Abstract
Inactivation by allelic exchange in clinical isolates of the emerging nosocomial pathogen Enterococcus faecium has been hindered by lack of efficient tools, and, in this study, transformation of clinical isolates was found to be particularly problematic. For this reason, a vector for allelic replacement (pTEX5500ts) was constructed that includes (i) the pWV01-based gram-positive repAts replication region, which is known to confer a high degree of temperature intolerance, (ii) Escherichia coli oriR from pUC18, (iii) two extended multiple-cloning sites located upstream and downstream of one of the marker genes for efficient cloning of flanking regions for double-crossover mutagenesis, (iv) transcriptional terminator sites to terminate undesired readthrough, and (v) a synthetic extended promoter region containing the cat gene for allelic exchange and a high-level gentamicin resistance gene, aph(2′′)-Id, to distinguish double-crossover recombination, both of which are functional in gram-positive and gram-negative backgrounds. To demonstrate the functionality of this vector, the vector was used to construct an acm (encoding an adhesin to collagen from E. faecium) deletion mutant of a poorly transformable multidrug-resistant E. faecium endocarditis isolate, TX0082. The acm-deleted strain, TX6051 (TX0082Δacm), was shown to lack Acm on its surface, which resulted in the abolishment of the collagen adherence phenotype observed in TX0082. A mobilizable derivative (pTEX5501ts) that contains oriT of Tn916 to facilitate conjugative transfer from the transformable E. faecalis strain JH2Sm::Tn916 to E. faecium was also constructed. Using this vector, the acm gene of a nonelectroporable E. faecium wound isolate was successfully interrupted. Thus, pTEX5500ts and its mobilizable derivative demonstrated their roles as important tools by helping to create the first reported allelic replacement in E. faecium; the constructed this acm deletion mutant will be useful for assessing the role of acm in E. faecium pathogenesis using animal models.
In today's world, enterococci, especially some strains of Enterococcus faecium, are best known in the clinical setting as multidrug-resistant opportunists causing difficult-to-treat hospital-acquired infections, including infective endocarditis (32, 33). While in the past clinical isolates of Enterococcus faecalis outnumbered those of E. faecium by approximately 9:1, this ratio has changed in some U.S. hospitals to ∼6:4, and this increase parallels the increase in vancomycin resistance of E. faecium (20, 33, 58). An important theme relating to strains causing E. faecium infections in hospitals today is that, in addition to acquiring antibiotic resistances, they seem to have lost the harmless, commensal nature of the strains colonizing healthy individuals in the community and often contain either new genes (e.g., espfm, encoding a potential enterococcal surface protein, or hyl, encoding a potential hyaluronidase [16, 25, 44, 62]) or a functional form of genes (e.g., acm, encoding a collagen adhesin [37]), thus presumably enhancing their ability to survive and/or cause infection in the clinical setting. While espfm and hyl genes were found to be rare in isolates from healthy volunteers and in isolates from animal origin (44), the acm gene was present in these isolates as an inactive gene (37 and S. R. Nallapareddy and B. E. Murray, unpublished results). To study virulence traits of E. faecium, it is necessary to have appropriate genetic methods. However, tools for constructing targeted mutations in E. faecium are rudimentary or contain resistant genes frequently present in clinical isolates.
Several methods have been previously developed for targeted mutagenesis of enterococci and found to be useful for genetic manipulation of E. faecalis. One of these makes use of plasmids that are suicidal in gram-positive hosts (13, 15, 43, 51). A second method of allelic replacement, involving electroporation of E. faecalis with DNA from a pool of mini-γδ-mutagenized cosmid clones, is applicable for genes with detectable phenotypes (28, 51). The disadvantage of these two techniques is the requirement for efficient transformation, since selection depends on both successful bacterial transformation and successful integration in the same step. However, the problem with enterococci is that, although some strains are readily transformable using electroporation, others are not. A third method of delivery integration is based on pORI19/pG+host3 and requires individual transformation with two different plasmids (45). Other methods based on conjugation systems (30, 56) and temperature-sensitive (ts) plasmids (1, 12, 47) have also been used intermittently for E. faecalis mutagenesis to circumvent the difficulty of electroporation. However, these vectors were constructed for specialized uses and lack many characteristics associated with cloning vectors, such as availability of unique recognition sites and appropriate markers that would be useful in mutagenesis of clinical isolates of E. faecium. Additionally, the ts-based plasmids used in mutagenesis of other gram-positive bacteria usually contain remnants of enterococcal transposons Tn916 and Tn917, thus posing potential problems with recombination into these elements which are common in clinical isolates.
In spite of a number of available vector systems, due to low transformability of the host as well as some limitation in each of the vector system(s) described above, we and others have had limited success in generating targeted mutations in E. faecium genes. The two E. faecium genes that were interrupted, albeit with low efficiency, in our laboratory were generated in the moderately transformable E. faecium strain TX1330 (also known as SE-34), a fecal isolate, isolated from a healthy community volunteer (50, 55). It is important to note that this E. faecium fecal strain does not contain espfm or hyl, nor does it express acm (the acm gene of this strain is a pseudogene), the three potential virulence-related genes described to date for this species (37, 44).
In order to circumvent the limitations described above, in the present study we engineered Escherichia coli-enterococcal cloning vectors that are conditionally suicidal in enterococci at nonpermissive growth temperatures and which harbor antibiotic genes that are selectable in both Escherichia coli and enterococci. Appropriate acm gene fragments were cloned into these new vectors and then used to introduce allelic replacement or disruption mutations in the acm gene of clinical strains of E. faecium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli, E. faecalis, and E. faecium strains used in this study are listed in Table 1. Enterococcal isolates initially identified to the species level by biochemical tests were confirmed by colony hybridization (49) using an intragenic ace probe for E. faecalis (9, 36) and an aac(6′)-Ii probe for E. faecium (9). E. coli strains were grown in Luria-Bertani media (Difco Laboratories, Detroit, Mich.). Enterococci were grown in either brain heart infusion (BHI) or Todd-Hewitt (TH) broth or agar (Difco Laboratories) at 37°C, unless a different growth temperature is specified. The following antibiotic concentrations were used with enterococci: chloramphenicol, 10 μg/ml; erythromycin, 15 μg/ml (for selection of transformants); erythromycin, 200 μg/ml [for selection of naturally resistant E. faecium strains due to msrC (50) plus erm(B)]; gentamicin, 125 μg/ml; and tetracycline, 10 μg/ml. With E. coli, the concentrations used were chloramphenicol, 10 μg/ml; erythromycin, 200 μg/ml; and gentamicin, 25 μg/ml. All antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.). All plasmids used in this study are listed in Table 1. All constructs were given TX numbers as shown in Table 1. Plasmids from these constructs were assigned respective pTEX numbers.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. faecium | ||
| TX1330 | Fecal isolate from community volunteer; Amps, Chls, Erys, Gens, Kanr, Strs, Tets; Cn-Adh− | 8, 37 |
| TX2466 | Blood isolate; Eryi, Kanr, Gens, Vanr | This study |
| TX0082 | Endocarditis isolate; Ampr, Chls, Eryr, Kanr, Gens, Vanr; Cn-Adh+ | This study |
| TX2555 | Perineal wound isolate; Chls, Eryr, Kanr, Gens, Vanr; Cn-Adh+ | 37 |
| TX6051 | TX0082Δacm, acm deletion mutant of TX0082, Chlr; Cn-Adh− | This study |
| TX6054 | TX2555acm::pTEX6052, acm insertion disruption mutant of TX2555, Genr, Cn-Adh− | This study |
| E. faecalis | ||
| JH2Sm::Tn916 | JH2 harboring Tn916; Gens, Strr, Tetr | 6 |
| TX6053 | JH2Sm::Tn916(pTEX6052); Strr, Tetr, Genr | This study |
| OG1RF | Laboratory strain; Rifr, Fusr, Chls, Gens | 35 |
| E. casseliflavus | ||
| UC73 | aph(2″)-Id containing strain; Genr | 59 |
| E. coli | ||
| DH5α | E. coli host strain for routine cloning | Stratagene |
| TX5500 | DH5α(pTEX5500ts); Chlr, Genr | This study |
| TX5501 | DH5α(pTEX5501ts); Chlr, Genr | This study |
| TX6050 | DH5α(pTEX6050); Chlr, Genr | This study |
| TX6052 | DH5α(pTEX6052); Genr | This study |
| Plasmids | ||
| pAM401 | Shuttle plasmid; Chlr and, in E. coli, Tetr | 64 |
| pAT18 | Shuttle plasmid; Eryr | 60 |
| pTV1-OK | pWV01 replicon-based ts vector; Kanr, Eryr | 14 |
| pTV1-ts | pE194 replicon-based ts vector; Chlr, Eryr | 66 |
| pAM401ts | pIP501 replicon-based ts vector; Chlr | 60 |
| pFW14 | Streptococcal integration vector; Chlr | 42 |
| pHS1 | E. faecalis mobilizable ts plasmid containing oriTTn916; Kanr, Genr | 1 |
| pTEX5500ts | Shuttle plasmid, ts in gram-positive hosts; Chlr, Genr | This study |
| pTEX5501ts | Mobilizable ts vector containing oriTTn916; Chlr, Genr | This study |
| pTEX6050 | Plasmid for acm deletion with flanking regions of the acm binding domain cloned into pTEX5500ts; Chlr, Genr | This study |
| pTEX6052 | acm insertion construct, intragenic fragment of acm cloned into pTEX5501ts with deletion of cat; Genr, Chls | This study |
Chl, chloramphenicol; Ery, erythromycin; Fus, fusidic acid; Gen, gentamicin; Kan, kanamycin; Rif, rifampin; Tet, tetracycline; Str, streptomycin; Van, vancomycin; and ts, temperature sensitive. Superscript “s” designates sensitivity, “r” designates resistance, and “i” designates intermediate susceptibility (MIC of 2 to 4 μg/ml for Ery); for aminoglycosides “r” is defined for enterococci as MIC > 500 for Gen and > 2,000 for Kan. Cn-Adh+, adherence to collagen types I and IV; Cn-Adh−, no adherence to collagen types I and IV.
