Abstract
Methyl fluoride is frequently used to specifically inhibit acetoclastic methanogenesis, thus allowing determination of the relative contribution of acetate versus H2/CO2 to total CH4 production in natural environments. However, the effect of the inhibitor on growth of the target archaeal population has not yet been studied. Therefore, we incubated rice roots as an environmental model system under anoxic conditions in the presence and absence of CH3F, measured the activity and Gibbs free energy (ΔG) of CH4 production, and determined the abundance of individual archaeal populations by using a combination of quantitative (real-time) PCR and analysis of terminal restriction fragment length polymorphism targeting the 16S rRNA gene. It was shown that CH3F specifically inhibited not only acetoclastic methanogenic activity but also the proliferation of Methanosarcina spp, which were the prevalent acetoclastic methanogens in our environmental model system. Therefore, inhibition experiments with CH3F seem to be a suitable method for quantifying acetoclastic CH4 production. It is furthermore shown that the growth and final population size of methanogens were consistent with energetic conditions that at least covered the maintenance requirements of the population.
Atmospheric CH4 significantly contributes to global warming (4, 35). Among the dominant sources of CH4 are wetlands and flooded rice fields, which are responsible for a third of the atmospheric CH4 budget (4). To understand the processes responsible for methane production and consumption in anoxic environments, it is useful to study the flow of carbon and electrons. For this purpose, metabolic inhibitors are frequently used to allow quantification of single processes (31). Methyl fluoride (CH3F), when applied at the appropriate concentration, was found to be a rather specific inhibitor of acetoclastic methanogenesis, while the operation of CH4 production from H2/CO2 was unaffected (8, 16). Consequently, CH3F has been used to monitor changes in carbon flow in methanogenic systems (9). However, a crucial requirement for unbiased application of inhibitors is specificity, meaning that they must not influence any other process or reaction, while the target reaction or process must be completely inactivated. Methyl fluoride is also known to affect processes other than acetoclastic methanogenesis. For example, it also inhibits aerobic oxidation of CH4 and, to a lesser extent, oxidation of ammonia (32, 33). With respect to CH4 production, the optimized concentration of CH3F for complete inhibition of acetoclastic methanogenesis must not be exceeded, since at too-high concentrations hydrogenotrophic methanogenesis can also be partially inhibited (16). On the other hand, CH3F is apparently not inhibitory for acetotrophic sulfate reducers (e.g., Desulfotomaculum spp.) and for acetogenic fermenting bacteria (e.g., Acetobacterium spp.) (16). However, many archaeal and bacterial phylogenetic clusters that are found in natural environments are yet uncultured, so inhibiting effects on them cannot be excluded. Although the inhibition of acetoclastic methanogenesis by CH3F in anoxic environments has been well established (1, 20), the effect of the inhibitor on the metabolically active microbial community has not yet been investigated.
Here we tested the effect of CH3F on a natural archaeal community by using molecular techniques. For our study we chose anoxically incubated rice roots, which are a well-studied model system with respect to the archaeal community structure (2, 22, 38) and biogeochemical processes (7). Growth of archaeal populations was monitored by a combination of quantitative PCR (qPCR) and terminal restriction fragment length polymorphism (T-RFLP) analysis and compared to the maximum population densities that are feasible by applying maintenance theory.
MATERIALS AND METHODS
Growth and incubation conditions.
Rice plants (Oryza sativa, var. Roma, type japonica) were grown in a greenhouse as described by Lehmann-Richter et al. (22), using soil obtained from rice fields in Vercelli, Italy (15). After 90 days the plants were removed from the growth container and roots were carefully washed, maintaining anoxic conditions (22). Freshly collected and washed rice roots (30 g per incubation) were placed into glass bottles (1,000 ml; Müller and Krempel, Bülach, Switzerland) filled with 500 ml anoxic deionized water and 50 g marble grains, giving a neutral carbonate-buffered aqueous phase. The bottles were closed with latex stoppers and gassed with N2. Acetoclastic methanogenesis was inhibited by addition of 1.3% methyl fluoride (CH3F) (99%; ABCR, Karlsruhe, Germany). Methyl fluoride treatments and controls without inhibitor were incubated in triplicate at 25°C in the dark.
Extraction of RNA/DNA and PCR amplification of archaeal 16S rRNA genes.
The incubation vessels were shaken vigorously to detach microorganisms from the roots before liquid samples were taken from each replicate. Nucleic acids were extracted from 10 ml of the liquid phase. After centrifugation (26,000 × g, 15 min, 4°C), the cell pellets were resuspended in 0.5 ml sterile distilled water and extracted according to a cell lysis protocol involving bead beating in the presence of the denaturant sodium dodecyl sulfate, phenol-chloroform-isoamyl alcohol extraction, and polyethylene glycol precipitation as previously described (30).
