Abstract
Mixed anaerobic microbial subcultures enriched from a multilayered aquifer at a former chlorinated solvent disposal facility in West Louisiana were examined to determine the organism(s) involved in the dechlorination of the toxic compounds 1,2-dichloroethane (1,2-DCA) and 1,1,2-trichloroethane (1,1,2-TCA) to ethene. Sequences phylogenetically related to Dehalobacter and Dehalococcoides, two genera of anaerobic bacteria that are known to respire with chlorinated ethenes, were detected through cloning of bacterial 16S rRNA genes. Denaturing gradient gel electrophoresis analysis of 16S rRNA gene fragments after starvation and subsequent reamendment of culture with 1,2-DCA showed that the Dehalobacter sp. grew during the dichloroelimination of 1,2-DCA to ethene, implicating this organism in degradation of 1,2-DCA in these cultures. Species-specific real-time quantitative PCR was further used to monitor proliferation of Dehalobacter and Dehalococcoides during the degradation of chlorinated ethanes and showed that in fact both microorganisms grew simultaneously during the degradation of 1,2-DCA. Conversely, Dehalobacter grew during the dichloroelimination of 1,1,2-TCA to vinyl chloride (VC) but not during the subsequent reductive dechlorination of VC to ethene, whereas Dehalococcoides grew only during the reductive dechlorination of VC but not during the dichloroelimination of 1,1,2-TCA. This demonstrated that in mixed cultures containing multiple dechlorinating microorganisms, these organisms can have either competitive or complementary dechlorination activities, depending on the chloro-organic substrate.
1,2-Dichloroethane (1,2-DCA) and 1,1,2-trichloroethane (1,1,2-TCA) are regulated chloro-organic compounds that have been used extensively in industrial processes, 1,2-DCA primarily as a feedstock for plastics production (3) and 1,1,2-TCA as a degreasing agent (2). Due to past storage and disposal practices, these compounds have entered into the environment, contaminating groundwater; 1,2-DCA was found in at least 570 of 1,585 National Priorities List (NPL) sites identified by the U.S. Environmental Protection Agency (3), whereas 1,1,2-TCA was found in at least 45 of the NPL sites (2). This raises concern because of the potential effects of these compounds on the nervous system, the liver, the kidneys, and the lungs (2, 3). Fortunately, it has been observed that both compounds are susceptible to biodegradation, under aerobic (5, 23, 28, 32, 47), cometabolic (6, 11, 12, 17, 20, 24, 30, 48), and anaerobic (9, 10, 18, 33, 36, 38, 44, 52) conditions. Therefore, bioremediation is potentially a viable option for removing these compounds from contaminated sites.
Anaerobic processes are attractive for in situ bioremediation because oxygen does not need to be introduced into the subsurface. Although the anaerobic degradation of chlorinated ethenes (e.g., tetrachloroethene [PCE] and trichloroethene [TCE]) has been intensely studied over the last decade, less work regarding the degradation of chlorinated ethanes has been reported, despite their widespread presence in the environment. Nonetheless, investigations into the organisms involved in the anaerobic removal of 1,2-DCA and 1,1,2-TCA have begun to reveal bacterial populations that can dechlorinate these compounds through a respiratory process, whereby dechlorination is linked to growth. Dehalococcoides sp. strains 195 (39) and BAV1 (25) and Desulfitobacterium dichloroeliminans strain DCA1 (9) can grow during the dichloroelimination of 1,2-DCA, whereas only strain DCA1 has been shown to grow through the dichloroelimination of 1,1,2-TCA (9). Since many other bacteria can respire with various chlorinated compounds (see, for example, references 1, 4, 13, 21, 29, 37, 40, 49-51, and 56), it is likely that there exist other organisms that can respire with 1,2-DCA and 1,1,2-TCA. However, their identification may be limited by culturing and isolation challenges. A means to overcome these challenges is to use culture-independent molecular techniques to study the organisms involved in dechlorination.
