Abstract
The heat resistance of Campylobacter jejuni strains AR6 and L51 and the heat resistance of Campylobacter coli strains DR4 and L6 were measured over the temperature range from 50 to 60°C by two methods. Isothermal measurements yielded D55 values in the range from 4.6 to 6.6 min and z values in the range from 5.5 to 6.3°C. Dynamic measurements using differential scanning calorimetry (DSC) during heating at a rate of 10°C/min yielded D55 values of 2.5 min and 3.4 min and z values of 6.3°C and 6.5°C for AR6 and DR4, respectively. Both dynamic and isothermal methods yielded mean D55 values that were substantially greater than those reported previously (0.75 to 0.95 min). DSC analysis of each strain during heating at a rate of 10°C/min yielded a complex series of overlapping endothermic peaks, which were assigned to cell wall lipids, ribosomes, and DNA. Measurement of the decline in the numbers of CFU in calorimetric samples as they were heated showed that the maximum rate of cell death occurred at 56 to 57°C, which is close to the value predicted mathematically from the isothermal measurements of D and z (61°C). Both estimates were very close to the peak m1 values, 60 to 62°C, which were tentatively identified with unfolding of the 30S ribosome subunit, showing that cell death in C. jejuni and C. coli coincided with unfolding of the most thermally labile regions of the ribosome. Other measurements indicated that several essential proteins, including the α and β subunits of RNA polymerase, might also unfold at the same time and contribute to cell death.
Thermophilic Campylobacter species, particularly Campylobacter jejuni and to a lesser extent Campylobacter coli, are the most important cause of bacterial gastroenteritis in the developed world (5, 11). This is surprising because campylobacters are considered to be poor survivors in the environment. In particular, they are considered to be more heat and radiation sensitive than most bacteria, including other gram-negative food-borne pathogens, such as Salmonella spp. and Escherichia coli (8). They are also considered to be sensitive to drying and are unable to multiply at temperatures below 31°C (6, 22). However, there have been few detailed studies that have determined D and z values. In this study our goals were to examine the heat resistance of several strains of C. jejuni and C. coli over the temperature range from 50 to 60°C and to investigate the mechanism of heat damage by use of differential scanning calorimetry (DSC).
MATERIALS AND METHODS
Strains.
Two strains of C. jejuni and two strains of C. coli were examined. C. jejuni AR6, an isolate from poultry feces, was supplied by D. Newell (Food and Environmental Safety, Veterinary Laboratories Agency, Weybridge, United Kingdom). C. jejuni L51 and C. coli L6 were isolated from broiler chickens on a slaughter line. C. coli DR4, which was isolated from retail chicken, was obtained from T. Humphrey (University of Bristol, Bristol, United Kingdom) and was found previously to be particularly heat resistant when it was inoculated onto chicken breasts and submerged in hot water.
Atmosphere for incubation.
All incubation was done in a microaerobic atmosphere, which was produced by partially evacuating anaerobic jars (without a catalyst) to one-third atmospheric pressure and then adding a mixture of 10% H2, 10% CO2, and 80% N2 to obtain an atmosphere containing approximately 6% O2, 6% H2, 6% CO2, and 82% N2.
Methods of growth and enumeration.
Strains were stored at −80°C on beads, subcultured onto blood agar (Oxoid CM 271 plus 7% defibrinated sheep blood), and incubated for 48 h in a microaerobic atmosphere.
Inocula for testing heat resistance were subcultured from blood agar into heart infusion broth (HIB) (Difco 238400) with campylobacter FBP growth supplement (Oxoid SR 084) and incubated for 24 h at 42°C. The broth was preequilibrated in the microaerobic atmosphere at 42°C before inoculation. One milliliter of the culture was used to inoculate biphasic medium in 25-cm2 cell culture flasks with vents (plastic disposable; Appleton Woods); this medium consisted of 4 ml Colombia blood agar (Oxoid CM 331 with 5% sheep blood) overlaid with 6 ml of HIB containing the FBP supplement (24). The flasks were incubated for 24 h at 42°C, which yielded approximately 1010 CFU per ml.
Samples for calorimetry were prepared as described above for the biphasic culture. The optical density at 600 nm (>1) was checked, and the suspensions were centrifuged at 5,000 × g for 15 min to obtain pellets. In the viability test, a suspension was used without centrifugation. Alternatively, Campylobacter strains were subcultured onto blood agar, incubated at 42°C for 24 or 48 h, and harvested by gently scraping the growth from the surface of the agar using a spatula.