For DNA analysis (targeting 16S rRNA genes), 5 μl was removed from the primary extract of each replicate of control and inhibition incubations. The remaining nucleic acid extracts of the control and the inhibition experiment were pooled and subsequently used for preparation of rRNA. The RNAs in the two composite extracts were purified by digestion of coextracted DNA with RQ1 RNase-free DNase (Promega, Hilden, Germany) according to the manufacturer's instructions and subsequent reextraction with phenol-chloroform-isoamyl alcohol (see above). Aliquots of raw nucleic acid extracts and RNA preparations were visualized by standard agarose gel electrophoresis to verify the quality of extracted total nucleic acids and RNA preparations. First-strand synthesis of cDNA from RNA was done as follows. An 8.5-μl portion of RNA extract, 1 μl of 10× Hexanucleotide Mix (diluted 1:50; Roche, Mannheim, Germany), and 20 U of RNasin RNase inhibitor (Promega) were incubated for 10 min at 70°C. After cooling on ice, Moloney murine leukemia virus reverse transcriptase 5× reaction buffer, 100 pmol of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech, Freiburg, Germany), and 200 U Moloney murine leukemia virus reverse transcriptase (Promega) were added to a final volume of 25 μl and incubated at 37°C for 1 h.
Archaeal 16S rRNA genes were amplified using the forward primer A109f (5′-ACKGCTCAGTAACACGT-3′) (12) and the 5-carboxyfluorescein-labeled (5′-terminal) backward primer A915b (5′-GTGCTCCCCCGCCAATTCCT-3′) (40). In a total volume of 50 μl, the PCR mixture contained 10× PCR buffer (Invitrogen GmbH, Karlsruhe, Germany), 1.25 U of Taq DNA polymerase (Invitrogen GmbH), 2.5 nmol of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech), 75 nmol MgCl2, 4 μg of bovine serum albumin (Roche), and 16.5 pmol of each primer (MWG Biotech, Ebersberg, Germany). A volume of 1 μl DNA or cDNA solution was added as template. Amplification was performed by using a Gene Amp system 9700 (Applied Biosystems, Weiterstadt, Germany) with an initial denaturation step (4 min, 94°C) followed by 30 cycles of denaturation (45 s, 94°C), annealing (1 min, 55°C), and extension (1 min, 72°C) and a terminal extension step (7 min, 72°C).
Real-time PCR.
The archaeal 16S rRNA gene copy number in DNA extracts was determined by qPCR assays based on real-time PCR as previously described (36, 42). PCR was carried out in an iCycler IQ thermocycler (Bio-Rad, Munich, Germany) using the primer pair A109f/A915b described above. Each 25-μl PCR mixture contained 12.25 μl SYBR Green Jumpstart Taq Ready Mix (Sigma-Aldrich, Taufkirchen, Germany), 37.5 nmol MgCl2, (Invitrogen GmbH), 8.25 pmol of each primer (MWG Biotech), and 5 μl of DNA or H2O as a negative control. The assay was performed with the following thermal profile: DNA denaturation (40 s, 94°C), primer annealing (30 s, 55°C), and elongation (90 s, 72°C). Fluorescence data were collected during the elongation step. Quantification of archaeal templates was done with a serial dilution of a cloned 16S rRNA gene sequence amplified with vector primers (17). The standard DNA was fluorimetrically quantified using the PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Invitrogen). iCycler software (version 3.0a; Bio-Rad) was used for data analysis, and calculation of target molecules (16S rRNA gene copies) was done as described earlier (19, 41).
Terminal restriction fragment length polymorphism analysis.
The principle of the T-RFLP analysis has been described by Liu et al. (23). Fluorescently labeled 16S rRNA gene amplicons were purified by use of the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) according to the instructions of the manufacturer. DNA concentrations of purified 16S rRNA gene fragments were determined by standard UV photometry (Biophotometer; Eppendorf, Hamburg, Germany). Restriction digestion was performed in a total volume of 10 μl containing ≈80 ng of 16S rRNA gene amplicons. 16S rRNA gene amplicons were restricted with 5 U of enzyme TaqI (Fermentas, St. Leon-Rot, Germany) and 1 μl of the appropriate incubation buffer and incubated for 3 h at 65°C. The digested amplicons were mixed with an internal lane standard and analyzed by polyacrylamide gel electrophoresis as previously described (3). Analysis was performed for the DNA extract of each replicate. The relative abundance of a detected terminal restriction fragment (T-RF) within a given T-RFLP pattern was calculated as the respective signal area of the peak divided by the peak area of all peaks of the T-RFLP pattern, starting from a fragment size of 56 bp to exclude T-RFs caused by primers. Standard errors for averaged relative abundances of T-RFs were ≤2% of the total or 10% of the relative abundance of the particular peak. Changes in the absolute 16S rRNA gene copy numbers of each T-RF were calculated by multiplication of its relative abundance by the total 16S rRNA gene copy number from that particular time point. Note that the same primer set was used for both T-RFLP analysis and qPCR.
Cloning and sequencing.
Three clone libraries of archaeal 16S rRNA or 16S rRNA gene amplicons were created using samples from day 28 of the incubation (cDNA and DNA from the control and DNA from the CH3F treatment). Amplicons (A109f/A915b) were cloned in Escherichia coli JM109 by using the pGEM-T Vector System II cloning kit (Promega) according to the manufacturer's instructions. Clones were selected randomly and checked for correct insert size by vector-targeted PCR and agarose gel electrophoresis. DNA sequences were determined on an ABI Prism 377 DNA sequencer with Big Dye terminator chemistry as specified by the manufacturer (Applied Biosystems).