In the present study, we examined a set of previously uncharacterized anaerobic enrichment cultures (7), with the purpose of determining the microorganisms implicated in the biodegradation of 1,2-DCA and 1,1,2-TCA. These enrichment cultures were derived from a former chlorinated solvents disposal facility that was contaminated with large amounts of 1,2-DCA and 1,1,2-TCA. 16S rRNA gene cloning, denaturing gradient gel electrophoresis (DGGE), and real-time quantitative PCR (qPCR) were used to delineate the roles of two putative dechlorinating organisms: a Dehalobacter-like species and a Dehalococcoides-like species. Differences in dechlorination-dependent growth on 1,2-DCA and 1,1,2-TCA were demonstrated for these two species. This report adds to the understanding of the organisms involved in the degradation of chlorinated ethanes, an understanding that will be required for the successful application of bioremediation for removing 1,2-DCA and 1,1,2-TCA from the environment.
MATERIALS AND METHODS
Establishment of enrichment cultures and subcultures.
Anaerobic microcosms were constructed in 1999 with groundwater and solids from a multilayered aquifer at a former chlorinated solvent disposal facility in West Louisiana (WL), USA (7). This site was contaminated with 1,2-DCA and 1,1,2-TCA at high concentrations (>0.30 mM), as well as various other chlorinated ethenes and methanes. Aquifer material and groundwater were anaerobically transferred to sterile bottles, purged with N2/CO2 (80%/20%) to remove volatile organic compounds, and then amended with an electron donor mixture of methanol, ethanol, acetate, and lactate and 1,2-DCA and 1,1,2-TCA as electron acceptors. Microcosms and subsequent cultures were maintained statically in an anaerobic chamber (Coy Laboratory Products, Madison, WI) filled with a CO2/H2/N2 (10%/10%/80%) gas mix. After dechlorination activity was observed, transfer cultures were prepared in defined anaerobic mineral medium containing trace minerals and vitamins (16). These transfer cultures were amended with either 1,2-DCA only (WL/1,2-DCA,), 1,1,2-TCA only (WL/1,1,2-TCA), or both (WL/Mix) at initial aqueous concentrations ranging from 0.08 to 0.20 mM. Ethanol was provided to these cultures as electron donor at concentrations ranging from 0.13 to 0.67 mM, representing approximately 10 times the electron equivalents required for dechlorination, assuming two electron equivalents of donor are required per dechlorination step and that ethanol provides 12 electron equivalents per mole. When dechlorination ceased, 20 to 50% of the culture was replaced with fresh mineral medium. These subcultures have been maintained in butyl rubber-stoppered 2-liter bottle batch culture since 2002.
Substrate range study.
The ability of the three WL subcultures to degrade selected chlorinated ethanes and ethenes over a 6-week period was tested. A series of duplicate screw-top vials (45 ml) with Mininert septa (VICI Precision Sampling, Baton Rouge, LA) were filled with 10 ml of mineral medium and 10 ml of appropriate WL subculture. Vials were amended with appropriate chlorinated electron acceptor at an aqueous concentration of 0.06 to 0.10 mM and ethanol as electron donor (0.45 mM). Tested chlorinated compounds included: 1,2-DCA, 1,1,2-TCA, 1,1,1-trichloroethane (1,1,1-TCA), tetrachloroethene (PCE), trichloroethene (TCE), cis-dichloroethene (cDCE), and vinyl chloride (VC). Uninoculated controls were prepared for each electron acceptor.
16S rRNA gene cloning and PCR-DGGE.