The numbers of CFU in the inoculum and in suspensions after heat treatment or heating in the calorimeter were determined by dilution in maximum recovery diluent (MRD) (Oxoid CM 733) and plating onto modified cefoperazone charcoal deoxycholate agar (Oxoid CM 739) without a supplement (cefoperazone and amphotericin B), either by using a modified Miles-Misra method (spreading 0.01-ml portions of decimal dilutions on quarters of agar plates) or by using an automated spiral plater (Don Whitley, Yorkshire, United Kingdom) using duplicate plates at each dilution. The preparations were incubated microaerobically at 42°C for 48 h. In the Miles-Misra method, the mean number of CFU in the original suspension was calculated by the weighted means method (4).
Isothermal measurements of heat resistance.
Isothermal heat resistance was measured at 50, 55, and 60°C as follows. Duran bottles (100 ml) containing 45 ml sterile HIB were immersed in a well-stirred, temperature-controlled water bath so that the fluid level in the bottles was 4 cm below the fluid level of the bath and heated for at least 1 h to equilibrate the bottles to the required temperature before addition of 5 ml of the cell suspension to each bottle. The temperatures in the water bath and broth were measured with thermocouples to ±0.1°C. Thermocouples were placed in a bottle containing control sterile broth to monitor the temperature over the experimental period. At regular intervals, 1-ml samples were removed and diluted in 9 ml of MRD at 5 to 8°C, and the numbers of survivors were determined as described above.
Log10 numbers of surviving bacteria were plotted against time and D values calculated from the reciprocal of the slope, while z values were the reciprocal of the slope of the plot of log10 D against temperature. The heat resistance tests were repeated in order to obtain three D values at each temperature. For each determination, the reduction in the number of CFU was at least 3 logs; usually it was 5 to 6 logs.
Calorimetry.
Samples (10 to 15 mg) were weighed to ±0.01 mg and sealed in aluminum pans, which were then heated in a differential scanning calorimeter (Perkin-Elmer DSC-7) at a rate of 10°C per min to temperatures from 5 to 130°C, using an empty pan as the reference. The dry box and DSC head were purged with zero-grade nitrogen provided via an in-line silica gel dryer. The heating rate was chosen arbitrarily because initial tests demonstrated that it produced well-separated and clearly defined thermogram peaks. Samples were weighed before and after calorimetric measurements to check for loss of mass, and results for samples that showed signs of leakage were discarded.
Without adjustment of the DSC or disturbance of the pans, the samples were cooled rapidly to the initial temperature at a nominal rate of 200°C per min and then rescanned to investigate the recovery of the thermograms. The sample dry mass was determined by piercing the pan, drying it overnight in an oven at 105 ± 1°C, and weighing it to ±0.01 mg.
Data were collected and the scans were controlled and analyzed with a personal computer using Perkin-Elmer software. Only peaks that were consistently present in repeated runs were selected for analysis. Initial transient peaks and some initial step-like features that appeared to be instrument artifacts were ignored.
Temperature and power scales were calibrated in accordance with the manufacturer's instructions, using the melting of indium and ice as standards. Thermograms of blood agar, obtained as controls, showed that slight contamination of the samples with agar had only a minor effect on the results.
Viability study.
Two strains, C. jejuni AR6 and C. coli DR4, were used for a viability study. Aluminum pans were filled with 15-μl bacterial suspensions using micropipettes, sealed, and heated at a rate of 10°C per min from 20°C to one of the following temperatures: 30, 40, 50, 52.5, 55, 57.5, 60, 65, or 70°C. Then the pans were immediately cooled to 20°C at a nominal rate of 200°C per min. Pans, including controls, were opened in 5 ml MRD (CM 733; Oxoid), and the contents were dispersed by vigorous vortex mixing. Viable counts were determined on modified cefoperazone charcoal deoxycholate agar as described above.
Dynamic measurements of heat resistance.
Dynamic D and z values were calculated using the analysis of Miles and Mackey (17), which showed that if the natural logarithm of the negative natural logarithm of the fraction of survivors in a population subjected to a temperature that rises at a constant rate (r) is plotted against temperature, the slope is 2.303/z:
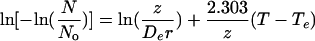 |
(1) |
where De is the D value at any arbitrary temperature, Te. The slope yielded a z value, and the D value at Te was calculated using this value of z with the measured intercept.
Statistical analysis.