Standard molecular techniques.
Specific primers were purchased either from Invitrogen (Carlsbad, Calif.) or from Sigma-Genosys (The Woodlands, Tex.). Restriction enzymes and DNA modification enzymes were purchased mostly from Invitrogen, and reactions were carried out under the recommended conditions. Bovine collagen type I was purchased from Cohesion Technologies, Inc. (Palo Alto, Calif.), collagen type IV was purchased from Sigma Chemical Co., and fibrinogen was purchased from Enzyme Research Laboratories (South Bend, Ind.). Tran35S label and bovine serum albumin (BSA) were purchased from MP Biomedicals Inc. (Irvine, Calif.). All other chemicals used in the investigation were of molecular biology grade. Chromosomal DNA from E. faecium isolates was prepared following the hexadecyltrimethyl ammonium bromide method described earlier (63). Plasmid DNA isolation from E. coli used the Wizard Plus SV minipreps DNA purification system (Promega Corporation, Madison, Wis.) and, from enterococci, by a previously described method (65). General recombinant DNA techniques such as ligation and agarose gel electrophoresis were performed using standard methods (46). When necessary, DNA fragments were purified with low-melting-point agarose gels followed by purification using QIAquick-gel extraction kits (QIAGEN, Inc., Valencia, Calif.). PCRs were performed with a Perkin-Elmer GeneAmp PCR system 9700 using the optimized buffer B (1× buffer: 60 mM Tris-HCl [pH 8.5], 15 mM ammonium sulfate, and 2 mM MgCl2) obtained from Invitrogen. PCR-generated fragments were purified using the Wizard PCR DNA Cleanup System (Promega Corporation). Recombinant plasmids were generated in E. coli DH5α. Agarose plugs containing genomic DNA were digested with SmaI, and pulsed-field gel electrophoresis (PFGE) was performed using previously described methods (34) but with different ramped pulse times, 2 s and 28 s. Southern blotting was performed using Hybond-N+ nylon membranes and a 0.4 N NaOH solution. The RadPrime DNA Labeling System (Invitrogen) was used to label the probes with [α-32P]dCTP (GE Healthcare, Piscataway, NJ), and hybridizations were carried out using high-stringency conditions (35). DNA sequencing reactions were performed by the Taq dye-deoxy terminator method and an automated ABI Prism sequencer (Applied Biosystems, Foster City, Calif.). Sequences were assembled using SeqMan program of DNASTAR software (Lasergene; Madison, Wis.).
Bacterial electrotransformation and conjugation.
Electroporation of E. coli, E. faecalis, and E. faecium was carried out using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) as described previously (28, 50). Broth-mating experiments were performed as previously described (57), except for a donor:recipient ratio of 2:1. In brief, overnight cultures of 0.5 ml of donor and 0.25 ml of recipient were added to 4.25 ml of fresh BHI broth, and the mixture was incubated without shaking at 28°C overnight. After vortexing, cells were harvested by centrifugation at 4,000 rpm and 4°C and resuspended in 200 μl of saline followed by plating on BHI agar plates with 200 μg/ml erythromycin and 125 μg/ml gentamicin. Plates were checked for colonies for up to 96 h (at 24-h intervals) of incubation at 28°C. For some strains, in an effort to increase the conjugation frequency, filter-mating experiments were also performed as previously described (57), except for a donor/recipient ratio of 20:1. In brief, 1 ml of an overnight culture of donor (grown at 28°C in BHI broth containing 125 μg/ml gentamicin and 10 μg/ml tetracycline to increase the transfer frequency [6]) and 50 μl of an erythromycin-resistant recipient were added to 4.5 ml of fresh BHI broth, and the mixture was then passed through a 0.45-μm membrane filter (Millipore, Bedford, Mass.). The cells trapped on the filters were incubated at 28°C overnight and were then suspended in 1 ml of saline. Appropriate dilutions of the mixture were transferred to BHI agar plates containing 200 μg/ml erythromycin and 125 μg/ml gentamicin. Plates were assessed for colonies after 24 and 48 h of incubation at 28°C.
Determination of plasmid segregation rates.
TX0082 transformed with pTEX5500ts (construction of this plasmid is described in Results and Discussion) was grown at 28°C with 10 μg of chloramphenicol and 125 μg of gentamicin per ml until the optical density at 600 nm (OD600) reached approximately 1. The culture was then diluted 1:1,000 with fresh BHI broth and grown overnight (16 h) without antibiotic selection at 28, 37, and 42°C. Initial and final numbers of TX0082 cells with and without pTEX5500ts were determined by quantitative plate counting on agar plates with and without antibiotics after growth at 28°C. Plasmid segregation rates were derived from these CFU using the following formula described by Framson et al. (11): percent loss per generation = [1 − (Ff/Fi)1/g] × 100, where F is the frequency of the plasmid-mediated resistance phenotype in the population, f is the final CFU, i is the initial CFU, and g is the number of generations intervening between the final and initial CFU (determined on nonselective media) according to the formula g = [loge (final CFU/initial CFU)]/loge (2).
Growth curves.
Overnight cultures were inoculated in BHI broth at a dilution of 1:100. The cultures were then grown at 37°C with shaking in an orbital shaker, and the aliquots were removed hourly from 0 to 12 h and at 24 h for determining CFU on BHI agar and also to measure the absorbance at 600 nm (OD600) with a spectrophotometer.
Cell wall-associated protein extraction and Western blotting.