Sequence data and phylogenetic analysis.
Sequences were assembled and checked with the Lasergene software package (DNASTAR, Madison, WI). 16S rRNA gene sequences (approximately 800 bp) were compared by a BLAST search to sequences of the EMBL database (www.ebi.ac.uk). Sequence alignment (Fast Aligner tool version 1.03), calculation of distance matrices, and construction of phylogenetic trees were accomplished with the ARB software package (version Linux Beta 030822; http://www.arb-home.de) (26). Sequences closely related to the cloned 16S rRNA gene sequences were obtained from the GenBank database (http://www.ncbi.nih.gov/GenBank) and integrated into the 16S rRNA gene database (released June 2002, ARB).
The terminal sequence positions at the 5′ and 3′ends of the 16S rRNA gene sequences (300 bp for partial sequences and 500 bp for full-length sequences) were also subjected to a separate treeing analysis (“fractional treeing” [25]) to identify chimeric sequences. Differences in the phylogenetic placement of a fragment pair were considered indicative of chimera formation. For in silico determination of T-RFs, the ARB-implemented TRF-CUT tool was used (37).
Quantification of gaseous and dissolved compounds.
Liquid samples (2.5 ml) were taken with a sterile syringe, membrane filtered (0.2 μm), and stored frozen (−20°C) until analysis. Gas samples (0.25 to 1.0 ml) were taken with a gas-tight pressure lock syringe (Dynatech, Baton Rouge, LA), after the bottles were vigorously shaken by hand, and analyzed immediately by gas chromatography. CH4 and CO2 were analyzed by gas chromatography using a flame ionization detector (Shimadzu, Kyoto, Japan). CO2 was detected after conversion to CH4 with a methanizer (Ni catalyst at 350°C; Chrompack, Middelburg, The Netherlands). H2 was analyzed by gas chromatography using a thermal conductivity detector (Shimadzu) and an HgO-to-Hg conversion detector (RGD2; Trace Analytical, Menlo Park, CA) (39). Acetate, ethanol, formate, and fatty acids were measured by high-pressure liquid chromatography (Sykam, Gilching, Germany) with a refraction index and UV detector, having a detection limit of 3 to 5 μM (21).
Calculations.
Gibbs free energies (ΔG) of the production of CH4 were calculated from the respective standard Gibbs free energies (ΔG°) and the actual concentrations of reactants and products by using Nernst's equation. The values of ΔG° were calculated from the standard Gibbs energies of formation (44) using the reactions 4H2 + CO2 → CH4 + 2 H2O (ΔG° = −130.7 kJ mol−1) and CH3COO− + H+ → CH4 + CO2 (ΔG° = −75.7 kJ mol−1).
The potential capacity for maintaining a particular population size (N) of methanogens was calculated from the thermodynamic data and from the CH4 production rates measured in the experiment by using the equation Nmc = −ΔGvCH4mE−1γ−1 (5), where Nmc = number of (hydrogenotrophic or acetoclastic) methanogenic archaea (cells milliliter−1), ΔG = Gibbs free energy (kilojoules mole−1) of the (hydrogenotrophic or acetoclastic) methanogenic reaction determined for the particular incubation condition, vCH4 = rate of (hydrogenotrophic or acetoclastic) methanogenesis (moles CH4 hour−1 milliliter−1), mE = maintenance energy (kilojoules hour−1 mol carbon [C-mol] of methanogenic biomass−1), and γ = molar mass of a methanogenic cell (C-mol). The value of γ was assumed to be 8 × 10−15 C-mol, using the equivalence of 25 g microbial dry mass for 1 C-mol biomass (45) and assuming that a microbial cell had a mass of about 2 × 10−13 g (34, 46). The value of mE was reported to be constant for anaerobic microorganisms, amounting to 3.3 kJ h−1 C-mol biomass−1 at 25°C (45). The vCH4 values for hydrogenotrophic and acetoclastic methanogeneses were calculated from the measured rates of total CH4 production (vtot) times fmc and (1 − fmc), respectively, with fmc being the fraction of methane formed from H2/CO2. Values of fmc were determined from measurement of δ13C in CH4, CO2, and acetate as described previously (6). Detailed results of the measurements will be presented elsewhere. The calculation of N is not very sensitive to the accuracy of fmc but is linearly influenced by the other parameters, i.e., ΔG, mE, vtot, and γ.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences generated from the control (control rice root) and from the CH3F incubation (inhibition rice root) were deposited in the EMBL database under accession numbers AM050403 to AM050425.
RESULTS
Process data for rice root incubations.