Data for D and z values were analyzed by one-way analysis of variance and Fisher's pairwise comparisons to examine any differences in heat resistance among the four strains.
RESULTS
Isothermal measurements showed that there was an approximately exponential decline in CFU with time (Fig. 1a). The mean D values ranged from 4.6 to 6.6 min at 55°C with z values of 5.5 to 6.3°C (Table 1). Statistical analysis showed that there were some small but statistically significant differences between the strains (Table 1). As the D values were higher than those recorded previously for other strains (3, 21, 26), further measurements were obtained using the coil method of Cole and Jones (2, 29). These measurements yielded the same D values within the limits of experimental uncertainty, confirming the reliability of the results shown in Table 1.
FIG. 1.
Linear regressions of the loss of CFU during isothermal heating at 55°C (data for AR6) (a) and a temperature scan from 5°C to different temperatures at a heating rate of 10°C/min (b). Circles and dotted line, data for AR6; triangles and solid line, data for DR4. The lines are least-squares regression lines. The reciprocal of the slope in panel a gives the D value (mean values are shown in Table 1). In panel b the slopes and intercepts (means ± standard errors) are as follows: for AR6, 0.365 ± 0.0442°C−1 and −20.54 ± 2.515, respectively; and for DR4, 0.354 ± 0.0358°C−1 and −20.246 ± 2.039, respectively. The corresponding D55 and z values calculated from these slopes and intercepts using equation 1 are as follows: for AR6, 2.5 min and 6.3°C, respectively; and for DR4, 3.4 min and 6.5°C, respectively.
TABLE 1.
D and z values of four Campylobacter strains in HIBa
| Strain |
D value (min)
|
z value (°C)
|
||||||
|---|---|---|---|---|---|---|---|---|
| 50°C
|
55°C
|
60°C
|
|
|||||
| Mean | SE | Mean | SE | Mean | SE | Mean | SE | |
| AR6 (C. jejuni) | 36 A | 1.5 | 5.3 D | 0.4 | 0.7 F | 0.1 | 5.8 DE | 0.1 |
| L51 (C. jejuni) | 39 A | 0.8 | 4.6 D | 0.4 | 0.8 F | 0.1 | 5.9 DE | 0.2 |
| DR4 (C. coli) | 60 C | 0.4 | 6.6 E | 0.5 | 0.9 F | 0.0 | 5.5 D | 0.0 |
| L6 (C. coli) | 51 B | 1.8 | 6.6 E | 0.2 | 1.4 G | 0.1 | 6.3 E | 0.2 |
Each value is the mean of three determinations. Within columns, means followed by different letters are significantly different (as determined by Fisher's pairwise comparisons) at the following levels: A to C, P < 0.001; D and E, P < 0.05; F and G, P < 0.01.
Dynamic measurements of D and z using the decline in viable counts during temperature scanning at a rate of 10°C/min (Fig. 1b) yielded D55 values of 2.5 min and 3.4 min and z values of 6.3°C and 6.5°C for strains AR6 and DR4, respectively, confirming that in dynamic tests these strains of campylobacter were more heat resistant than the strains examined previously.
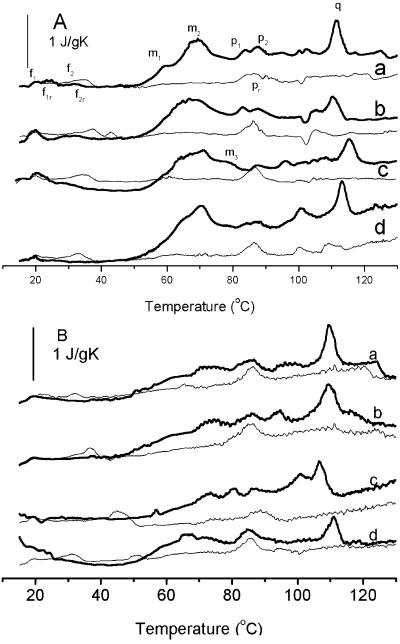
The DSC thermograms of the four strains of campylobacter (Fig. 2) showed a sequence of endothermic transitions that were similar in form to those observed for other vegetative cells and in particular for E. coli, in which the major events have been identified by cell fractionation (14). It was possible to assign some peaks to cellular components. This was done by (i) analogy with previous identifications (13, 14, 18); (ii) comparison of the changes in thermogram shape (Fig. 2) with the stage of growth (and the concomitant decline in ribosome content [9]); (iii) observation of the thermal recovery by rescanning after the initial heating (18); and (iv) comparison of the sizes and positions of the peaks with the likely concentration and enthalpy of the process (18).