Protein extracts from E. faecium strains were prepared using mutanolysin (Sigma Chemical Co.) as described previously (37). Protein concentrations were estimated by bicinchoninic acid assay (Pierce, Rockford, Ill.). Equal concentrations of mutanolysin extracts were electrophoresed in 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) under reducing conditions in morpholinepropanesulfonic acid buffer and were transferred to a polyvinylidene difluoride membrane according to the protocol supplied by Invitrogen. Membranes were then probed with anti-AcmA polyclonal antiserum or preimmune serum (antibody I) (37) followed by Protein A horseradish peroxidase conjugate (antibody II) and developed with 4-chloronaphthol in the presence of H2O2.
Adherence assay.
Adherence of E. faecium to collagen, fibrinogen, and BSA was determined in four independent experiments using Tran35S-labeled bacteria by a previously described assay (37).
Nucleotide sequence accession numbers.
Nucleotide sequences of pTEX5500ts and pTEX5501ts were submitted to GenBank under accession numbers DQ208936 and DQ208937.
RESULTS AND DISCUSSION
The recent increase in multidrug-resistant E. faecium infections and its emergence as a nosocomial pathogen highlight the need for genetic studies addressing the mechanism of bacterial pathogenesis of this species. Although we and others have reported the presence of homologues of known virulence factors among E. faecium strains (e.g., efaAfm, espfm, hyl, and acm) (25, 37, 44, 49, 62), the relative contributions of these genes to E. faecium pathogenicity have not been assessed, largely due to the lack of tools for replacement of a wild-type gene with a mutated gene. Our repeated attempts to generate targeted mutations in some E. faecium clinical strains with suicide, temperature-sensitive, and/or mobilizable vector systems (1, 51, 56) developed for E. faecalis mutagenesis were unsuccessful. Hence, the aims of this study were to develop a delivery and screening system of E. faecium and to validate the applicability of this system for generating targeted mutations in the acm gene of E. faecium clinical isolates. As a first step, some clinical strains were screened for their ability to acquire plasmid pAM401.
Clinical strains of E. faecium are poorly transformable.
We and others previously found that E. coli-E. faecalis shuttle plasmids (e.g., pAM401, pWM401, and pAT18) can be transformed into and will replicate in selected strains of E. faecium, although they do so with some instability in the absence of selection pressure. However, the three E. faecium strains that we previously reported as transformable (37) either lack the virulence factors of interest (44) or contain a pseudogene (37). In this study, diverse E. faecium strains containing potential virulence genes were screened for their transformation capability using electroporation conditions optimized for E. faecium (50) and a pIP501-derived high-copy-number shuttle plasmid, pAM401 (64). Results from this preliminary experiment using a total of 11 human-derived E. faecium strains, including 8 clinical isolates, revealed that strains varied markedly in their ability to acquire this plasmid, with most failing to do so (Table 2). As is evident from Table 2, among the tested strains, the community-derived fecal E. faecium isolate TX1330 exhibited high transformation efficiency, in the range of 1,200 to 4,000 CFU/μg of plasmid. Similar transformation efficiencies were noted for pAT18 (with the pAMβ1 replicon) with this strain. Two other strains that were found to be moderately transformable were D344-S (a laboratory recipient isolate and spontaneous mutant of D344 [48]) and an American Type Culture Collection (ATCC 51558) recipient isolate, GE-1 (10). Their efficiencies of transformation were 100-fold less than that of pAM401 transformation into E. faecalis OG1X (64). While some clinical strains exhibited low-level transformation with pAM401 (e.g., an endocarditis isolate TX0082 yielded 33 ± 8 CFU/μg of plasmid), others appeared to be nontransformable (e.g., wound isolate TX2555) (Table 2). Additionally, in the absence of selective pressure, ca. 6 to 16% of overnight cultures were found to have lost pAM401 in three independent experiments. Poor transformability increases the difficulty of using suicidal vectors and motivated us to explore conditional replicons that may be applicable for E. faecium.
TABLE 2.
Transformation efficiencies of various strains of E. faecium with high-copy-number shuttle vector pAM401
| Strain (reference) | Source | No. of transformants/μga |
|---|---|---|
| TX1330 (8, 37) | Fecal isolate from community volunteer | 2,113 ± 1,313 |
| D344-S (37, 48) | Laboratory recipient | 955 ± 503 |
| GE-1 (10) | Recipient (ATCC 51558) | 222 ± 86 |
| TX0054 (37) | Endocarditis | 0 |
| TX0082 (this study) | Endocarditis | 33 ± 8 |
| TX2535 (37) | Endocarditis | 0 |
| TX2699 (this study) | Endocarditis | 0b |
| TX2095c (this study) | Catheter tip | 0b |
| TX2092 (this study) | Rectum | 9 ± 3 |
| TX2084 (this study) | Urine | 0 |
| TX2555 (37) | Wound | 0 |
All experiments were carried out with 500 ng of pAM401 DNA isolated from E. coli DH5α. Electroporation was performed with a Bio-Rad Gene Pulser apparatus with the following conditions: 12.5 kV (per 1-cm cuvette), 25 mF capacitance, and 400 Ω resistance. These values represent the means and standard deviations of three independent competent cell preparations.
Sporadically, some breakthrough colonies were found on chloramphenicol plates. These colonies did not contain pAM401 and, upon restreaking, were unable to grow on chloramphenicol plates.
Transformation and stability of conditional replication based ts plasmids in E. faecium.
At least two thermosensitive broad-host-range vector families (derived from pWVO1 and pE194) have been shown to be useful for high-efficiency gene inactivation and replacement in gram-positive bacteria (2, 3, 5, 17, 41, 54). In order to test the applicability of these systems in E. faecium, the ability of three established ts plasmids, pTV1-OK (14), pTV1-ts (66), and pAM401ts (60), derived from pWV01, pE194, and pIP501 replicons, respectively, were assessed to transform and replicate in the transformable E. faecium strain, TX1330, at permissive temperature (established as 30°C for other gram-positive bacteria). All three plasmids were found to transform, although poorly, and replicate in TX1330 at 28°C (Table 3). At 30°C, recovery of transformants was found to be reduced to approximately half of what we recovered at 28°C. Among these three ts plasmids, pTV1-OK showed maximum recovery, followed by pAM401ts, and pTV1-ts being the least (Table 3). Restriction digestions of plasmids analyzed from transformants grown at 28°C identified that pTV1-OK and pTV1-ts were relatively stable in E. faecium at permissive temperature and maintained their original molecular sizes. In contrast, pAM401ts in E. faecium was unstable with multiple deletions and different colonies showed different digestion patterns. Since pTV1-OK is not suitable for E. faecium clinical strains (due to lack of appropriate markers and the presence of Tn917), we selected pHS1 (a pWV01ts-based E. faecalis suicidal vector [1]) and introduced it in two clinical strains (TX0082 and TX2466) at 28°C. We next cloned an acm intragenic fragment into pHS1 and introduced this plasmid into E. faecium TX0082 by electroporation, selected at 28°C, with subsequent growth of an individual colony at 42°C. In addition to the expected single-crossover recombination into acm at the nonpermissive temperature (42°C), persistence of the plasmid was noticed in TX0082 even after >10 serial passages at 42°C. Upon sequencing, we found that RepA of this construct was not that of the more ts version found in pVE6004 (29) but rather is like the parental plasmid pWV01 (27), which is able to replicate at higher temperatures. This observation warranted the need for a vector that would be more reliably nonreplicative at elevated temperatures.
TABLE 3.
Transformation efficiencies of E. faecium TX1330 with various plasmids
| Plasmid (replicon, type)a | Size (kb) | Antibiotic markerb | No. of transformantsc/μg |
|---|---|---|---|
| pAM401 (pIP501, θ) | 10.4 | Chl | 2,113 ± 1,313 |
| pTV1-OK (pWV01, RC) | 11 | Ery | 64 ± 6 |
| pTV1-ts (pE194, RC) | 12.4 | Chl | 13 ± 3 |
| pAM401ts (pIP501, θ)d | 10.4 | Chl | 19 ± 2 |
The replicon is derived from the replication apparatus of the original plasmid. The replication mechanism used by the plasmid is RC, rolling circle, or θ, theta.
