml) of 0.5% soluble starch in 0.05 M phosphate buffer (pH 6.0) was mixed with 0.1 ml of enzyme solution. After incubation for 15 to 45 min at 40°C, a 20-μl aliquot of the reaction mixture was added to 0.5 ml of 0.2 mM l2-Kl solution, and the optical density at 700 nm was measured in a spectrophotometer. One unit of enzyme was defined as the amount necessary to hydrolyze 0.1 mg of soluble starch in 1 min. Protease activity was assayed as described by Shimizu et al. (27). One unit of enzyme was defined as the amount of the enzyme which solubilized a 1-μg equivalent of tyrosine in 1 min.
Incorporation of [3H]leucine.
Strains were grown for various periods in diluted (1/8) NG medium, after which [3H]leucine (0.2 μCi, 200 μM) was added to 10-ml samples of culture and incubated for 0, 10, 20, or 30 min. One-milliliter aliquots were then collected, mixed with 1 ml of cold 10% (wt/vol) trichloroacetic acid, and kept on ice for 30 min to precipitate the protein from the solution. The precipitated protein was collected by filtration on a nitrocellulose filter (pore size, 0.45 μm) and washed with 10 ml of 5% (wt/vol) trichloroacetic acid. The filters (containing protein) were then dried and their radioactivity measured using a liquid scintillation counter.
In vitro translation assay.
poly(U)-directed cell-free synthesis of polyphenylalanine was carried out as described by Legault-Demare and Chambliss (16) with slight modifications. Bacillus subtilis cells grown to various growth phases in NG medium were collected by centrifugation and washed with standard buffer (10 mM Tris-HCl [pH 7.7], 10 mM magnesium acetate, 30 mM ammonium acetate, and 6 mM 2-mercaptoethanol) containing 2 mM phenylmethylsulfonyl fluoride. The cell paste was broken by grinding with aluminum oxide powder (2 g for each 1 g of cell paste; Wako) for 10 min, after which the ground paste was suspended in standard buffer containing 2 mM phenylmethylsulfonyl fluoride plus 10% (wt/vol) glycerol. The resultant lysate was treated with RNase-free DNase I (10 U/ml; Takara) for 10 min on ice and then centrifuged at 30,000 × g for 30 min to remove cell debris. The resultant supernatant was further fractionated into the S-150 fraction (supernatant) and the ribosomes (precipitant) by centrifugation at 150,000 × g for 3 h. The ribosomes were washed once more with standard buffer plus 10% (wt/vol) glycerol, after which both the ribosomes and the S-150 fraction were dialyzed against 60 volumes of standard buffer plus 10% (wt/vol) glycerol for 6 h, divided into small aliquots, frozen in liquid nitrogen, and stored at −80°C until use. The reaction mixture for polyphenylalanine synthesis (100 μl) consisted of 55 mM HEPES-KOH (pH 7.5), 1 mM dithiothreitol, 210 mM potassium acetate, 27.5 mM ammonium acetate, 10.7 mM magnesium acetate, 68 μM folinic acid, 5 mM spermidine, 1.2 mM ATP, 0.8 mM GTP, 0.64 mM 3′,5′-cyclic AMP, 80 mM creatine phosphate, 0.25 mg/ml creatine kinase, 200 units/ml RNase inhibitor (recombinant solution; Wako), 0.45 mg/ml Escherichia coli total tRNA, 0.4 mM concentrations of each l-amino acid (except phenylalanine, depending on the labeled amino acid used), 0.22 μM [14C]phenylalanine (0.01 μCi), 0.5 mg/ml S-150 fraction, and 10 A260 units/ml ribosome fraction. The reaction was initiated by adding 75 μg of poly(U), after which the mixture was incubated at 37°C for the appropriate time; 1 ml of 10% (wt/vol) trichloroacetic acid was then added to stop the reaction, after which the mixtures were boiled for 15 min. Precipitated proteins were collected on nitrocellulose filters, and the incorporation of [14C]phenylalanine into the acid-insoluble fraction was determined using a liquid scintillation counter. To reduce levels of endogenous mRNA, ribosomes plus the S-150 were preincubated just prior to use at 37°C for 10 min in the reaction mixture as described above without the amino acids, energy-generating reagents, or poly(U).