Bacterial 16S rRNA genes were cloned from WL subcultures to determine the present community members. The UltraClean Soil DNA Kit (Mo Bio Laboratories, Inc., Solana Beach, CA) was used to extract total genomic DNA from pelleted culture according to the manufacturer's alternative protocol for maximum yields. Bacterial 16S rRNA genes were selectively amplified from the purified DNA by PCR using the forward primer 27f and the reverse primer 1492R (55). PCR was performed in triplicate 50-μl reactions containing 1× ThermoPol PCR buffer (New England Biolabs, Mississauga, Ontario, Canada), 0.5 μM concentrations of each primer, 0.5 mM deoxynucleoside triphosphates, 1.5 U of Taq DNA polymerase (New Englands Biolabs), and 50 ng of DNA. The conditions used for PCR amplification were as follows: initial denaturation at 94°C for 5 min and then 25 cycles of (denaturation 94°C for 30 s, primer annealing at 52°C for 30 s, and chain extension for 1 min at 72°C), followed by a final extension at 72°C for 10 min. A PTC-200 thermocycler (MJ Research, Inc., Waltham, MA) was used for PCR. Triplicate reactions were combined and cloned with the TOPO TA cloning kit (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's protocol. Positive clones (42 clones) were sequenced by the University Health Network Research DNA Sequencing Facility (Toronto, Ontario, Canada) with the primer 27f, and then the sequence closest match was identified with the blastn utility of GenBank.
PCR for DGGE of amplified bacterial 16S rRNA gene fragment was performed according to the protocol described by Muyzer et al. (43), except that 30 PCR cycles were carried out per reaction, and each subculture DNA template was amplified in triplicate reactions. The same PCR conditions were used to amplify the internal fragment of cloned 16S rRNA genes (from cloning study above) to putatively identify bands in the DGGE gel. Triplicate reactions were pooled and amplicons were separated by DGGE as previously described (15), except that the gradient ranged from 30 to 60% denaturant.
Starvation and reamendment experiment.
WL/1,2-DCA subculture was centrifuged for 30 min at 4°C at 8,100 × g in an Avanti-J-20 XP centrifuge (Beckman-Coulter Canada, Mississauga, Ontario, Canada) equipped with a JCA-8.1000 rotor. The supernatant was anaerobically decanted, and the pellet was resuspended in fresh mineral medium. Aliquots of this suspension were transferred to a sterile, anaerobic media bottle (1 liter). The suspension was diluted with additional mineral medium, and the bottle was sealed. This was purged for 1 h with N2 and CO2 to remove residual H2 from the anaerobic chamber atmosphere and stored in the dark in the chamber. After 170 days without amendment with carbon source, electron donor, or electron acceptor, 66-ml aliquots were transferred to each of three screw-top bottles (250 ml), which were purged for 30 min with N2 and CO2. Two bottles were amended with neat 1,2-DCA (initial aqueous concentration 0.3 mM), whereas the third bottle was left unamended (no electron acceptor control). After a 4-h equilibration period, sodium acetate (from a sterile aqueous stock, to an initial concentration of 5 mM) and 1.5 kPa of H2 and CO2 (80 and 20%, respectively) were added to all bottles. After the 1,2-DCA was ∼80% degraded in the 1,2-DCA-amended bottles (285 h after 1,2-DCA amendment), 10 ml of culture was removed from each bottle, centrifuged for 40 min at 2000 × g, and the DNA was extracted from the pellet with UltraClean Soil DNA Kit. The bacterial community was then analyzed by PCR-DGGE.
Time course experiments.
Growth of putative dechlorinating microorganisms during degradation of 1,1,2-TCA was monitored. This experiment was conducted twice (October 2004 and January 2005). Screw-top bottles (250 ml) with Mininert septa were filled with 150 ml of mineral medium. Replicate bottles were amended with neat 1,1,2-TCA (initial aqueous concentration of 0.3 mM) and ethanol (initial aqueous concentration of 0.26 mM). Other bottles were amended with ethanol only, as a no-electron-acceptor control. WL/1,1,2-TCA culture was added at a 1 or 0.1% (vol/vol) inoculum. At this time point (T = 0), 50 ml of culture was removed for DNA extraction. DNA was subsequently extracted from 50-ml samples from all bottles when all of the 1,1,2-TCA was degraded to VC (time point T = 1) in the 1,1,2-TCA-amended bottles, when the VC was ∼75% degraded (time point T = 2), and when the VC was fully degraded (time point T = 3). For the October 2004 experiment, DNA was only extracted at the start of the experiment (T = 0), when 1,1,2-TCA had degraded to VC (T = 1), and when the VC was completely degraded (T = 3). All bottles were reamended twice with ethanol during VC degradation to prevent electron donor shortage.