Antibiotic markers used to select for the plasmid transformants. Ery, erythromycin; Chl, chloramphenicol.
These values represent the means and standard deviations of three independent competent cell preparations.
This plasmid is highly unstable in E. faecium.
Construction of pTEX5500ts and its mobilizable derivative.
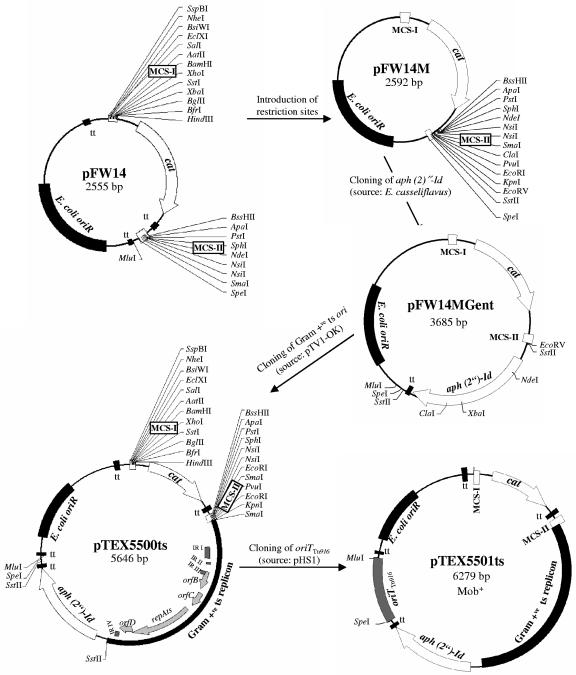
Since currently available vectors often contain regions of DNA from enterococci as markers, which may lead to undesired recombination, the minimal functional regions were amplified using specific primers containing rare restriction sites. The streptococcal integration vector pFW14, containing two multiple-cloning sites (MCS), was used as the backbone for pTEX5500ts construction (Fig. 1). The vector pFW14 is a derivative of pFW8 (42) in which the aad9 promoter driving cat gene expression has been replaced by a synthetic promoter region from pFW11 (42) that contains two extended −10 and −35 boxes. To clone a minimal gram-positive ts replicon region by restriction digestion and ligation (and also to add commonly used restriction sites to MCS-II of pFW14), we first introduced ClaI, PvuI, EcoRI, KpnI, EcoRV, and SstII restriction sites into the MCS-II. This was done by treating the appropriately designed synthetic primer duplexes with T4 DNA polymerase and T4 polynucleotide kinase (52) and subsequent cloning into the SmaI site of pFW14. This intermediate construct was designated pFW14M (Fig. 1). The gentamicin resistance gene aph(2′′)-Id was then amplified from Enterococcus casseliflavus strain UC73 (59) using primers GentF and GentR (Table 4). This PCR-amplified fragment containing the aph(2′′)-Id gene and its promoter and terminator regions was digested with SstII and then ligated to similarly digested pFW14M, and this construct was designated pFW14MGent (Fig. 1). The gentamicin resistance gene of this vector functions as a spacer as well. We next ligated the ts replicon (obtained by filling in the eluted ∼2.5-kb XbaI fragment of pTV1-OK with T4 DNA polymerase, followed by treating with T4 polynucleotide kinase and finally digesting with KpnI) and the previously KpnI- and EcoRV-digested pFW14MGent. This construct was designated pTEX5500ts (Fig. 1). To construct plasmid pTEX5501ts, the PCR-amplified oriT fragment (using primers oriTF and oriTR) from the E. faecalis mobilizable vector pHS1 (1) was digested with SpeI and MluI and then ligated into similarly digested pTEX5500ts (Fig. 1).
FIG. 1.
Construction of pTEX5500ts, a temperature-sensitive (ts) delivery vector, and its mobilizable derivative, pTEX5501ts. Both vectors contain a ts version of the broad-host-range pWV01 replicon for replication in gram-positive hosts at permissive temperatures and oriR derived from pUC18 to replicate in E. coli. Arrow directions in each plasmid indicate the direction of transcription. Only relevant restriction sites are shown. Maps of pTV1-OK and pHS1, not shown here, have been previously published (1, 14). MCS, multiple cloning sites; cat, the chloramphenicol acetyltransferase gene from pC194; tt, transcriptional terminator sites (tts from pFW14 are not shown in pFW14M and pFW14Mgent constructs); E. coli oriR, origin of replication derived from pUC18; aph(2′′)-Id, gentamicin resistance gene from E. casseliflavus; gram-positive ts replicon, temperature-sensitive origin of replication from pTV1-OK; IR, inverted repeats; oriTTn916, origin of transfer region of Tn916; and Mob+, mobilizable by trans-activation from strains harboring Tn916.
TABLE 4.
Primers used in this study
| Primer name | Type | Sequence (5′ → 3′)a |
|---|---|---|
| GentF | Forward | TTCCCCGCGGAGATAGGTTATGCAAGATTTTTATATG |
| GentR | Reverse | TTCCCCGCGGATTCCGGATTCTAAAAAAGGATTGCTA |
| 5500-MCS-IFb | Forward | GACAATACTGATAAGATAATATATC |
| 5500-MCS-IRb | Reverse | TTACGTTACGTTATTAGTTATAG |
| 5500-MCS-IIFb | Forward | TTTGTTTTATTATTTAATATTTGG |
| 5500-MCS-IIRb | Reverse | GCCGATAACTAAACGAAATAAACG |
| oriTF | Forward | GGACTAGTTTTTATACTCCCCTTGTACCAAAAG |
| oriTR | Reverse | GGACGCGTCAAAGGACGAATATGTCGCCTAAAG |
| AcmDelF1 | Forward | CGCGGATCCTATCTCTTGAAACGTTCTTAGAATTGC |
| AcmDelR1 | Reverse | CCCAAGCTTACTATATCATCTCTCAAAAATCCTTTC |
| AcmDelF4 | Forward | AAAACTGCAGAATGAGAACCAAGACGGCAAACGAC |
| AcmDelR6 | Reverse | TCCCCCGGGCCAGCAGGTACAGTGACTCCATTAGC |
| AcmUp11 | Forward | TTTGTAAATAATTGGATGGCAGATT |
| CmR | Reverse | AAAGCATTTTCAGGTATAGGTG |
| CmF | Forward | GAATGACTTCAAAGAGTTTTATG |
| AcmDownR11 | Reverse | AACCGAACAAGAAGCTTATGAACTA |
| AcmF2 | Forward | CAGGCAGAGATATCAGCAG |
| AcmR1 | Reverse | ATTCTCATTTGTAACGACTAGC |
| AcmInsF | Forward | CGCGGATCCACGAATATTACAGATGGAGGAAACA |
| Acm19R | Reverse | CGGGGTACCTTAATTTTTAACTGTATGATTGAAACTTTC |
Introduced restriction sites are underlined.
These primers flanking each of the MCS will facilitate DNA sequencing across the vector-insert junctions.
With the aim of improving the selection efficiency of these vectors, we sought to utilize a counter selection based on sucrose sensitivity, proven to be functional in some other gram-positive bacteria (4, 21, 39, 40). Cloning of sacB derived from Bacillus subtilis (encoding levansucrase) under the extended synthetic promoter of pFW14 followed by electroporation in E. faecium strain TX1330 resulted in normal growth of the transformant even in medium containing sucrose. At this stage, it is unknown whether the sacB product is nonfunctional in E. faecium or whether the toxicity due to the sacB product was complemented by an alternate mechanism of sucrose resistance. Similarly, with the aim of facilitating detection of allelic exchange events, blue-white selection as a reporter system was also investigated. However, growth of E. faecium on plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside rendered a light-blue colonial phenotype indistinguishable from colonies containing the lacZ construct we made from pORI280 (26). Additionally, use of a new marker encoded by emtA (31) was also found to be ineffective due to the observed background growth of avilamycin-susceptible E. faecium when electrocompetent cells (∼1 × 1010 CFU) were plated.