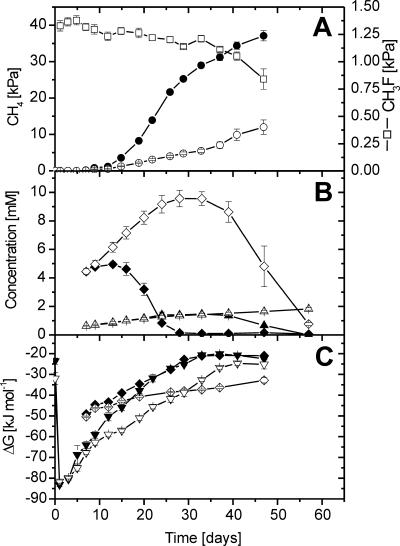
During the anoxic incubation of excised rice roots, CO2 and CH4 accumulated steadily, whereas H2, acetate, and propionate accumulated only transiently and later decreased again (Fig. 1) (data for H2 and CO2 not shown). In the control incubations, CH4 production started immediately, albeit at a low rate, and strongly increased on day 14 (Fig. 1A). Similar activity patterns have been observed before (7, 38). In the CH3F incubations, on the other hand, CH4 accumulated very little until day 24, while acetate accumulated linearly, reaching about 10 mM (Fig. 1B), indicating that acetoclastic methanogenesis was inhibited. Complete inhibition of acetoclastic methanogenesis was confirmed by stoichiometric calculations. Until day 24 the sum of the rate of acetate production in the CH3F incubation and the rate of acetate consumption in the control equaled the difference in methane production rates between control and CH3F incubations. After day 24, accumulation of acetate ceased, and it turned into a net decrease after day 33, while the CH4 production rate increased, indicating resumption of acetoclastic methanogenesis. This resumption was due to the decrease of the CH3F concentration below the threshold (about 1.0 to 1.2%) of inhibition (Fig. 1A). The loss of CH3F was accounted for by gas and liquid sampling.
FIG. 1.
Change in gas partial pressures, concentrations, and Gibbs free energies of control (closed symbols) and CH3F (open symbols) rice root incubations: (A) CH4 (•, ○) and CH3F (□); (B) acetate (♦, ⋄) and propionate (▴, ▵); (C) Gibbs free energies of the conversion of CO2 and H2 to CH4 (▾, ▿) and of the conversion of acetate to CH4 and CO2 (♦, ⋄). Values are means ± standard errors; n = 3.
Propionate accumulated to 1.45 mM in both the control and CH3F treatments until day 33 (Fig. 1B). Thereafter propionate further increased in the CH3F incubation, while it was completely consumed in the control. Low concentrations of ethanol (≤150 μM), butyrate (≤70 μM), valerate (≤50 μM), and caproate (≤30 μM) were also detected. Although their maximum concentrations were comparable in the control and CH3F incubations, the compounds accumulated in the CH3F incubation until the end, whereas none of them was detectable in the control after day 33.
In the control, Gibbs free energies for CH4 production from H2/CO2 and acetate were strongly exergonic in the beginning but then increased with incubation time, and they finally reached about −20 kJ mol−1 after day 40 (Fig. 1C). These values are close to the thermodynamic threshold of methanogenic activity (14, 39, 47). In the CH3F treatments, the ΔG of hydrogenotrophic methanogenesis was relatively more negative throughout the experiment, since H2 partial pressures were always slightly higher in the CH3F treatments than in the control, but finally also reached relatively high values of −25 kJ mol−1, close to the thermodynamic threshold (Fig. 1C). Acetoclastic methanogenesis, on the other hand, was always thermodynamically feasible in the CH3F incubations.
Diversity of archaea in rice root incubations.
T-RFLP analysis targeting the 16S rRNA gene was done using samples collected from control and CH3F treatments over the entire incubation time. The T-RFLP patterns of the control were similar to those observed before (2), revealing a dynamic change of the different phylogenetic groups of archaea. The T-RFLP patterns are not shown explicitly but were used in calculation of 16S rRNA gene copy numbers for the individual archaeal populations as described below.
Three clone libraries of archaeal 16S rRNA fragments were constructed from samples taken on day 28: (i) 16S rRNA (RNA based) from the control, (ii) 16S rRNA gene (DNA based) from the control, and (iii) 16S rRNA gene (DNA based) from the CH3F incubation. From a total of 90 clones, no chimeras were identified. Phylogenetic analysis of clones showed that all sequences were affiliated with the same eury- and crenarchaeotal lineages described before (2). These included the methanogenic families of Methanobacteriaceae and Methanosarcinaceae, as well as uncultured archaea designated rice clusters I, III, IV, and V (3, 13). The sequence dissimilarities of Methanosarcinaceae- and Methanobacteriaceae-related sequences were <3 and <2% with respect to Methanosarcina barkeri and Methanobacterium bryantii, respectively. Rice cluster I (RC-I) clone sequences were >97% similar to clone AS08-16 from rice field soil (28), RC-III sequences were >97% similar to ARR16 from rice roots (13) and AS08-11 from rice field soil (28), and RC-IV sequences were >94% similar to AS01-06 and AS08-25 from rice field soil (28). Clones from RC-V were >82% similar to the closest relatives UniArc49 (43) and WCHD3-30 (10).
The relative abundance of the different lineages on day 28 of incubation differed between the different clone libraries (Table 1). In the control, Methanosarcinaceae strongly dominated on rRNA gene level and were the exclusive archaea on the rRNA level. By contrast, the clone library of 16S rRNA genes from the CH3F incubation was much more diverse, representing all detected groups. RC-V accounted for the largest number of clones (25%), and RC-I and RC-III represented 17.5% each, followed by the other groups. In silico determination of T-RFs of the archaeal 16S rRNA fragments using TaqI (37) yielded uniform and specific lengths for each archaeal lineage (Table 1), indicating that the combination of the chosen primer set and restriction enzyme was suitable for differentiating the individual T-RFs as phylogenetic lineages. We compared the relative abundances of archaeal T-RFs in the clone library with the T-RFLP pattern of the same replicate, from which the clone library was constructed (Table 1). The relative abundances were in fairly good agreement, indicating that all major T-RFs could be phylogenetically assigned and that cloning bias did not play a serious role.