Similar time course experiments were prepared for the degradation of 1,2-DCA in January and April 2005, except that bottles initially contained 200 ml of mineral medium, replicates were amended with neat 1,2-DCA (initial aqueous concentration of 0.4 mM) and ethanol (initial aqueous concentration of 0.85 mM), and WL/1,1,2-TCA was added at an 0.5% (vol/vol) inoculum. Ethanol-only controls were also used. DNA was extracted at the beginning of the experiment (T = 0), at two points during degradation (T = 1 and T = 2), and after degradation of 1,2-DCA was complete (T = 3). If degradation stalled, ethanol was added to all treatments.
For DNA extractions, culture was transferred to conical centrifuge tubes (50 ml) (Fisher Scientific, Toronto, Ontario, Canada) and centrifuged at 2,300 × g for 50 min at 4°C. The pellet was collected and DNA extracted with the UltraClean Soil DNA Kit according to the manufacturer's alternative protocol, except that the DNA was finally eluted with 5 mM Tris-HCl (pH 8.0). The copies of Dehalobacter and Dehalococcoides spp. 16S rRNA genes in the extracted DNA were analyzed by qPCR (see below).
The growth yield of each organism was determined by assuming a near-100% DNA extraction efficiency (14). Yield was calculated by first determining how many moles of the chlorinated compound were degraded between two time points considering both liquid and headspace in the bottle and taking into account mass removed with DNA extraction. Changes in 16S rRNA gene copy number were calculated for the same time periods. A yield of gene copies per mole of compound degraded was then determined.
qPCR.
qPCR for enumerating copies of Dehalobacter sp. and Dehalococcoides sp. 16S rRNA genes in extracted DNA was conducted with an Opticon 2 (MJ Research) thermocycler and SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich Co., St. Louis, MO). Each 30-μl reaction contained 15 μl of SYBR Green JumpStart Taq ReadyMix, 11.8 μl of sterile water, 2 μl of DNA template, and each forward and reverse primer at 0.5 μM. For Dehalococcoides-specific qPCR, the primers 1F and 264R (27) were used, and the thermocycling program was as follows: initial denaturation for 10 min at 94°C; 45 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 30 s; and a final melting curve analysis from 72 to 95°C, measuring fluorescence every 0.5°C. Primers specific to the Dehalobacter sequence identified in the culture were designed by aligning this sequence with Dehalobacter 16S rRNA gene sequences from the GenBank database. For Dehalobacter-specific qPCR, similar conditions were used, except the primers used were DHB477f (5′-GATTGACGGTACCTAACGAGG-3′) and DHB647r (5′-TACAGTTTCCAATGCTTTACGG-3′), and the annealing temperature was 63°C. Calibration was performed with serial dilutions of a known quantity of one of either Dehalococcoides or Dehalobacter 16S rRNA gene-containing plasmids generated in the cloning study described above. The detectable range for qPCR for both targeted 16S rRNA genes was 4 × 103 to 4 × 108 16S rRNA gene copies/reaction. DNA concentrations were determined with UV absorbance or with Picogreen (Molecular Probes, Eugene, OR). Differences between 16S rRNA gene copies/ml at different sampling time points were compared with a one-tailed Student t test. Significant differences had a P value of <0.05.
Analytical procedures.