Properties of the newly derived ts replicon vector, pTEX5500ts.
pTEX5500ts contains (i) E. coli oriR of pUC18 derived from the streptococcal integration vector pFW14 (42); (ii) a gram-positive repAts replicon from pTV1-OK (14) which is functional in E. faecium at a permissive temperature; (iii) two distinct multiple cloning sites (MCSs) on either side of the cat gene to provide several alternative sites for cloning of flanking regions for double-crossover mutagenesis; (iv) a synthetic functional promoter region (with two extended −35 and −10 boxes) to overcome relatively low expression levels of the pC194-based cat gene (19, 42, 61); (v) a gentamicin resistance gene aph(2′′)-Id from E. casseliflavus (59); (vi) transcriptional terminator sites (tts) to terminate undesired readthrough and to prevent destabilizing effects of cloned inserts; and (vii) primer sites flanking each of the MCSs (5500-MCS-IF, 5500-MCS-IR, 5500-MCS-IIF, and 5500-MCS-IIR [Table 4]) that facilitate screening the vector by PCR or for DNA sequencing across the vector-insert junctions. The cat gene encoding chloramphenicol resistance can be used as a marker for selecting chromosomal integration. The gentamicin resistance marker located on the vector can be used to distinguish double-crossover recombinants after plasmid excision. Both resistance markers are functional in E. coli and enterococcal backgrounds. Restriction sites adjacent to each of these markers will be helpful for replacing them with alternative markers, as needed. Although the tt positioned downstream of the cat gene may have some disadvantages due to a possible polar effect, in this study, it was found to be necessary for cloning of the AcmDn fragment (see below) in MCS-II of pTEX5500ts. A similar observation of the necessity of introduction of tts for efficient cloning of streptococcal DNA fragments was reported in previous studies (7). However, it is possible to use this vector to generate a nonpolar mutation of polycistronic genes by cloning the flanking regions contiguously in-frame followed by deletion of the cat cassette including its terminator.
Upon complete sequencing of pTEX5500ts, an additional SmaI site in the 5′ region of the ts replicon and creation of an EcoRI site due to a single mutation in MCS-II were noticed. Since these restriction sites are also part of MCS-II, these sites still can be used in cloning procedures. However, XbaI of MCS-I and NdeI and ClaI of MCS-II are unusable because of their secondary occurrence in aph(2′′)-Id. These plasmids should be high copy number in E. coli due to the pUC18-based replicon, are stable after growth at 37°C, and should be low copy number (the pGK12 parent of the pWV01 replicon is known to occur in three copies per cell in lactococci [24]) in gram-positive hosts at permissive temperatures.
Replication, temperature sensitivity, and segregation stability of pTEX5500ts.
To ensure replication, both postelectroporation incubation and growth on selective media (48 to 96 h) were performed at 28°C. When the primary selection plates contained more than one antibiotic (i.e., both chloramphenicol and gentamicin), either no transformants were found or they were very rare. Using gentamicin for selection, transformation with successful replication was tested with TX1330 and four clinical isolates. Among these, TX1330 yielded more colonies, as anticipated from the above results, in the range of transformation efficiencies noted with pTV1-OK (ca. 80 CFU/μg of plasmid). Two clinical strains that we were able to transform with pTEX5500ts are TX0082 and TX2466, albeit with poor efficiency, i.e., 13 and 9 CFU/μg of plasmid, respectively. The trend of these results is consistent with those obtained with pAM401 (Table 2), suggesting that the transformation variability is a property of the specific strain and not plasmid instability. Colonies that appeared on selective plates were grown in BHI broth with gentamicin, and plasmid DNA isolated from overnight cultures was detected by agarose gel electrophoresis (data not shown).
In E. faecium, pTEX5500ts was found to be relatively stable, showing 48% retention (stability seen in three independent experiments was 60, 35, and 49%) of the chloramphenicol resistance phenotype following 16 h of growth at 28°C in the absence of antibiotics. In order to confirm the permissive and nonpermissive temperatures for the replication of pTEX5500ts in E. faecium, the stability of this plasmid was analyzed at different temperatures. The ratio of the number of CFU on selective plates to the number of CFU on nonselective plates dropped by ca. 3,100 and ca. 4,700-fold at 37 and 42°C, respectively. The segregation rates of the chloramphenicol resistance phenotype from TX0082 in broth culture in the absence of antibiotic selection at different temperatures were as follows: at 37°C, 59% per generation; at 42°C, 64% per generation; and at 28°C, 5% per generation. These results confirm that pTEX5500ts exhibits a dramatic loss in plasmid replication between 28 and 37 or 42°C, as observed with pWV01ts-based plasmids in other gram-positive pathogens. In subsequent experiments, a culture temperature of 42°C completely cured the plasmid from TX0082 after five serial passages. The nonpermissive temperature of 42°C found for pTEX5500ts in E. faecium does not limit the applicability of this system, since these organisms are characterized by high heat tolerance and grow relatively well up to ≥45°C.
Construction of a deletion mutation in the acm gene of the collagen-binding E. faecium endocarditis isolate, TX0082.
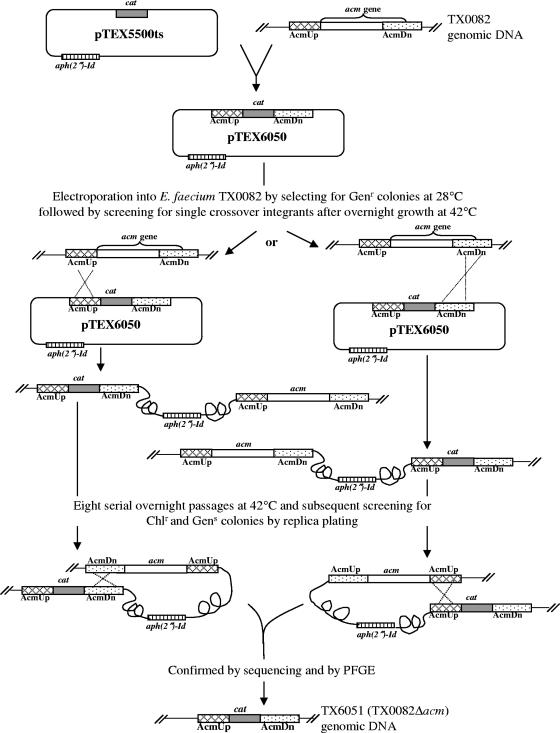
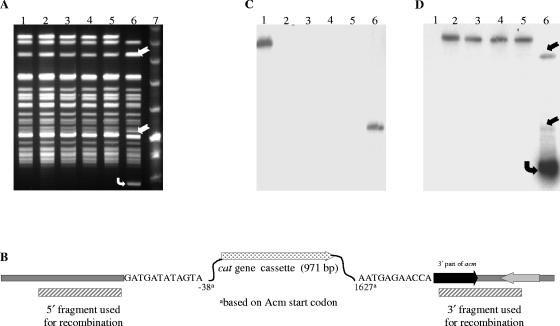
To demonstrate the applicability of our new vector for targeted gene disruption, the acm gene was chosen. The choice of acm is because of our hypothesis that Acm plays a role in E. faecium pathogenesis, including endocarditis (37). This is based on our earlier observation of the predominance of a functional acm gene in clinical isolates but a pseudogene in fecal and animal isolates (37 and S. R. Nallapareddy and B. E. Murray, unpublished results) and also data for the importance of the Staphylococcus aureus collagen adhesin Cna (an Acm homologue) in animal models, including endocarditis (18, 23, 38). Figure 2 outlines the protocol that was used for acm gene replacement in E. faecium. A 1,106-bp DNA fragment designated AcmUp (Fig. 2), encompassing the region upstream of acm, was amplified from TX0082 genomic DNA template using primers AcmDelF1 and AcmDelR1 (Table 4), digested with BamHI and HindIII and ligated with similarly digested pTEX5500ts. Similarly, a 1,031-bp DNA fragment designated AcmDn (Fig. 2), encompassing some of the coding region of the 3′ end of acm and the downstream region, was amplified from the same genomic DNA template using primers AcmDelF4 and AcmDelR4 (Table 4). The AcmDn PCR product digested with PstI and SmaI was ligated to similarly digested pTEX5500ts::AcmUp. This in vitro-ligated construct for generating an acm deletion (designated pTEX6050 [Fig. 2]) was transformed into E. coli to obtain TX6050 and was then introduced into electrocompetent cells of TX0082 which were allowed to recover at room temperature for 2 h; the latter