Sucrose density gradient centrifugation of ribosomes.
Crude ribosomes prepared from mid-exponential-phase cells (see above) were precipitated by centrifugation at 150,000 × g for 3 h and resuspended in standard buffer containing the specified concentration of magnesium acetate. The ribosomes were then laid onto a 10% to 30% (wt/vol) linear sucrose gradient prepared in buffer containing the same concentration of magnesium acetate and centrifuged in a SW41Ti rotor at 38,000 rpm for 4 h at 4°C. The profiles of the ribosomes were observed at 254 nm using an ATTO Bio-Mini UV Monitor equipped with a Biocomp Piston Gradient Fractionator (Towa Kagaku).
RESULTS
Isolation of B. subtilis rpsL mutants.
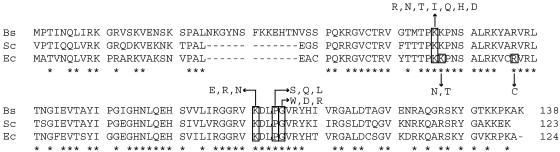
We previously isolated various rpsL mutants that developed spontaneously on plates containing streptomycin (10, 14). In addition, to examine an even wider variety of rpsL mutations with the aim of improving α-amylase production, we also prepared three additional rpsL mutants, WL6 (K56H), WL10 (G105W), and WL15 (K101E), which are known to confer streptomycin resistance in E. coli (30) (Table 1). Although strains WL4 and WL5 grew at a slightly slower rate than parental strain 168, all of the other strains harboring rpsL mutations grew as well as strain 168 and sporulated well, producing about 109 spores/ml in sporulation medium. Strains WL1, WL2, WL3, WL4, WL5, and WL6 all exhibited a high degree of resistance to streptomycin, tolerating levels of >500 μg/ml, while strains WL9, WL10, and WL15 showed an intermediate resistance, tolerating levels of 50 to 100 μg/ml. By contrast, strain 168 tolerated streptomycin at only 1 μg/ml. The rpsL mutations in B. subtilis, together with the previously reported rpsL mutations in E. coli and S. coelicolor, are illustrated in Fig. 1.
FIG. 1.
Alignment of amino acid sequences of the ribosomal S12 proteins from several bacteria and location of mutations that confer resistance to streptomycin. Data are from the present work and references 9, 14 and 22. Bs, Sc, and Ec represent Bacillus subtilis, Streptomyces coelicolor, and Escherichia coli, respectively. Asterisks indicate identical amino acids.
Enhanced production of α-amylase in some rpsL mutants.
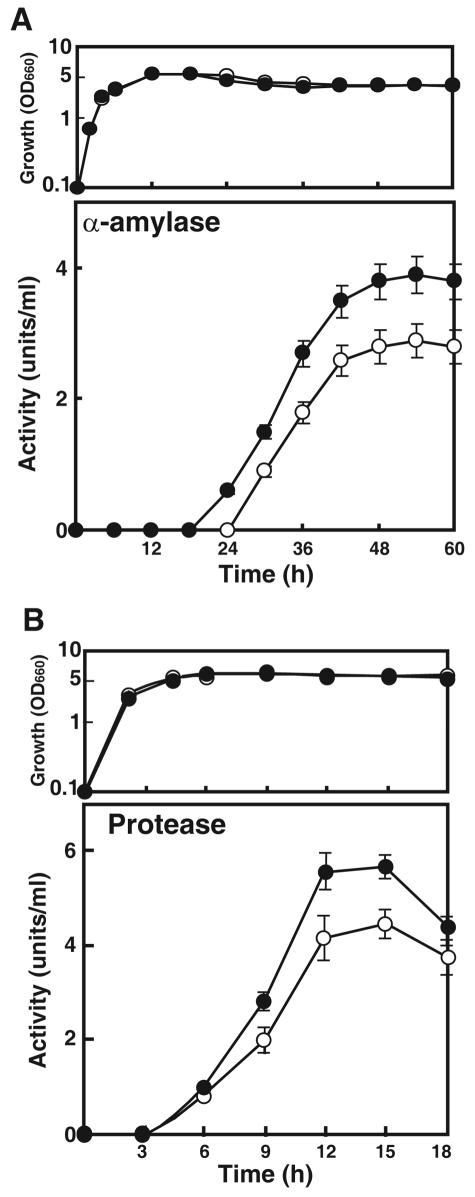
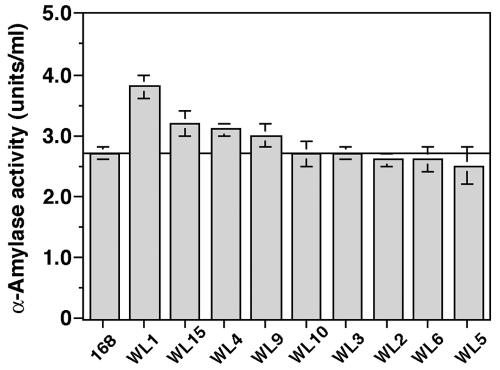
We first monitored growth and α-amylase production in strain 168 (Fig. 2A). In NG medium, the production of α-amylase commenced during the stationary phase (24 h) and reached a maximum by 54 h. We next examined the capacity of rpsL mutation to enhance α-amylase production using the rpsL mutant strains listed in Table 1. It was found that the K56R mutation led to the greatest increase in α-amylase production but that increases were also seen with the