TABLE 1.
Relative abundances of phylogenetic groups in rice root incubations on day 28, based on frequencies of 16S rRNA or 16S rRNA genes in clone libraries and T-RFLP analysis
| Phylogenetic group (T-RF length, bp)a | Relative abundance (%)
|
|||||
|---|---|---|---|---|---|---|
| Control incubation (rRNA gene; n = 24)
|
CH3F incubation (rRNA gene; n = 35)
|
Control incubation (rRNA; n = 31)
|
||||
| Clone library | T-RFLPb | Clone library | T-RFLPb | Clone library | T-RFLPb | |
| MB (92) | 0.9 | 5.0 | 1.8 | |||
| MS (186) | 87.5 | 92.8 | 12.5 | 14.0 | 100.0 | 99.2 |
| RC-I (393) | 2.7 | 17.5 | 21.5 | |||
| RC-III (381) | 0.7 | 17.5 | 9.0 | 0.8 | ||
| RC-V (689) | 4.2 | 1.3 | 25.0 | 35.7 | ||
| RC-IV (810) | 8.3 | 1.0 | 10.0 | 11.0 | ||
MB, Methanobacteriaceae; MS, Methanosarcinaceae; RC, rice cluster.
The same replicate as for construction of the clone library was used.
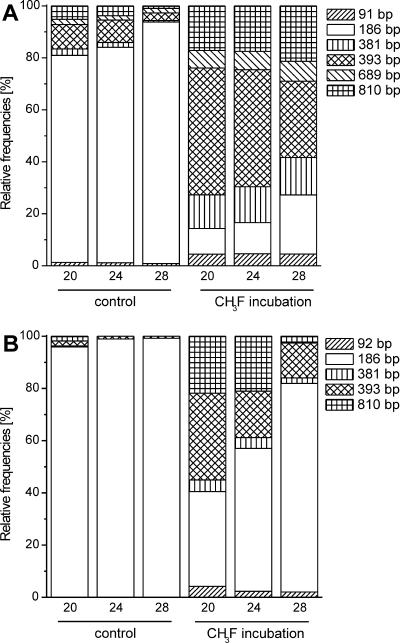
T-RFLP analysis of 16S rRNA (RNA based) was performed only for days 20, 24, and 28 and is shown together with the relative abundance patterns of the 16S rRNA gene (DNA based) at the same time points (Fig. 2). Comparing the course of T-RFs on the RNA and DNA levels shows that the 186-bp T-RF always had a higher relative abundance on the RNA level, whereas the other T-RFs had either a similar or lower relative abundance. The 689-bp T-RF was not even detected on the RNA level. The relative increase of the 186-bp T-RF and the relative decrease of the other T-RFs on the RNA level was seen about 8 days earlier than on the DNA level in the control and the CH3F treatments.
FIG. 2.
Course of archaeal population dynamics determined by (A) 16S rRNA gene-targeted and (B) 16S rRNA-targeted T-RFLP analysis for incubation days 20, 24, and 28 for control and CH3F incubations. Numbers in base pairs indicate the fragment length of the T-RF. (A) Average values of triplicate incubations; (B) single value from three pooled replicates.
Population dynamics of archaea.
To address the dynamics of archaeal groups, we performed T-RFLP analysis and qPCR targeting 16S rRNA genes (DNA level).
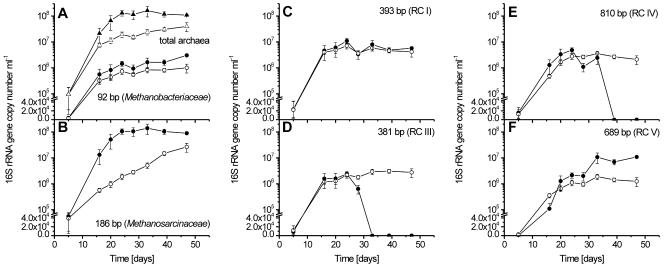
The numbers of 16S rRNA gene copies (Ncopy) determined by qPCR increased rapidly in the control until day 24 and then stabilized at 108 ml−1 (Fig. 3A). In the CH3F incubation, on the other hand, Ncopy reached only 1.5 × 107 ml−1 at days 20 to 28 but then increased further, reaching 4 × 107 ml−1 at day 47.
FIG. 3.
Temporal change of archaeal 16S rRNA gene copy numbers (Ncopy) per milliliter. (A) Total archaea (triangles) and the 92-bp T-RF (circles); (B) 186-bp T-RF; (C) 393-bp T-RF; (D) 381-bp T-RF; (E) 810-bp T-RF; and (F) 689-bp T-RF. All panels show temporal changes of control (closed symbols) and CH3F (open symbols) incubations (means ± standard deviations; n = 3).