For culture maintenance and time course experiments, chlorinated ethanes, ethenes, methane, and ethene were measured by injecting a 300-μl headspace sample onto a Hewlett-Packard 5890 Series II gas chromatograph fitted with a GSQ column (30-m-by-0.53-mm [inner diameter] PLOT column; J&W Scientific, Folsom, CA) and a flame ionization detector as described in Duhamel et al. (14), except that to resolve chlorinated ethanes the oven temperature was programmed to hold at 50°C for 90 s and then to increase to 180°C at 60°C/min with a final hold at 180°C for 5 min.
For substrate range experiments, chlorinated ethanes, ethenes, methane, and ethene were analyzed in a 1-ml liquid sample that was mixed with 5 ml of acidified water in a headspace vial (10 ml) and crimp-sealed with a Teflon-coated silicone septum (Agilent, Mississauga, Ontario, Canada). Samples were analyzed with an HP 7694 headspace sampler (Hewlett-Packard, Mississauga, Ontario, Canada) connected to a HP 5890A gas chromatograph (Hewlett-Packard) fitted with the same GSQ column and a flame ionization detector. Headspace sampler settings were as follows: oven temperature at 70°C, loop temperature at 80°C, transfer line at 90°C, gas chromatograph cycle time of 35 min, vial equilibration time of 45 min, pressurization time of 0 min, loop fill time of 0.2 min, loop equilibration of 0 min, injection time of 3 min, vial pressure at 17.3 lb/in2, and carrier pressure at 9.4 lb/in2. The gas chromatograph oven temperature was programmed to hold at 35°C for 2 min, then to increase to 100°C at 10°C/min, then to increase to 185°C at 6°C/min, and finally to hold at 185°C for 1.34 min. Calibration of 1,2-DCA, 1,1,2-TCA, 1,1,1-TCA, 1,1-dichloroethane (1,1-DCA), monochloroethane (CA), cDCE, TCE, and PCE was performed with aqueous external standards prepared gravimetrically from neat or methanolic stock solutions. VC was added to these external standards via a gastight syringe. Ethene and methane were calibrated with a 1% gas mixture (Scotty II; Alltech Associates, Inc.).
Accession numbers.
The cloned Dehalobacter and Dehalococcoides 16S rRNA gene sequences identified in these cultures were deposited in GenBank with the following accession numbers: Dehalobacter sp. strain WL, DQ250129; and Dehalococcoides sp. strain WL, AY882434.
RESULTS
Subculture degradation characteristics and community analysis.
Three methanogenic WL subcultures have been maintained for >2.5 yrs with the addition of only the chlorinated ethane(s) and ethanol. In the WL/1,2-DCA subculture 1,2-DCA is transformed to ethene via dichloroelimination, whereas in the WL/1,1,2-TCA subculture 1,1,2-TCA is first transformed to VC via dichloroelimination, and then VC is reductively dechlorinated to ethene. In the WL/Mix subculture, 1,2-DCA and 1,1,2-TCA are simultaneously transformed via dichloroelimination to ethene and VC, respectively. In mid-2003 this subculture lost the ability to degrade VC; thus, VC accumulates in these bottles in molar amounts equivalent to the 1,1,2-TCA added.
Substrate range studies were performed on these three subcultures to test their ability to degrade select chlorinated ethanes and ethenes. WL/1,2-DCA and WL/1,1,2-TCA performed similarly, degrading 1,2-DCA, 1,1,2-TCA, PCE, TCE, cDCE, and VC completely to ethene at the tested concentrations. In contrast, the WL/Mix subculture could dichloroeliminate 1,2-DCA to ethene and 1,1,2-TCA to VC but otherwise could only reductively dechlorinate TCE to cDCE. This culture did not dechlorinate PCE, cDCE, or VC. None of the three subcultures could degrade 1,1,1-TCA at the tested concentration during the observed time period.