step was found to be essential for transformation of plasmids into poorly transformable strains. The cells were then plated on gentamicin plates at the permissive temperature (28°C) to select for transformants. After overnight growth at elevated temperature (42°C), the cells were plated on chloramphenicol plates and incubated at 37°C. The single-crossover integration (TX0082::pTEX6050) was tested by PCR (primers sets AcmUp11 and CmR as well as AcmDownR11 and CmF were used for verifying integration into AcmUp and AcmDn regions, respectively [Table 4]), which confirmed that integration was specific and occurred either in AcmUp or in AcmDn regions in different colonies. Also, 40 of 40 gentamicin-resistant colonies tested showed integration. However, PFGE followed by Southern blot probing showed persistence of free plasmid, in addition to single-crossover integration, after overnight growth at 42°C. One of the integrants was picked and was grown for eight serial passages at a culture temperature of 42°C to completely cure the plasmid. The cultures from the fifth passage onward were serially diluted and plated at 37°C on nonselective media to select for plasmid excision by double-crossover recombination. These master plates were then replica plated to chloramphenicol plates and gentamicin plates to identify colonies that retained the cat gene and not the vector. Double crossovers were expected to be chloramphenicol resistant and gentamicin sensitive, as shown in Fig. 2. We found 4 of ∼5,000 colonies in which the acm gene replacement had occurred from the plates from the eighth passage. One of these colonies was designated TX6051 (TX0082Δacm). The deletion was indicated by PCR with primers AcmUpF11 and AcmDownR11, and the strain's identity was confirmed by PFGE (Fig. 3A). Sequencing of this PCR product confirmed the correct deletion of acm from −38 to +1677 (including the ribosome-binding site, signal sequence, complete A domain [binding domain], and 13 amino acids of the B domain) and is replaced by cat (Fig. 3B). Southern hybridizations using the deleted fragment of acm (amplified from TX0082 using primers AcmF2 and AcmR1 [Table 4]), the cat gene (amplified from pTEX5500ts using primers 5500-MCS-IF and 5500-MCS-IIR [Table 4]), and pTEX5500ts as probes confirmed the absence of unintegrated plasmid (Fig. 3C and D). Since in silico analysis as well as mRNA analysis of the acm locus (data not shown) confirmed the monocistronic nature of acm (and thus predicts absence of a polar effect with this system), we used a replacement strategy in this study to introduce the cat marker into the mutant to take advantage of the discriminatory capability; this will facilitate distinguishing between wild-type and mutant bacteria after in vivo animal experiments with mixed cultures.
FIG. 2.
Protocol used for replacing the acm wild-type sequence on the TX0082 chromosome with the cat gene. The gene replacement construct (pTEX6050) carrying in vitro-altered sequences (AcmUp, the region upstream of acm [shown by the cross-hatched box], and AcmDn, part of the acm 3′ region as well as the downstream region [shown by the dotted box]) was transformed into E. coli. At permissive temperature (28°C), pTEX6050 was introduced into TX0082 by electroporation. Colonies were screened for an integration event when the temperature was shifted to 42°C. One of the integrants was grown for eight serial overnight passages at 42°C to completely cure the plasmid. The culture from the eighth passage was serially diluted and plated at 37°C on nonselective media to select for double crossover recombination. Upon replica plating to chloramphenicol plates and gentamicin plates, the colonies that retained the cat gene only were identified. One of these colonies was designated TX6051 (TX0082Δacm). The gray box represents the cat gene coding for chloramphenicol resistance, and the striped box represents the aph(2′′)-Id gene coding for gentamicin resistance.
FIG. 3.
Confirmation of allelic replacement of the acm gene of TX6051. (A) PFGE analysis of SmaI-digested genomic DNA of TX0082 (lane 1), four double-crossover colonies that retained the chloramphenicol resistance (lanes 2 to 5), a colony resulting from a single-crossover integration event (TX0082AcmUp::pTEX6050) (lane 6), and a molecular weight marker (lane 7). Two fragments marked with straight arrows in lane 6 are due to a SmaI site in the inserted plasmid. The extrachromosomal (nonintegrated) plasmid band is marked with a bent arrow. (B) Illustration of a 4,474-bp Δacm region of E. faecium TX6051 (TX0082Δacm). Sequencing confirmed that 1,663 bp of the acm locus is replaced with a 971-bp cat cassette. The 5′ and 3′ regions that were used for recombination events are shown by cross-striped boxes. (C and D) Southern blot analysis of the digests shown in lanes 1 to 6 of panel A. Panel C was probed with the deleted fragment of acm, and panel D was probed with the cat gene. Hybridization results obtained with pTEX5500ts are identical to those obtained with its cat gene, as shown in panel D.
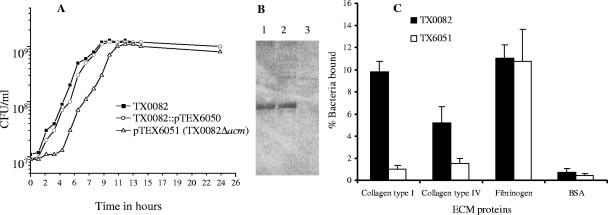
Characterization of the acm mutant. (i) Growth characteristics.
TX6051 (TX0082Δacm) colonies appeared smaller in size compared to the wild type (TX0082) when grown overnight either on BHI or TH agar. However, no abnormality in cell shape was noticed when the acm mutant was observed by light microscopy at various time points of growth. To further characterize the behavior of the acm mutant, growth was monitored by CFU and OD600. As shown in Fig. 4A, the doubling time of the acm mutant (TX6051) was longer than that of the wild type (TX0082) in the lag and early log phases. Also, the final culture density of the acm mutant in BHI was reproducibly two times lower than that of the wild type. The growth of a single-crossover construct (TX0082AcmUp::pTEX6050), which does not cause an interruption of acm, was similar to the wild type (Fig. 4A), suggesting that the presence of cat had no effect on the observed slow growth of acm mutant. Similar observations were made when growth was monitored by OD600. Southern hybridization results showed localization of a single copy of the cat gene to the acm locus without evidence of a second integration elsewhere in the genome of TX6051 (Fig. 3D).
FIG. 4.
Characterization of TX6051. (A) Growth curve of wild-type E. faecium strain TX0082, its isogenic acm deletion mutant (TX6051), and a colony resulting from a single-crossover integration event (TX0082AcmUp::pTEX6050). Aliquots of culture were withdrawn every hour through the growth cycle for the measurement of CFU. (B) Western blots of mutanolysis extracts of E. faecium isolates (lane 1, TX2535; lane 2, TX0082; and lane 3, TX6051). Samples were probed with polyclonal antiserum raised against recombinant AcmA (37). (C) Adherence of wild-type E. faecium TX0082 and its isogenic acm deletion mutant (TX6051) to immobilized collagen types I and IV, fibrinogen, and BSA. Adherence was tested in wells coated with 1 μg of extracellular matrix (ECM) proteins. Bars represent the mean percentages of cells bound ± standard deviations for eight wells. Results are from four independent experiments. BSA was used as a negative control.