K101E, K56I, and P104S mutations (Fig. 3). The other mutations (G105W, K56T, K56N, K56H, and K56Q) were without effect. The α-amylase production by strain WL1 (Fig. 2A), which harbored the K56R mutant ribosome, was about 40% greater than that seen with the parental strain. It is notable that the K56R mutation was effective also in improving protease productivity (30% greater than that of the parental strain) (Fig. 2B). Thus, certain rpsL mutations effectively improved the production of extracellular enzymes, α-amylase, and protease.
FIG. 2.
(A) Time course of growth (upper panel) and α-amylase production (lower panel) by B. subtilis in NG medium at 45°C. (B) Time course of growth and protease production in PM medium at 45°C. The values shown in each panel are the means of the results from three independent experiments. Error bars represent standard deviations. Symbols: ○, 168 (parental strain); •, Smr mutant WL1 (rpsL1). OD660, optical density at 660 nm.
FIG. 3.
Comparison of α-amylase production in the parental strain (168) and rpsL mutant strains (WL1 to WL15). Cultivation was carried out in NG medium for 54 h at 45°C. The values are means of the results from four independent experiments. Error bars represent standard deviations.
The WL1 rpsL mutant strain exhibits enhanced protein synthesis activity.
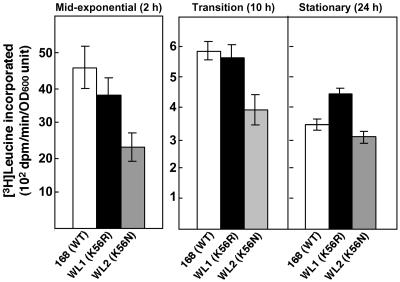
Recent work in our laboratory revealed that cells harboring the K88E (in S. lividans and E. coli) or K88R (in Streptomyces albus) rpsL mutation exhibited enhanced protein synthesis during the stationary phase, as determined by both in vivo and in vitro translation assays (9, 22, 29). We therefore reasoned that the enhanced α-amylase production by B. subtilis strain WL1 harboring the K56R rpsL mutation might reflect a similar effect on protein synthesis. To test that idea, we first monitored the abilities of wild-type and mutant cells to synthesize protein in vivo. Strains 168, WL1, and WL2, which served as a reference strain, were grown to various growth phases in diluted (1/8) NG medium (to promote the incorporation of [3H]leucine), after which [3H]leucine was added and the cells were incubated for an additional 30 min. In parental strain 168, protein synthesis was maximal during the mid-exponential phase (2 h) and then declined sharply (by 13-fold) once cells entered the transition or stationary phase (Fig. 4). By contrast, strain WL1 (K56R mutant) sustained a higher level of protein synthesis during the stationary phase than was seen with strain 168: protein synthesis by strain WL1 was only reduced eightfold during the stationary phase. Strain WL2 (K56N mutant), which did not show enhanced α-amylase production, also did not show enhanced protein synthesis during the stationary phase (Fig. 4).