Experiments by Lueders and Friedrich (29) showed that our T-RFLP assay allows the determination of the relative abundances of the individual T-RFs within the total archaeal population. However, even if the amplification efficiencies for the investigated archaeal groups were different, this would not affect our interpretation, since the inhibition effects are compared within the same archaeal groups and not among them. The qPCR assay, which uses the same primers and PCR conditions, quantifies the total 16S rRNA gene copy numbers of the archaeal population. The combination of both data sets thus allows the determination of the temporal change of the 16S rRNA gene copy numbers of individual archaeal lineages (Fig. 3). The 92-bp T-RF, representing Methanobacteriaceae, steadily increased with incubation time in both the control and the CH3F incubations, while absolute numbers were slightly higher in the control (significant only on day 39 and 47) (Fig. 3A). Maximum copy numbers were 3 × 106 and 1 × 106 ml−1 in control and CH3F incubations, respectively. The 186-bp T-RF, representing Methanosarcinaceae, exponentially increased in the control until day 28 and then stayed at about 1 × 108 ml−1 until the end (Fig. 3B). By contrast in the CH3F incubation, the Ncopy was always much lower than that in the control and increased much more slowly. The 393-bp T-RF, representing methanogenic RC-I, initially increased but reached a constant level of about 5 × 106 ml−1 after about 20 days in both control and CH3F incubations (Fig. 3C). A different time course was observed for the 381-bp T-RF, representing euryarchaeotal RC-III (Fig. 3D), and for the 810-bp T-RF, representing crenarchaeotal RC-IV (Fig. 3E). The copy numbers of both T-RFs increased in the control as well as in the CH3F incubation until day 24. Thereafter, Ncopy stabilized in the CH3F incubations (3 × 106 ml−1) but decreased to zero in the control. A value of zero means that no peak of the particular T-RF was detected in the T-RFLP analysis. The 689-bp T-RF, representing euryarchaeotal RC-V, showed a similar increase in both incubations until day 24, with a slightly higher Ncopy in the control (Fig. 3F). Then, however, it increased further in the control (reaching about 107 ml−1) but remained constant in the CH3F incubation (about 106 ml−1).
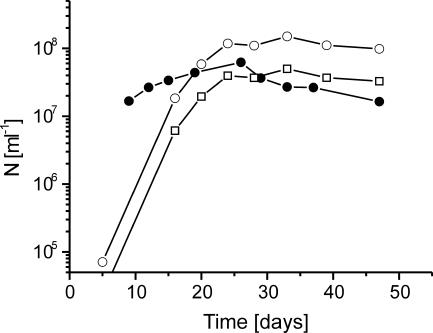
Number of theoretically maintained cells.
The CH4 production rates and Gibbs free energies of methanogenesis (Fig. 1) were used to calculate the number of methanogens (Nmc; see equation in Materials and Methods) that can be maintained at steady state by the respective energy conditions (Fig. 4). Values of Nmc were in a range of 2 × 107 to 6 × 107 ml−1 and compared fairly well with the measured numbers (Nmm). Values of Nmm were determined using 16S rRNA gene copy numbers (Ncopy) of methanogenic Methanobacteriaceae (92-bp T-RF), Methanosarcinaceae (186-bp T-RF), and RC-I (393-bp T-RF), which increased with time from <105 ml−1 to about 108 ml−1. Since Methanosarcina spp., the dominant methanogenic population, have three rRNA gene copies per genome (18), final values of Nmm were around 4 × 107 ml−1, i.e., similar to the Nmc (Fig. 4).
FIG. 4.
Comparison of experimentally determined 16S rRNA gene copy numbers (Ncopy) (○) and derived cell numbers (Nmm) (□) with the theoretically derived maximum number (Nmc) (•) of methanogenic archaea in the control incubation.
DISCUSSION
Our experiments with rice roots as a model system confirmed that CH3F inhibited acetoclastic CH4 production and resulted in the accumulation of acetate (8). The inhibition was released around day 24, when the CH3F concentration decreased below a threshold of 1.0 to 1.2%, resulting in resumption of methanogenic acetate utilization. Simultaneous T-RFLP analysis and qPCR of archaeal 16S rRNA genes showed that CH3F specifically inhibited proliferation of acetoclastic methanogenic Methanosarcinaceae. In the uninhibited control, Methanosarcinaceae grew exponentially mainly on transiently produced acetate and reached titers compatible with maintenance theory.
The temporal changes of the population sizes (i.e., 16S rRNA copy numbers) of six prevalent archaeal groups were quantified (Fig. 3). The presumed target organisms for the inhibitor CH3F are the archaea performing acetoclastic methanogenesis, i.e., Methanosarcinaceae and Methanosaetaceae. We detected neither a clone sequence of Methanosaetaceae nor a T-RF characteristic for this group, which is in agreement with earlier rice root studies, where this group was hardly detectable (2, 13). Methanosarcinaceae (186-bp T-RF), on the other hand, were abundant and active as shown by the T-RFLP patterns based on rRNA genes (abundance) and rRNA (activity) (Fig. 2). The abundance and activity of Methanosarcinaceae were strongly decreased in the CH3F versus the control incubation. Instead, RC-I methanogens became relatively more abundant and active (Fig. 2) confirming previous experiments by Lu et al. (24). Nevertheless, growth of Methanosarcinaceae was not completely abolished in the CH3F treatment (Fig. 3B). Although the acetoclastic methanogenic pathway was inhibited, Methanosarcinaceae were probably able to gain energy from conversion of H2/CO2 to CH4. After cessation of inhibition by CH3F, growth could again be sustained by acetoclastic methanogenesis in addition to or instead of hydrogenotrophic methanogenesis. The cessation of growth in the control after day 24 was probably due to energetic limitation, as the Gibbs free energy of both hydrogenotrophic and acetoclastic methanogenesis approached the thermodynamic threshold. This threshold is believed to be around −20 kJ mol−1, equivalent to synthesis of about 1/3 ATP (14, 39, 47). By contrast, in the CH3F incubation, the Methanosarcinaceae population also grew after day 28, when the CH3F inhibition ceased, since acetate was still available to provide sufficient energy.