(ii) Lack of Acm in surface preparations of TX6051 and lack of adherence of this acm mutant to collagen(s).
Western blots of cell wall-associated proteins released by mutanolysin were probed with anti-Acm polyclonal antibodies, which we previously raised against the A domain of Acm (37). A single immunoreactive protein of ∼86 kDa was detected in an endocarditis isolate TX2535 (37) and in TX0082 but not in TX6051 (Fig. 4B). Probing of mutanolysin extracts of these isolates with preimmune rabbit serum showed no reactive bands in all three isolates (data not shown).
E. faecium TX0082 and its isogenic Δacm strain TX6051 were tested for their ability to adhere to immobilized collagen types I and IV, fibrinogen, and BSA. Strain TX0082 adhered to collagen(s) and fibrinogen, whereas the acm mutant (TX6051) was completely defective in adhering to collagen types I and IV but not to fibrinogen (Fig. 4C). This genetic evidence corroborates our earlier paper indicating that Acm mediated collagen adherence of E. faecium based on results with recombinant Acm and complementation of acm-pseudogene containing E. faecium isolates (37). Using this acm deletion mutant, the role of acm in E. faecium pathogenesis will be tested in animal models including endocarditis.
Use of a mobilizable derivative of pTEX5500ts to interrupt the acm gene of a nonelectroporable E. faecium strain.
A mobilizable derivative of pTEX5500ts, designated pTEX5501ts was created by adding oriT from Tn916 in order to transfer it by conjugation from E. faecalis JH2Sm::Tn916 (6). To generate the plasmid for insertional inactivation, an 846-bp DNA fragment designated AcmIns, which is an intragenic region of acm, was amplified from TX2555 genomic DNA template using primers AcmInsF and Acm19R. The AcmIns PCR product was digested with BamHI and KpnI and ligated to a similarly digested 5.24-kb BamHI and KpnI gel-eluted fragment from pTEX5501ts, yielding pTEX6052 (pTEX5501ts::acm). This plasmid was electroporated into E. coli DH5α to obtain TX6052, and from there it was introduced into JH2Sm::Tn916 by electroporation, selecting for gentamicin resistance after growth at 28°C; a resulting colony was chosen and designated TX6053. Using the principle of trans-activation (Tn916 presence in the donor strain activates conjugal transfer of coresident plasmids carrying oriTTn916 [1, 22]), pTEX6052 from E. faecalis JH2Sm::Tn916 was mobilized to the nonelectroporable (in our hands) E. faecium strain TX2555 by broth mating. Transconjugants were selected for resistance to 125 μg/ml gentamicin and 200 μg/ml erythromycin (present in the recipient). PFGE analysis confirmed the strain identity of the transconjugant (data not shown). Selected gentamicin-resistant colonies were shown to stably maintain pTEX6052 after growth at 28°C. Subsequently, a culture of TX2555(pTEX6052) was shifted to 42°C (to cause plasmid loss), grown for 10 serial passages, serially diluted, and plated on gentamicin at 37°C. PFGE analysis followed by probing of Southern blots showed complete loss of detectable plasmid in some colonies (data not shown). This insertional mutant strain of acm was designated TX6054.
Growth patterns and adherence characteristics of wild-type TX2555 versus TX6054 (data not shown) were essentially the same as we observed with wild-type TX0082 versus TX6051. We also showed the stability of the acm insertion of TX6054, without evidence of excised plasmid DNA by Southern blots using pTEX6052 as a probe, after two overnight in vitro passages at 37°C and without antibiotic (data not shown). Our ability to disrupt the acm gene in a nontransformable E. faecium strain using mobilization and recombination events indicate that the same technique could be used for allelic replacement, as described above with pTEX5500ts and TX0082.
Conjugal transfer of a pTEX5501ts-derived plasmid from E. faecalis JH2Sm::Tn916 to diverse E. faecium clinical strains.
By broth mating, the transfer of pTEX6052 (pTEX5501ts::acm intragenic fragment) was successful only with the recipient strain TX2555 (1 × 10−8), and no transconjugants were seen with TX2535 and TX2466. Subsequent experiments with filter matings demonstrated the transferability of pTEX6052 into diverse E. faecium clinical strains of different PFGE types, and the transfer frequencies varied with different strains (Table 5). From every mating, randomly selected transconjugants were analyzed for plasmid content and the strain identity was confirmed by PFGE (data not shown). These results suggest that mobilization from an E. faecalis donor is a convenient method for introducing pTEX5501ts derivatives into diverse clinical strains of E. faecium and even into the strains that have been recalcitrant to standard electroporation.
TABLE 5.
Transferability of plasmid pTEX6052 (a pTEX5501ts derivative) from E. faecalis JH2Sm::Tn916 to diverse E. faecium clinical strains by trans-activation
| Recipient (source of isolation, place of origin) | Antibiotic selection (μg/ml) | Transfer frequencye |
|---|---|---|
| TX2442 (blood, Belgium)a | Gen, 125; Ery, 200 | 3 × 10−9 |
| TX2535 (endocarditis, Houston, Tex.)b,c,d | Gen, 125; Ery, 200 | 3.8 × 10−7 |
| TX2067 (clinical isolate, Boston, Mass.)c | Gen, 125; Ery, 200 | 2.0 × 10−7 |
| TX2492 (blood, Houston, Tex.)b,c,d | Gen, 125; Ery, 200 | 3.6 × 10−8 |
Vancomycin resistant, VanA type.
Vancomycin resistant, VanB type.
Ampicillin resistant.
Isolates TX2535 and TX2492 were from different hospitals.
Transfer frequencies are expressed as number of transconjugants per donor CFU.
Replication of pTEX5500ts in E. faecalis.
To assess the functionality of this plasmid in E. faecalis mutagenesis, transformation and stability of pTEX5500ts was also tested with E. faecalis OG1RF. This plasmid was transformed with better efficiency (420 CFU/μg of plasmid) into E. faecalis OG1RF than E. faecium. The permissive temperature as well as plasmid loss rates were similar to what we noted for E. faecium. Since pWV01-based repAts has been used for allelic replacement of several gram-positive bacteria, including lactococci (3), streptococci (41), staphylococci (17, 54), and Desulfitobacterium (53), these new vectors should be applicable for mutagenesis of a broad range of hosts.
In summary, to overcome the limitation of poor transformability of clinical E. faecium isolates, a ts replicon derived from the low-copy-number plasmid pWV01 was used to construct a thermosensitive vector which is functional in E. faecium at permissive temperature and suicidal at nonpermissive temperature. Using this vector, the acm gene of an endocarditis-derived E. faecium strain was successfully deleted, and this is the first example to our knowledge of allelic replacement in this species. More importantly, the transferability of mobilizable derivatives of this vector was demonstrated with nonelectroporable E. faecium clinical strains. These vectors will be particularly useful for genes that do not confer easily screenable phenotypes. The improved cloning vector properties of this system should also be helpful for mutagenesis of other gram-positive bacteria as well.
Acknowledgments
We thank M. Arthur for providing plasmid pHS1 before it was published. pFW14 was kindly provided by Andreas Podbielski, University of Rostock, Germany. We thank Karen Jacques-Palaz for her technical assistance.
This work was supported by NIH grants R01/R56 AI 42399 and R37 AI 47923 from the Division of Microbiology and Infectious Diseases to B. E. Murray.