FIG. 4.
Capacity of wild-type (168) and mutant (WL1 and WL2) strains to synthesize proteins during the indicated growth phases. Incorporation of [3H]leucine into the total protein fraction was determined as described in Materials and Methods. The values are the means of the results from three independent experiments. Error bars represent standard deviations. OD600, optical density at 600 nm.
K56R mutant ribosomes exhibit enhanced translational activity.
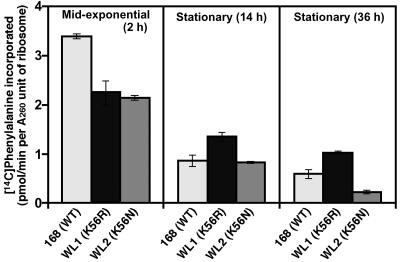
To confirm the results of the in vivo experiments, we measured the in vitro translational activity of ribosomes isolated from wild-type and mutant cells at various growth stages using a poly(U)-directed cell-free translation system (polyphenylalanine synthesis) (see Materials and Methods). For ribosomes isolated from wild-type (strain 168) cells, the rate of polyphenylalanine synthesis was maximal when the organelles were extracted during the mid-exponential phase and was markedly lower when they were extracted during the stationary phase (Fig. 5). Notably, K56R mutant ribosomes isolated from strain WL1 cells during the stationary phase (36 h) exhibited twice as much activity as those from the wild-type strain. No such increase in ribosomal activity was seen with the strain WL2 K56N mutant, indicating the enhanced translational activity during late growth was a specific characteristic of the K56R mutant ribosome.
FIG. 5.
In vitro translation activities of ribosomes harvested from cells during the indicated growth phases. Strains were grown in NG medium at 45°C. Samples were taken during the indicated growth phases, and in vitro translation was carried out using a poly(U)-directed cell-free translation system (see Materials and Methods). The values are the means of the results from three independent experiments. Error bars represent standard deviations.
K56R mutant ribosomes form more stable 70S complexes.
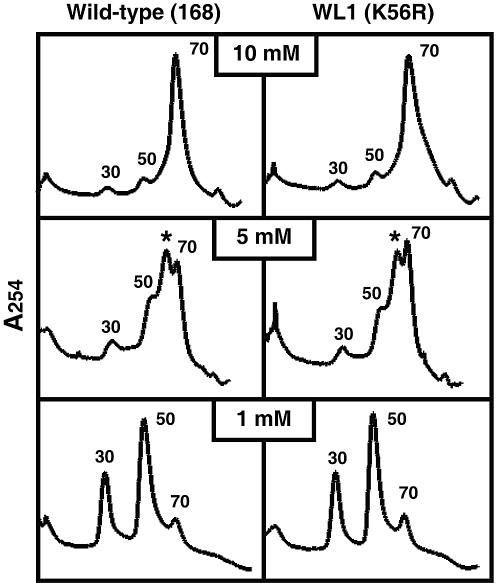
Starvation for an essential amino acid increases the spatial separation between the two ribosomal subunits (21, 34). Indeed, ribosomes from methionine-starved cells are reported to be structurally unstable in the presence of low concentrations of Mg2+, which is indicative of the structural instability of open-form ribosomes (25). We therefore reasoned that K56R mutant ribosomes from strain WL1, which actively synthesize protein even under starvation conditions, such as those encountered during the late stationary phase, may maintain more stable intersubunit interactions. To test this hypothesis, we prepared ribosomes from wild-type and mutant strains grown to mid-exponential phase in NG medium and examined the stability of the 70S complex in the presence of selected concentrations of Mg2+. At a high Mg2+ concentration (10 mM), almost all ribosomes were recovered in the 70S form (Fig. 6). However, dissociation of 70S ribosomes into the 30S and 50S subunits, along with formation of an intermediate (detected as a peak between the 50S and 70S peaks), occurred when the Mg2+ concentration was reduced to 5 mM or less. In that regard, a notably larger fraction (about 1.5-fold size) of the K56R mutant ribosomes from strain WL1 remained in the 70S form in the presence of low concentrations (5, 2, and 1 mM) of Mg2+, as confirmed by four independent experiments. A typical result at 5 mM and 1 mM Mg2+ concentrations is presented as Fig. 6. The K56N mutant ribosomes from strain WL2 displayed a pattern similar to that of strain 168 (data not shown). These findings suggest that the K56R (but not K56N) ribosomes are structurally more stable under stressful conditions, which could account for the enhanced protein synthesis observed in strain WL1.