FIG. 2.
Thermograms of four thermophilic Campylobacter strains grown at 42°C for 24 h (A) and for 48 h (B). The thick line for each organism is the initial scan, and the thin line is the rescan. Scan a, C. jejuni AR6; scan b, C. jejuni L51; scan c, C. coli DR4; scan d, C. coli L6. All scans were at a rate of 10°C/min.
In ascending temperature order, peaks occurred as follows (unless indicated otherwise, the temperatures are those recorded for strain AR6 grown for 24 h at 42°C by the biphasic method; data for other strains and conditions are shown in Table 2). Peaks f1 (21°C) and f2 (32°C) corresponded to lipid melting transitions, showing recovery with cooling and rescanning at slightly higher temperatures (typical of lipid transitions).
TABLE 2.
Numerical description of Campylobacter thermograms
| Medium and growth conditions | Strain | Temp at which peaks occurred (°C)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run
|
Rerun
|
|||||||||||
| Peak f1 | Peak f2 | Peak m1 | Peak m2 | Peak m3 | Peak p1 | Peak p2 | Peak q | Peak f1r | Peak f2r | Peak pr | ||
| 24 h at 42°C in biphasic medium (broth)b | AR6 (C. jejuni) | 22 | 32 | 60 | 70 | 84 | 88 | 112 | 20 | 34 | 86 | |
| L51 (C. jejuni) | 20 | 32 | 60 | 68 | 83 | 87 | 110 | 20 | 37 | 87 | ||
| DR4 (C. coli) | 21 | 31 | 62 | 71 | 76 | 88 | 96 | 115 | 21 | 36 | 87 | |
| L6 (C. coli) | 20 | 33 | 66 | 70 | 77 | 84 | 87 | 113 | 20 | 33 | 87 | |
| 24 h at 42°C on blood agarc | AR6 (C. jejuni) | 24 | 32 | 63 | 69 | 84 | 87 | 111 | 20 | 32 | 87 | |
| L51 (C. jejuni) | 23 | 34 | 64 | 71 | 83 | 85 | 109 | 21 | 38 | 87 | ||
| DR4 (C. coli) | 20 | 33 | 64 | 70 | 77 | 87 | 94 | 113 | 21 | 31 | 87 | |
| L6 (C. coli) | 21 | 30 | 64 | 70 | 79 | 84 | 87 | 111 | 20 | 30 | 86 | |
| 48 h at 42°C on blood agard | AR6 (C. jejuni) | 21 | 31 | 63 | 70 | 85 | 109 | 18 | 30 | 85 | ||
| L51 (C. jejuni) | 65 | 73 | 89 | 106 | 19 | 31 | 85 | |||||
| DR4 (C. coli) | 19 | 34 | 61 | 73 | 86 | 108 | 19 | 36 | 86 | |||
| L6 (C. coli) | 19 | 42 | 63 | 72 | 86 | 110 | 20 | 32 | 87 | |||
All scans were performed at a rate of 10°C/min.
DSC thermograms were run three times for each strain.
DSC thermograms were run twice for each strain.
DSC thermograms were run at least twice for each strain except the L51 strain (once).
This was followed a period of calm between f2 (32°C) and the onset of irreversible denaturation at about 50°C, in which there were no appreciable thermal perturbations above the baseline level. This corresponded to the approximate temperature range for growth, indicating that the cell envelope lipids were substantially in a liquid-like state during growth and that heat denaturation of macromolecules was relatively slight.
There then was a large irreversible multicomponent endotherm comprising peaks m1 at 60°C, m2 at 70°C, and (in only the C. coli strains) m3 at 76°C. The tallest of these peaks was peak m2, corresponding to peak m of Miles et al. (18). These peaks were caused by denaturation of the ribosome. In E. coli, peak m1 corresponds to unfolding of the 30S component and peak m2 corresponds to the 50S component (14).
Smaller peaks followed at 84°C (peak p1), 88°C (peak p2), and 112°C (peak q), and peak pr (86°C) was seen after cooling and rescanning. The latter peak was certainly due to the melting of reannealed DNA (13, 18), but peaks p1, p2, and q were also probably due in part to DNA, which in living organisms is thermally stabilized (14).