REFERENCES
- 1.Arbeloa, A., H. Segal, J. E. Hugonnet, N. Josseaume, L. Dubost, J. P. Brouard, L. Gutmann, D. Mengin-Lecreulx, and M. Arthur. 2004. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J. Bacteriol. 186:1221-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bramucci, M. G., and V. Nagarajan. 1996. Direct selection of cloned DNA in Bacillus subtilis based on sucrose-induced lethality. Appl. Environ. Microbiol. 62:3948-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. D., and D. A. Morrison. 1987. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene 55:179-187. [DOI] [PubMed] [Google Scholar]
- 8.Coque, T. M., R. C. Arduino, and B. E. Murray. 1995. High-level resistance to aminoglycosides: comparison of community and nosocomial fecal isolates of enterococci. Clin. Infect. Dis. 20:1048-1051. [DOI] [PubMed] [Google Scholar]
- 9.Duh, R. W., K. V. Singh, K. Malathum, and B. E. Murray. 2001. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb. Drug Resist. 7:39-46. [DOI] [PubMed] [Google Scholar]
- 10.Eliopoulos, G. M., C. Wennersten, S. Zighelboim-Daum, E. Reiszner, D. Goldmann, and R. C. Moellering, Jr. 1988. High-level resistance to gentamicin in clinical isolates of Streptococcus (Enterococcus) faecium. Antimicrob. Agents Chemother. 32:1528-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frere, J., A. Benachour, J. C. Giard, J. M. Laplace, S. Flahaut, and Y. Auffray. 1998. A new theta-type thermosensitive replicon from Lactococcus lactis as an integration vector for Enterococcus faecalis. FEMS Microbiol. Lett. 161:107-114. [DOI] [PubMed] [Google Scholar]
- 13.Giard, J. C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock, L. E., and M. Perego. 2004. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J. Bacteriol. 186:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington, S. M., T. L. Ross, K. A. Gebo, and W. G. Merz. 2004. Vancomycin resistance, esp, and strain relatedness: a 1-year study of enterococcal bacteremia. J. Clin. Microbiol. 42:5895-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartford, O., L. O'Brien, K. Schofield, J. Wells, and T. J. Foster. 2001. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147:2545-2552. [DOI] [PubMed] [Google Scholar]
- 18.Hienz, S. A., T. Schennings, A. Heimdahl, and J. I. Flock. 1996. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J. Infect. Dis. 174:83-88. [DOI] [PubMed] [Google Scholar]
- 19.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwen, P. C., D. M. Kelly, J. Linder, S. H. Hinrichs, E. A. Dominguez, M. E. Rupp, and K. D. Patil. 1997. Change in prevalence and antibiotic resistance of Enterococcus species isolated from blood cultures over an 8-year period. Antimicrob. Agents Chemother. 41:494-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager, W., A. Schafer, A. Puhler, G. Labes, and W. Wohlleben. 1992. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 174:5462-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaworski, D. D., and D. B. Clewell. 1995. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J. Bacteriol. 177:6644-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson, A., J. I. Flock, and O. Svensson. 2001. Collagen and fibronectin binding in experimental staphylococcal osteomyelitis. Clin. Orthop. 2001:241-246. [DOI] [PubMed] [Google Scholar]
- 24.Kok, J., J. M. van der Vossen, and G. Venema. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, J. van Embden, and R. J. Willems. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 27.Leenhouts, K. J., B. Tolner, S. Bron, J. Kok, G. Venema, and J. F. Seegers. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55-66. [DOI] [PubMed] [Google Scholar]
- 28.Li, X., G. M. Weinstock, and B. E. Murray. 1995. Generation of auxotrophic mutants of Enterococcus faecalis. J. Bacteriol. 177:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manganelli, R., R. Provvedi, C. Berneri, M. R. Oggioni, and G. Pozzi. 1998. Insertion vectors for construction of recombinant conjugative transposons in Bacillus subtilis and Enterococcus faecalis. FEMS Microbiol. Lett. 168:259-268. [DOI] [PubMed] [Google Scholar]
- 31.Mann, P. A., L. Xiong, A. S. Mankin, A. S. Chau, C. A. Mendrick, D. J. Najarian, C. A. Cramer, D. Loebenberg, E. Coates, N. J. Murgolo, F. M. Aarestrup, R. V. Goering, T. A. Black, R. S. Hare, and P. M. McNicholas. 2001. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol. Microbiol. 41:1349-1356. [DOI] [PubMed] [Google Scholar]
- 32.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 34.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nallapareddy, S. R., K. V. Singh, R. W. Duh, G. M. Weinstock, and B. E. Murray. 2000. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. Infect. Immun. 68:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nallapareddy, S. R., G. M. Weinstock, and B. E. Murray. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733-1747. [DOI] [PubMed] [Google Scholar]
- 38.Patti, J. M., T. Bremell, D. Krajewska-Pietrasik, A. Abdelnour, A. Tarkowski, C. Ryden, and M. Hook. 1994. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immun. 62:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J. Bacteriol. 178:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 42.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 43.Ramirez-Arcos, S., M. Liao, S. Marthaler, M. Rigden, and J. A. Dillon. 2005. Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology 151:1381-1393. [DOI] [PubMed] [Google Scholar]
- 44.Rice, L. B., L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang, and B. E. Murray. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508-512. [DOI] [PubMed] [Google Scholar]
- 45.Rince, A., J. C. Giard, V. Pichereau, S. Flahaut, and Y. Auffray. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh, K. V., T. M. Coque, G. M. Weinstock, and B. E. Murray. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323-331. [DOI] [PubMed] [Google Scholar]
- 50.Singh, K. V., K. Malathum, and B. E. Murray. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob. Agents Chemother. 45:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 52.Skryabin, B., and D. Vassilacopoulou. 1993. A simple and fast method for cloning and analyzing polymerase chain reaction products. Genet. Anal. Tech. Appl. 10:113-115. [DOI] [PubMed] [Google Scholar]
- 53.Smidt, H., J. van der Oost, and W. M. de Vos. 2001. Development of a gene cloning and inactivation system for halorespiring Desulfitobacterium dehalogenans. Appl. Environ. Microbiol. 67:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Supersac, G., Y. Piemont, M. Kubina, G. Prevost, and T. J. Foster. 1998. Assessment of the role of gamma-toxin in experimental endophthalmitis using a hlg-deficient mutant of Staphylococcus aureus. Microb. Pathog. 24:241-251. [DOI] [PubMed] [Google Scholar]
- 55.Teng, F., M. Kawalec, G. M. Weinstock, W. Hryniewicz, and B. E. Murray. 2003. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 71:5033-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng, F., B. E. Murray, and G. M. Weinstock. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182-186. [DOI] [PubMed] [Google Scholar]
- 57.Tomita, H., C. Pierson, S. K. Lim, D. B. Clewell, and Y. Ike. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J. Clin. Microbiol. 40:3326-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treitman, A. N., P. R. Yarnold, J. Warren, and G. A. Noskin. 2005. Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J. Clin. Microbiol. 43:462-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai, S. F., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2′′)-Id, in Enterococcus spp. Antimicrob. Agents. Chemother. 42:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver, K. E., K. D. Walz, and M. S. Heine. 1998. Isolation of a derivative of Escherichia coli-Enterococcus faecalis shuttle vector pAM401 temperature sensitive for maintenance in E. faecalis and its use in evaluating the mechanism of pAD1 par-dependent plasmid stabilization. Plasmid 40:225-232. [DOI] [PubMed] [Google Scholar]
- 61.Werner, G., B. Hildebrandt, I. Klare, and W. Witte. 2000. Linkage of determinants for streptogramin A, macrolide-lincosamide-streptogramin B, and chloramphenicol resistance on a conjugative plasmid in Enterococcus faecium and dissemination of this cluster among streptogramin-resistant enterococci. Int. J. Med. Microbiol. 290:543-548. [DOI] [PubMed] [Google Scholar]
- 62.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4. 2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. M. David, J. G. Scidman, J. A. Smith, and K. Struhl (ed.), Current Protocols in Molecular Biology. Green Publishing Associates, Brooklyn, N.Y.
- 64.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woodford, N., D. Morrison, B. Cookson, and R. C. George. 1993. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob. Agents Chemother. 37:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youngman, P. J., J. B. Perkins, and R. Losick. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. USA 80:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]