FIG. 6.
Sucrose gradient analysis of ribosome fractions. The patterns of dissociation of the 70S complex into 30S and 50S subunits at different concentrations of Mg2+ were determined using washed ribosomes harvested from cells grown to mid-exponential phase (2 h) in NG medium (see Materials and Methods). Asterisks indicate the peaks for the intermediates.
DISCUSSION
Working with B. subtilis rpsL mutations, we have been able to demonstrate the capacity of several (especially K56R) to improve production of α-amylase and protease. Although the observed increase of α-amylase or protease production was 1.3- to 1.4-fold, these values seem to be significant, since the increase of enzyme production caused by a single mutation is in general at most twofold in either wild-type strains and industrial strains that had been bred to produce a high level of enzymes (32; S. Itoh, personal communication). The present results show (i) that cells harboring the K56R mutant ribosome can sustain a higher level of gross protein synthetic activity than wild-type cells during the late growth phase, as determined by measuring the incorporation of [3H]leucine, (ii) that K56R mutant ribosomes isolated during the late stationary phase have a higher capacity for translating synthetic polynucleotide [poly(U)] than wild-type ribosomes, and (iii) that the mutant ribosomes are structurally more stable than wild-type ribosomes under stressful conditions. It is thus concluded that the enhanced protein synthesis seen with cells harboring the K56R mutant ribosome is likely due, at least in part, to the increased stability of the 70S particle. This proposal is consistent with earlier findings that the Streptomyces K88E and the E. coli K87E ribosomal mutants also show enhanced protein synthesis during the late phases of growth (9, 22, 29). Although much progress has been made toward increasing the productivity of amylase-producing strains (23), the present method is characterized by the host cell's amenability (generation of spontaneous drug resistance-producing mutations) and is thus applicable to a number of microorganisms (10, 11, 29). It should also be emphasized that rpsL mutations found in the present study caused no impairment of growth and sporulation under the conditions tested.
The effects of streptomycin on bacterial ribosomes have been studied in great detail (3, 4, 31). Among the numerous actions attributed to this drug, its ability to cause mRNA codons to be misread is the best characterized. In that regard, it is well known that S12 mutations that confer streptomycin resistance can increase the accuracy of protein synthesis, which was the case with B. subtilis rpsL mutants (14). However, working with S. coelicolor and S. lividans, it became apparent that the increased accuracy of the protein synthesis by rpsL mutants is unrelated to the increased production of antibiotic (22). It is therefore noteworthy that, in the present study, strain WL1 harboring the K56R mutant ribosome exhibited a higher degree of translational activity during the stationary phase (Fig. 4 and 5). This increased activity is likely the result of a more stable ribosomal structure (Fig. 6) and could explain why strain WL1 is capable of producing greater amounts of α-amylase and protease than the wild-type strain. Moreover, production of secondary metabolites, including α-amylase and protease, usually commences during the late growth phase (i.e., the transition or stationary phase); thus, the enhanced protein synthesis seen at that time would be expected to include increased production of these extracellular enzymes. Consistent with that idea, strain WL2 harboring the K56N mutant ribosome, which showed no increase in α-amylase production, did not exhibit increased translational activity during the stationary phase (Fig. 4 and 5). In E. coli, the rate of protein synthesis is reportedly reduced by more than 90% when cells are starved for amino acids (28). That the protein synthetic activity of strain WL1 persisted despite such conditions distinguishes it from previously studied wild-type strains.