When the decline in the number of CFU during scanning was superimposed on the corresponding DSC thermograms (Fig. 3), the peak death rate was estimated to occur at 56°C for C. jejuni AR6 and at 57°C for C. coli DR4, temperatures which were slightly lower than the temperature predicted (61°C) on the basis of the isothermal D and z values shown in Table 1. For both strains, the estimates were close to peak m1 (60°C for AR6 and 62°C for DR4). There are two possible explanations for the small difference between the experimental and theoretical values for the lethal temperature. First, the experimental pans were subjected to an additional cooling phase, which was not allowed for in the analysis, and there could have been appreciable loss of viability during this phase. Second, the experimental samples were subjected to temperature gradients and instrument response time lag. The response time of the Perkin-Elmer DSC is approximately 12 s (16). In 12 s at a scanning rate of 10°C per min, the temperature changes by 2°C. Uncertainties in the measurement of temperature are likely to be on this order under nonscanning conditions (e.g., at the end of the scan).
FIG. 3.
Decline in viability (N/N0) of campylobacters heated at a rate of 10°C/min in a DSC superimposed upon thermograms. All measurements were obtained from bacteria grown in biphasic medium. (a) C. jejuni AR6. (b) C. coli DR4. The thick solid line for each organism is the initial scan, and the thin solid line is the rescan. Squares, loss of CFU (experimental); dashed line, loss of viability (theoretically predicted); dotted line, death rate (theoretically predicted).
DISCUSSION
The results of this study clearly show that when campylobacters are subjected to a temperature that rises at a constant rate of 10°C/min, cells die in a temperature range that coincides with the thermal unfolding of cellular macromolecules (Fig. 3). Furthermore, the predicted peak of the rate at which cells die has a half-width that is comparable to typical macromolecular unfolding endotherms. This is consistent with a causal relationship between thermal unfolding and cell death, as proposed by Rosenberg et al. (25).
More specifically, heat killing occurs in the low-temperature region of the irreversible heat denaturation endotherm (between the onset of heat denaturation and the largest early peak, peak m2 in this study), a trait that campylobacters share with other vegetative cells for which there are data, including Listeria monocytogenes (1), Bacillus stearothermophilus, E. coli (15), Lactobacillus bulgaricus (28), and Lactobacillus plantarum (12). We know that in this region of the endotherm the ribosome starts to unfold, probably initially in a region of the thermally sensitive 30S subunit (14, 27).
Other data also associate ribosome unfolding with cell death. For example, comparisons of the thermal stabilities of isolated ribosomes showed that L. plantarum ribosomes were more thermally tolerant than E. coli ribosomes (12), which is consistent with the loss of viability of whole cells. Anderson et al. (1) found that increased salt in the heating medium stabilized the ribosomes of L. monocytogenes and that cell death during temperature scanning was delayed commensurately. This observation was confirmed by Stephens and Jones (27), who also found that stabilization of the 30S subunit and the resulting increased thermotolerance of L. monocytogenese were induced by a sublethal heat shock.
Tomlins and Ordal (31) further showed by biochemical assays that the death of Salmonella enterica serovar Typhimurium under mild heating conditions was accompanied by extensive degradation of 16S RNA (one of three RNA molecules in the bacterial ribosome), while cells grown in the presence of salt (which stabilizes the ribosome) were not killed under the same conditions and retained 16S RNA. 16S RNA is located specifically in the smaller, more thermally unstable (30S) subunit of the ribosome, so degradation of 16S RNA implies degradation of the 30S unit. Tolker-Nielsen and Molin (30) found that the death of heat-stressed S. enterica serovar Typhimurium coincided with a significant reduction in 16S RNA, while magnesium ions increased the stability of the ribosomes and increased the heat resistance of the cells. They concluded that degradation of rRNA was a direct cause of cell death.
However, the underlying log-linear kinetics observed here for isothermal measurements is generally interpreted as indicating that the key lethal target is present at a level of only one or a small number of copies per cell (7). There are numerous copies of the ribosome in each cell (9), and the absence of a very substantial initial shoulder is inconsistent with existing critical target theory. In an accompanying paper (19) one of us proposes a modification to the generally accepted theory which might explain the inconsistency, but further experiments are needed. Meanwhile, there are further complications.