Methanobacteriaceae (92-bp T-RF) and RC-I methanogens (393-bp T-RF) are the hydrogenotrophic methanogenic archaeal groups found in the rice root incubations. RC-I does not yet exist in pure culture, but genomic information on the methanogenic operons already exists (11, 27). Both groups are unable to perform acetoclastic methanogenesis and should therefore not be influenced directly by CH3F. Indeed, RC-I as the dominant hydrogenotrophic methanogenic group was not affected, and Methanobacteriaceae 16S rRNA gene copy numbers were only slightly decreased in the CH3F incubation. Hence, our study demonstrates that CH3F specifically inhibited the proliferation of acetoclastic methanogens in a natural model environment but did not affect hydrogenotrophic methanogens.
However, we observed an effect of CH3F on other nonmethanogenic archaeal populations present in our model environment. Thus, the 16S rRNA gene copy numbers of RC-III (381-bp T-RF) and RC-IV (810-bp T-RF) decreased in the control after day 24, finally reaching zero, but remained constant in the CH3F treatment. An enrichment culture of RC-III archaea, which are distantly related to Thermoplasmatales, was recently shown to grow anaerobically on yeast extract, peptone, and tryptone (17). However, nothing is known about the physiology of the crenarcheotal lineage RC-IV. The prevention of a decrease of RC-III and RC-IV populations by CH3F might be a secondary effect of the inhibition, e.g., due to the higher concentrations of acetate and H2 in the presence compared to the absence of CH3F. Analogously, ethanol and the fatty acids butyrate, valerate, and caproate were still detectable at the late phase of the CH3F incubation, whereas in the control they were below the detection limit. We speculate that CH3F inhibition leads to prolongation of favorable conditions for RC-III and RC-IV due such secondary effects of inhibition.
The 16S rRNA gene copy numbers of the euryarchaeotal lineage RC-V (689-bp T-RF), the physiology of which is unknown, still increased after day 24 in the control but not in the CH3F treatment. We do not know the mechanism behind this observation, but it is noteworthy that the T-RF of RC-V could not be detected in the T-RFLP pattern generated from the ribosomal fraction (RNA based) (Fig. 2B), suggesting that RC-V was not very active, at least not between days 20 and 28.
In the rice root environmental system, CH4 was produced from acetate and H2/CO2 by three different phylogenetic groups of methanogenic archaea (Methanobacteriaceae, RC-I, and Methanosarcinaceae). The numbers of methanogenic organisms that can be maintained by the Gibbs free energy and the CH4 production rate under the actual incubation conditions were found to be consistent with those actually observed (Fig. 4). The accuracy of the calculation depends linearly on the uncertainty of the input values. Gibbs free energies and CH4 production rates were determined from measured values and introduced only a relatively small error. However, the coefficient γ, which represents the C mass of a single cell, is highly uncertain and may vary within an order of magnitude. For our calculations we used a value of 2 × 10−13 g C per cell (34, 46). A larger cell mass would result in lower theoretical numbers of methanogens, and vice versa. The calculated number is the maximum of cells sustained without net growth at steady state. Our determinations showed that energetic conditions in the control incubation were permissive for net growth until about day 20 to 30 but then could not maintain more than about 2 × 107 to 4 × 107 methanogenic cells ml−1. At this time, the most prevalent methanogenic populations (i.e., Methanosarcinaceae and RC-I) had stopped growth (Fig. 3), and the numbers of methanogenic cells observed were similar to those calculated from maintenance theory (Fig. 4). This observation for the first time shows that the maintenance energy requirement of the cells is possibly important for determining microbial population size in a natural environment.
Acknowledgments
We thank Gesche Braker for helpful discussions and technical advice.
We thank Fonds der Chemischen Industrie, Germany, for financial support.