The ribosomal protein S12 is a component of the 30S subunit in bacteria. Best characterized is its role in determining the efficiency with which cognate tRNAs are selected, which contributes to the accuracy achieved when decoding mRNA sequences (3, 14, 15). Recent structural analyses of the 50S and 30S subunits, as well as the intact 70S ribosome, have greatly advanced our understanding of protein synthesis (24, 33). All of the Smr mutations found in the S12 protein are limited to two conserved regions: region 1, spanning amino acid residues 55 to 59 (in numbering system for B. subtilis), and region 2, spanning residues 101 to 107. Both of these regions comprise loop structures in the S12 protein, with region 1 projecting into the space between the 530 loop and the 1492 to 1493 strand of the decoding site (17). Most mutations within region 1 lead to a hyperaccurate phenotype (15), though they can weaken the interactions between the tRNA-mRNA complex and the 30S A site (33). Although it is difficult at present to explain how the K56R mutation in the S12 protein mediates ribosomal stabilization, it is possible that it affects interactions between the 16S and 23S rRNAs.
Our previous studies have shown that certain mutations within S12 confer resistance to streptomycin and enhance or activate antibiotic production (reviewed in reference 20). It is worth mentioning that the K56R S12 mutation found to enhance α-amylase production in B. subtilis also very effectively enhanced antibiotic production (80-fold) of this organism (10) and that the increased production of antibiotic is also attributable to the enhanced protein synthetic activity of the K56R mutant ribosome during the stationary growth phase. Recently, S. T. Jorgensen and colleagues (personal communication) determined that certain rpoB (the gene encoding the RNA polymerase β-subunit) mutations producing rifampin resistance in Bacillus species can give rise to a twofold increase of α-amylase production. This suggests that rpsL-rpoB double mutants might elicit an even greater capacity to produce α-amylase than either mutation alone, as was the case with antibiotic production by Streptomyces rpsL-rpoB double mutants (11, 12, 29).
Acknowledgments
This work was supported by grants from the Organized Research Combination System and the Effective Promotion of Joint Research of Special Coordination Funds (the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government)
We are grateful to S. T. Jorgensen (Novozymes, Denmark) and S. Itoh (Hokkaido University) for permitting us to cite their unpublished data.
REFERENCES
- 1.Ayusawa, D., Y. Yoneda, K. Yamane, and B. Maruo. 1975. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular α-amylase and protease in a Bacillus subtilis mutant. J. Bacteriol. 124:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoldo, C., and G. Antranikian. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 6:151-160. [DOI] [PubMed] [Google Scholar]
- 3.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 4.Cundliffe, E. 1990. Recognition sites for antibiotics within rRNA, p. 479-490. In A. D. W. E. Hill, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D. C.
- 5.Fujimoto, Z., K. Takase, N. Doui, M. Momma, T. Matsumoto, and H. Mizuno. 1998. Crystal structure of a catalytic-site mutant α-amylase from Bacillus subtilis complexed with maltopentaose. J. Mol. Biol. 277:393-407. [DOI] [PubMed] [Google Scholar]
- 6.Fuwa, H. 1954. A new method for microdetermination of amylase activity by the use of amylose as the substrate. J. Biochem. (Tokyo) 41:603. [Google Scholar]
- 7.Hagihara, H., K. Igarashi, Y. Hayashi, K. Endo, K. Ikawa-Kitayama, K. Ozaki, S. Kawai, and S. Ito. 2001. Novel α-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Appl. Environ. Microbiol. 67:1744-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hesketh, A., and K. Ochi. 1997. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J. Antibiot. (Tokyo) 50:532-535. [DOI] [PubMed] [Google Scholar]
- 9.Hosaka, T., N. Tamehiro, N. Chumpolkulwong, C. Hori-Takemoto, M. Shirouzu, S. Yokoyama, and K. Ochi. 2004. The novel mutation K87E in ribosomal protein S12 enhances protein synthesis activity during the late growth phase in Escherichia coli. Mol. Genet. Genomics 271:317-324. [DOI] [PubMed] [Google Scholar]
- 10.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, H., and K. Ochi. 2001. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 67:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, H., Q. Zhang, and K. Ochi. 2002. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase β subunit) of Streptomyces lividans. J. Bacteriol. 184:3984-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikawa, K., H. Araki, Y. Tsujino, Y. Hayashi, K. Igarashi, Y. Hatada, H. Hagihara, T. Ozawa, K. Ozaki, T. Kobayashi, and S. Ito. 1998. Hyperexpression of the gene for a Bacillus α-amylase in Bacillus subtilis cells: enzymatic properties and crystallization of the recombinant enzyme. Biosci. Biotechnol. Biochem. 62:1720-1725. [DOI] [PubMed] [Google Scholar]
- 14.Inaoka, T., K. Kasai, and K. Ochi. 2001. Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis. J. Bacteriol. 183:4958-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurland, C. G., F. Jorgensen, A. Richter, M. Ehrenberg, N. Bilgin, and A. M. Rojas. 1990. Through the accuracy window, p. 513-526. In W. E. Hill, A. E. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome; structure, function, and evolution. American Society for Microbiology, Washington, D. C.