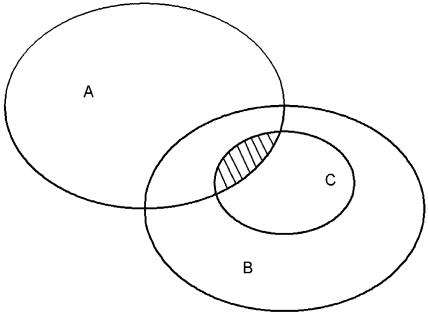
As ribosomes account for a large proportion of the cell's dry matter and ribosome denaturation is highly enthalpic, denaturation of the ribosome is the major cause of the largest peak in the multicomponent endotherm revealed by DSC of campylobacter and other vegetative bacterial cells. However, we know that other less energetic calorimetric events also occur simultaneously, at least in E. coli, because Mackey et al. (14) observed denaturation in the supernatant of fractionated cells when the dominating ribosomes were removed by centrifugation. Chemical analysis revealed that the source was heat-sensitive proteins. These proteins are present at much lower levels than ribosomes and exhibit a range of stabilities. They therefore give rise to a broad, multicomponent endotherm, the beginning of which overlapped with the rising edge of the endotherm of whole cells (14). We therefore deduce that numerous proteins unfold simultaneously with the 30S subunit of the ribosome (and with cell death in E. coli [15]). One or more of these proteins could be critical for cell survival and could also contribute to heat killing. As there are thousands of individual proteins in the cell, a search to identify potentially critical proteins has seemed overwhelmingly complex until now. Recently, other unrelated work, described below, has helped to narrow the field, but first we build on the concepts of Gould (7) and consider what constitutes a lethal target by using a Venn diagram (Fig. 4).
FIG. 4.
Venn diagram illustrating the logic for identifying lethal targets. Imagine a list of molecules (proteins, nucleic acids, etc.) and molecular assemblies (ribosomes, cell wall, etc.) of which a bacterium is composed. Each item on the list is a potential target. Group together all those targets that are essential for cell survival and draw a ring around them (region A), on the basis that a lethal target must be essential. Then draw a ring around another set of targets (region B) that are inactivated by the heat treatment. As all lethal targets must be inactivated by heat and must be essential, the number of potentially lethal targets is reduced to the overlap of regions A and B. Then introduce a further criterion, that the targets must be irrecoverably inactivated (region C). Unlike region A, regions B and C are not fixed; the numbers of targets increase with the duration and temperature of the heat treatment. The lethal targets are the targets in the area where regions A, B, and C overlap.
We imagine composing a complete list of all the proteins, nucleic acids, and molecular assemblies of which the cell is composed. First, a lethal target must be essential for the survival of the bacterium, in the sense that without that functioning molecule or molecular assembly (such as the ribosome or cell wall) the cell is not viable. We can therefore define a subset of the original very long list, and these components we place in region A of Fig. 4. Second, a lethal target must be sensitive to the heat treatment that results in the death of the cell. Only a subset of the original list satisfies this condition, and we imagine that all these heat-sensitive components are enclosed in region B of the Venn diagram. Region B, unlike region A, which is fixed, expands and contracts depending on the severity of the heating. Clearly, the components in the region where regions A and B overlap are heat sensitive and essential and are the only components that we may consider to be potentially lethal targets. However, there is a third condition, a lethal target, once heat denatured, must be irrecoverable within a time scale necessary for life. It is not sufficient that the thermally unfolded molecule is incapable of refolding; in addition, it must not be capable of being resynthesized or repaired within the required time frame. Such components are enclosed in region C, a subset of region B, which like region B also expands and contracts with the severity of the heat treatment. Lethal targets are thus located where regions A, B, and C overlap in Fig. 4. For a given heat treatment to be lethal, all copies of at least one lethal target must be destroyed by the heat treatment. It is not necessary that the heat treatment itself destroys all copies. It is only necessary that the heat treatment initiates a train of events that inevitably causes destruction of all functioning copies (e.g., by the release of degradation enzymes). It is obviously not necessary that the same critical target is lost in all cells; the heating is lethal if all copies of at least one lethal target are destroyed, so that the actual cause of death may be different in different cells. Equally, death may be due to the destruction of more than one lethal target.
Recent analysis of the 4,100 genes of Bacillus subtilis (10) revealed that only 271 of these genes are essential for a minimally viable cell; 52 of these genes are ribosomal proteins required (with three RNA molecules) to build ribosomes, which are essential for protein synthesis. Clearly, without working ribosomes, protein synthesis is impossible and the cell cannot maintain essential functions. Kobayashi et al. reported that “the average level at which homologues of essential B. subtilis genes are present in bacteria is rather high (approaching 80%).” Hence, we can assume that genes that are essential for B. subtilis are essential in E. coli also. Comparing the list of “essential genes” in B. subtilis with the list of aggregation-prone proteins in E. coli identified by Mogk et al. (20), we found only eight genes common to both lists (i.e., genes encoding essential soluble proteins that are also very heat sensitive).