REFERENCES
- 1.Chan, O. C., P. Claus, P. Casper, A. Ulrich, T. Lueders, and R. Conrad. 2005. Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment. Environ. Microbiol. 7:1139-1149. [DOI] [PubMed] [Google Scholar]
- 2.Chin, K. J., T. Lueders, M. W. Friedrich, M. Klose, and R. Conrad. 2004. Archaeal community structure and pathway of methane formation on rice roots. Microb. Ecol. 47:59-67. [DOI] [PubMed] [Google Scholar]
- 3.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicerone, R. J., and R. S. Oremland. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 2:299-327. [Google Scholar]
- 5.Conrad, R. 1999. Soil microorganisms oxidizing atmospheric trace gases (CH4, CO, H2, NO). Ind. J. Microbiol. 39:193-203. [Google Scholar]
- 6.Conrad, R. 2005. Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org. Geochem. 36:739-752. [Google Scholar]
- 7.Conrad, R., and M. Klose. 1999. Anaerobic conversion of carbon dioxide to methane, acetate and propionate on washed rice roots. FEMS Microbiol. Ecol. 30:147-155. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, R., and M. Klose. 1999. How specific is the inhibition by methyl fluoride of acetoclastic methanogenesis in anoxic rice field soil? FEMS Microbiol. Ecol. 30:47-56. [Google Scholar]
- 9.Conrad, R., and M. Klose. 2000. Selective inhibition of reactions involved in methanogenesis and fatty acid production on rice roots. FEMS Microbiol. Ecol. 34:27-34. [DOI] [PubMed] [Google Scholar]
- 10.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erkel, C., D. Kemnitz, M. Kube, P. Ricke, K. J. Chin, S. Dedysh, R. Reinhardt, R. Conrad, and W. Liesack. 2005. Retrieval of first genome data for rice cluster I methanogens by a combination of cultivation and molecular techniques. FEMS Microbiol. Ecol. 53:187-204. [DOI] [PubMed] [Google Scholar]
- 12.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosskopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 2001. Apparent minimum free energy requirements for methanogenic Archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol. Ecol. 38:33-41. [Google Scholar]
- 15.Holzapfel-Pschorn, A., and W. Seiler. 1986. Methane emission during a cultivation period from an Italian rice paddy. J. Geophys. Res. 91:11803-11814. [Google Scholar]
- 16.Janssen, P. H., and P. Frenzel. 1997. Inhibition of methanogenesis by methyl fluoride: studies of pure and defined mixed cultures of anaerobic bacteria and archaea. Appl. Environ. Microbiol. 63:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemnitz, D., S. Kolb, and R. Conrad. 2005. Phenotypic characterization of Rice Cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ. Microbiol. 7:553-565. [DOI] [PubMed] [Google Scholar]
- 18.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger, M., G. Eller, R. Conrad, and P. Frenzel. 2002. Seasonal variation in pathways of CH4 production and in CH4 oxidation in rice fields determined by stable carbon isotopes and specific inhibitors. Global Change Biol. 8:265-280. [Google Scholar]
- 21.Krumböck, M., and R. Conrad. 1991. Metabolism of position-labeled glucose in anoxic methanogenic paddy soil and lake sediment. FEMS Microbiol. Ecol. 85:247-256. [Google Scholar]
- 22.Lehmann-Richter, S., R. Grosskopf, W. Liesack, P. Frenzel, and R. Conrad. 1999. Methanogenic archaea and CO2-dependent methanogenesis on washed rice roots. Environ. Microbiol. 1:159-166. [DOI] [PubMed] [Google Scholar]
- 23.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, Y. H., T. Lueders, M. W. Friedrich, and R. Conrad. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326-336. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 28.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 31.Oremland, R. S., and D. G. Capone. 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 32.Oremland, R. S., and C. W. Culbertson. 1992. Evaluation of methyl fluoride and dimethyl ether as inhibitors of aerobic methane oxidation. Appl. Environ. Microbiol. 58:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oremland, R. S., and C. W. Culbertson. 1992. Importance of methane-oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor. Nature 356:421-423. [Google Scholar]
- 34.Paul, E. A., and F. E. Clark. 1989. Soil microbiology and biochemistry. Academic Press, New York, N.Y.
- 35.Prinn, R. G. 1994. Global atmospheric-biospheric chemistry, p. 1-18. In R. G. Prinn (ed.), Global atmospheric-biospheric chemistry. Plenum, New York, N.Y.
- 36.Raeymaekers, L. 2000. Basic principles of quantitative PCR. Mol. Biotechnol. 15:115-122. [DOI] [PubMed] [Google Scholar]
- 37.Ricke, P., S. Kolb, and G. Braker. 2005. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 71:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheid, D., S. Stubner, and R. Conrad. 2003. Effects of nitrate- and sulfate-amendment on the methanogenic populations in rice root incubations. FEMS Microbiol. Ecol. 43:309-315. [DOI] [PubMed] [Google Scholar]
- 39.Seitz, H. J., B. Schink, N. Pfennig, and R. Conrad. 1990. Energetics of syntrophic ethanol oxidation in defined chemostat cocultures. 1. Energy requirement for H2 production and H2 oxidation. Arch. Microbiol. 155:82-88. [Google Scholar]
- 40.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. A. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 41.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen (TM) detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, M. T., L. T. Taylor, and E. F. Delong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy-conservation in chemotropic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tijhuis, L., M. C. M. VanLoosdrecht, and J. J. Heijnen. 1993. A thermodynamically based correlation for maintenance Gibbs energy requirements in aerobic and anaerobic chemotrophic growth. Biotechnol. Bioeng. 42:509-519. [DOI] [PubMed] [Google Scholar]
- 46.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao, H., and R. Conrad. 1999. Thermodynamics of methane production in different rice paddy soils from China, the Philippines and Italy. Soil Biol. Biochem. 31:463-473. [Google Scholar]