- 16.Legault-Demare, L., and G. H. Chambliss. 1974. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J. Bacteriol. 120:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodmell, J. S., and A. E. Dahlberg. 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277:1262-1267. [DOI] [PubMed] [Google Scholar]
- 18.Machius, M., G. Wiegand, and R. Huber. 1995. Crystal structure of calcium-depleted Bacillus licheniformis α-amylase at 2.2 Å resolution. J. Mol. Biol. 246:545-559. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen, J. E., and T. V. Borchert. 2000. Protein engineering of bacterial α-amylases. Biochim. Biophys. Acta 1543:253-274. [DOI] [PubMed] [Google Scholar]
- 20.Ochi, K., S. Okamoto, Y. Tozawa, T. Inaoka, T. Hosaka, J. Xu, and K. Kurosawa. 2004. Ribosome engineering and secondary metabolite production. Adv. Appl. Microbiol. 56:155-184. [DOI] [PubMed] [Google Scholar]
- 21.Ofverstedt, L. G., K. Zhang, S. Tapio, U. Skoglund, and L. A. Isaksson. 1994. Starvation in vivo for aminoacyl-tRNA increases the spatial separation between the two ribosomal subunits. Cell 79:629-638. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto-Hosoya, Y., T. Hosaka, and K. Ochi. 2003. An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 149:3299-3309. [DOI] [PubMed] [Google Scholar]
- 23.Pandey, A., P. Nigam, C. R. Soccol, V. T. Soccol, D. Singh, and R. Mohan. 2000. Advances in microbial amylases. Biotechnol. Appl. Biochem. 31:135-152. [DOI] [PubMed] [Google Scholar]
- 24.Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108:557-572. [DOI] [PubMed] [Google Scholar]
- 25.Sells, B. H., and H. L. Ennis. 1970. Polysome stability in relaxed and stringent strain of Escherichia coli during amino acid starvation. J. Bacteriol. 102:666-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu, Y., T. Nishino, and S. Murao. 1984. Control of protease activity during sporulation of Bacillus subtilis. Agric. Biol. Chem. 48:3109-3114. [Google Scholar]
- 28.Sorensen, M. A., K. F. Jensen, and S. Pedersen. 1994. High concentrations of ppGpp decrease the RNA chain growth rate: implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J. Mol. Biol. 236:441-454. [DOI] [PubMed] [Google Scholar]
- 29.Tamehiro, N., T. Hosaka, J. Xu, H. Hu, N. Otake, and K. Ochi. 2003. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol. 69:6412-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timms, A. R., H. Steingrimsdottir, A. R. Lehmann, and B. A. Bridges. 1992. Mutant sequences in the rpsL gene of Escherichia coli B/r: mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol. Gen. Genet. 232:89-96. [DOI] [PubMed] [Google Scholar]
- 31.Wallace, B. J., P.-C. Tai, and B. D. Davies. 1979. Streptomycin and related antibiotics, p. 272-303. In F. E. Hahn (ed.), Antibiotics. V. Mechanism of action of antibacterial agents. Springer-Verlag, New York, N.Y.
- 32.Yoneda, Y., and B. Maruo. 1975. Mutation of Bacillus subtilis causing hyperproduction of α-amylase and protease, and its synergistic effect. J. Bacteriol. 124:48-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, K., L. Pettersson-Landen, M. G. Fredriksson, L. G. Ofverstedt, U. Skoglund, and L. A. Isaksson. 1998. Visualization of a large conformation change of ribosomes in Escherichia coli cells starved for tryptophan or treated with kirromycin. Exp. Cell Res. 238:335-344. [DOI] [PubMed] [Google Scholar]