Four of the gene products are involved in protein synthesis (RpoA and RpoB [the alpha and beta subunits of RNA polymerase] and LysS and AsnS [tRNA synthetases]), and the following four gene products are not involved in protein synthesis: Dxs (an enzyme used in isoprenoid synthesis [respiration]), Pgk (an enzyme used in glycolysis), FtsZ (a cell division initiation protein), and MurA (a protein used in cell wall [peptidoglycan] synthesis).
We emphasize that in the “thought experiment” described above the eight proteins that we have identified in E. coli are only potentially lethal targets (a subset of the proteins in the overlap of the two regions identified as regions A and B in Fig. 4). However, we suppose that irreversible denaturation of all copies of RNA polymerase or all ribosomes would be a lethal event, as these components could not be resynthesized by a cell lacking a single copy. Irreversible denaturation of any of the other proteins, however, might not be lethal, as potentially these proteins could be resynthesized.
The AsnS and Dxs genes are not on the list of genes in C. jejuni (23), but the genes encoding the remaining six proteins identified above are. They are likely to be essential and, by analogy with E. coli, might be thought to be heat sensitive. We therefore speculate that the thermal denaturation of RNA polymerase could occur simultaneously with early ribosome denaturation and may potentially contribute to heat killing of Campylobacter. Four other essential soluble proteins may also unfold in the same temperature range. To investigate this, we might subject Campylobacter to critical heat treatments by DSC and identify sensitive proteins using methods analogous to those of Mogk et al. (20).
To summarize, DSC and heat resistance measurements of campylobacters showed that cell death is coincident with heat denaturation of the most thermally sensitive parts of the ribosome. We speculate that within the same temperature range other proteins that are essential for the survival of a minimal cell also unfold. These proteins include proteins that are required for protein synthesis, particularly the alpha and beta subunits of RNA polymerase.
REFERENCES
- 1.Anderson, W. A., N. D. Hedges, M. V. Jones, and M. B. Cole. 1991. Thermal inactivation of Listeria monocytogenes studies by differential scanning calorimetry. J. Gen. Microbiol. 137:1419-1424. [DOI] [PubMed] [Google Scholar]
- 2.Cole, M. B., and M. V. Jones 1990. A submerged-coil heating apparatus for investigating thermal inactivation of micro-organisms. Lett. Appl. Microbiol. 11:223-235. [Google Scholar]
- 3.Doyle, M. P., and D. J. Roman 1981. Growth and survival of Campylobacter fetus. subsp. jejuni as a function of temperature and pH. J. Food Prot. 44:596-601. [DOI] [PubMed] [Google Scholar]
- 4.Farmiloe, F. J., S. J. Cornford, J. B. M. Coppock, and M. Ingram 1954. The survival of Bacillus subtilis spores in the baking of bread. J. Sci. Food Agric. 5:292-304. [Google Scholar]
- 5.Friedman, C. R., J. Niemam, H. C. Wegener, and R. V. Tauxe 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 6.Gill, C. O., and L. M. Harris 1982. Survival and growth of Campylobacter fetus subsp. jejuni on meat and in cooked foods. Appl. Environ. Microbiol. 44:259-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould, G. W. 1989. Heat-induced injury and inactivation, p. 11-42. In G. W. Gould (ed.), Mechanisms of action of food preservation procedures. Elsevier Science Publishers, Barking, United Kingdom.
- 8.ICMSF (International Commission of Microbiological Specification for Foods). 1996. Micro-organisms in food 5. Characteristics of microbial pathogens, p. 45-65. Blackie Academic, London, United Kingdom.
- 9.Kjeldgaard, N. O., and K. Gausing 1974. Regulation of biosynthesis of ribosomes, p. 369-392. In M. Nomura, A. Tissieres, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 10.Kobayashi, K., S. D. Ehrlich, A. Albertini, K. K. Amati, K. K. Andersen, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson, A. J., J. M. J. Logan, G. L. O'Neill, M. Desai, and J. Stanley 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 71:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J., and G. Kaletunc 2002. Evaluation of the heat inactivation of Escherichia coli and Lactobacillus plantarum by differential scanning calorimetry. Appl. Environ Microbiol. 68:5379-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackey, B. M., S. E. Parsons, C. A. Miles, and R. J. Owen 1988. The relationship between the base composition of bacterial DNA and its intracellular melting temperature as determined by differential scanning calorimetry. J. Gen. Microbiol. 134:1185-1195. [DOI] [PubMed] [Google Scholar]
- 14.Mackey, B. M., C. A. Miles, S. E. Parsons, and D. A. Seymour 1991. Thermal denaturation of whole cells and cell components of Escherichia coli examined by differential scanning calorimetry. J. Gen. Microbiol. 137:2361-2374. [DOI] [PubMed] [Google Scholar]
- 15.Mackey, B. M., C. A. Miles, D. A. Seymour, and S. E. Parsons 1993. Thermal denaturation and loss of viability in Escherichia coli and Bacillus stearothermophilus. Lett. Appl. Microbiol. 16:56-58. [Google Scholar]
- 16.Miles, C. A., T. V. Burjanadze, and A. J. Bailey 1995. The kinetics of the thermal denaturation of unrestrained rat tail tendon determined by differential scanning calorimetry. J. Mol. Biol. 245:437-446. [DOI] [PubMed] [Google Scholar]
- 17.Miles, C. A., and B. M. Mackey 1994. A mathematical analysis of microbial inactivation at linearly rising temperatures: calculation of the temperature rise needed to kill Listeria monocytogenes in different foods and methods for dynamic measurements of D and z values. J. Appl. Bacteriol. 77:14-20. [DOI] [PubMed] [Google Scholar]
- 18.Miles, C. A., B. M. Mackey, and S. E. Parsons 1986. Differential scanning calorimetry of bacteria. J. Gen. Microbiol. 132:939-952. [DOI] [PubMed] [Google Scholar]
- 19.Miles, C. A. 2006. Relating cell killing to inactivation of critical components. Appl. Environ. Microbiol. 72:914-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rudiger, D. Roder, H. Langen, and B. Bukau 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, J. E., and R. H. Madden 2000. The effect of thermal stress on Campylobacter coli. J. Appl. Microbiol. 89:892-899. [DOI] [PubMed] [Google Scholar]
- 22.Oosterom, J., G. J. A. De Wilde, E. De Boer, L. H. De Blaauw, and H. Karman. 1983. Survival of Campylobacter jejuni during poultry processing and pig slaughtering. J. Food Prot. 46:702-706. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., B. W. Wren, K. L. Mungall, J. M. Ketley, C. M. Churcher, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequencies. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 24.Rollins, D. M., J. C. Coolbaugh, R. I. Walker, and E. Weiss 1983. Biphasic culture system for rapid Campylobacter cultivation. Appl. Environ Microbiol. 45:284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, B., G. Kemeny, R. C. Switzer, and T. C. Hamilton 1971. Quantitative evidence for protein denaturation as the cause of thermal death. Nature 232:471-473. [DOI] [PubMed] [Google Scholar]
- 26.Sorqvist, S. 2003. Heat resistance in liquids of Enteroccus spp., Listeria spp., Escherichia coli, Yersinia enterocolitica, Salmonella spp. and Campylobacter spp. Acta Vet. Scand. 44:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens, P. J., and M. V. Jones 1993. Reduced ribosomal thermal denaturation in Listeria monocytogenes following osmotic and heat shocks. FEMS Microbiol. Lett. 106:177-182. [DOI] [PubMed] [Google Scholar]
- 28.Teixeira, P., H. Castro, C. Mohacsi-Farkas, and R. Kirby 1997. Identification of sites of injury in Lactobacillus bulgaricus during heat stress. J. Appl. Microbiol. 83:219-226. [DOI] [PubMed] [Google Scholar]
- 29.Thi Thu Tran, P. 2004. Investigation of some of the factors influencing the heat resistance of Campylobacter jejuni and Campylobacter coli. M.Sc. thesis. University of Bristol, Bristol, United Kingdom.
- 30.Tolker-Nielsen, T., and S. Molin 1996. Role of ribosome degradation in the death of heat-stressed Salmonella typhimurium. FEMS Microbiol. Lett. 142:155-160. [DOI] [PubMed] [Google Scholar]
- 31.Tomlins, R. I., and Z. J. Ordal 1976. Thermal injury and inactivation in vegetative bacteria, p. 153-190. In F. A. Skinner and W. B. Hugo (ed.), Inhibition and inactivation of vegetative microbes. Academic Press, London, United Kingdom.