To identify possible organisms directly involved in dechlorination through a dehalorespiratory process, bacterial 16S rRNA gene fragments were cloned for WL/1,2-DCA and WL/1,1,2-TCA with general bacterial primers. Only bacterial rRNA genes were targeted because our interest was to investigate organisms that degrade the chlorinated compounds through dehalorespiration, which is energy yielding and growth supporting. To date, no archaean has been shown to respire with chlorinated compounds, although cometabolic dechlorination has been observed. Therefore, it is possible that archaea are present in the culture and are contributing to dechlorination. Closest clone matches included: Clostridium (5 of 42 clones), Dehalobacter (25 of 42 clones), Dehalococcoides (4 of 42 clones), Spirochaeta (3 of 42 clones), Sedimentibacter (1 of 42 clones), Sporomusa (3 of 42 clones), and Syntrophomonas (1 of 42 clones). Strains of the genera Dehalobacter (29, 49, 56) and Dehalococcoides (8, 25, 26, 39) have previously been reported to reductively dechlorinate selected chlorinated ethanes and ethenes through a dehalorespiratory process. The Dehalobacter and Dehalococcoides sequences, in addition to being phylogenetically related to known dechlorinators (1,462/1,465-bp identity to Dehalobacter restrictus and 1,333/1,333-bp identity to D. ethenogenes strain 195, respectively), were among the most abundant in the clone libraries and were the brightest bands in DGGE analysis; therefore, subsequent analysis focused on these two phylotypes in the cultures.
A comparison of the bacterial community was performed for the three subcultures with PCR-DGGE using bacterial 16S rRNA gene-specific primers. The DGGE banding patterns differed between the subcultures (Fig. 1). Of note, WL/1,1,2-TCA had both strong Dehalococcoides and Dehalobacter bands, whereas WL/1,2-DCA had a strong Dehalococcoides band but only a very weak Dehalobacter band, and WL/Mix had a strong Dehalobacter band but no detectable Dehalococcoides band.
FIG. 1.
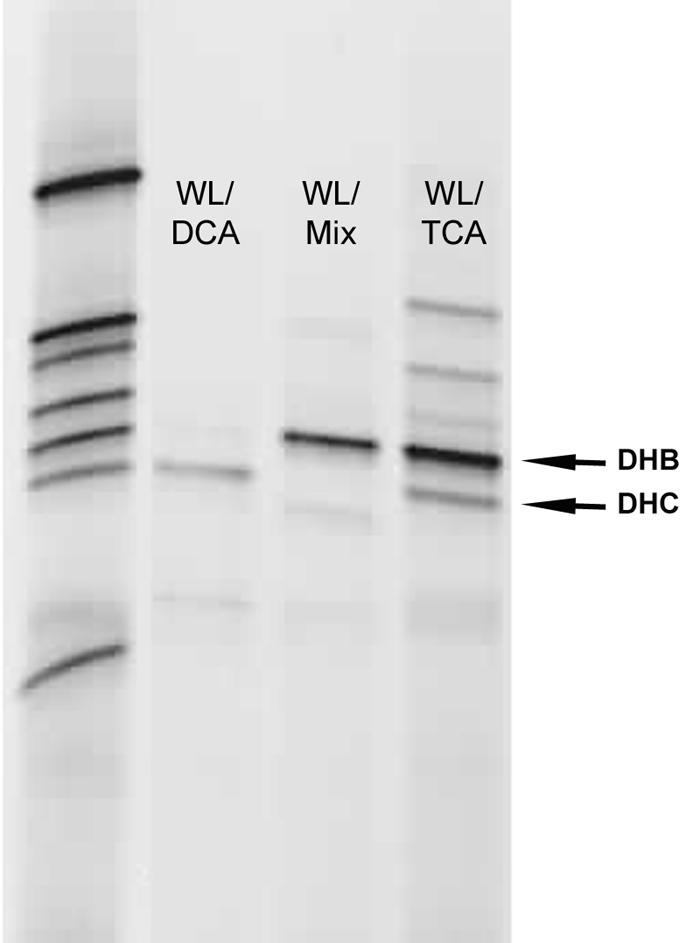
Bacterial 16S rRNA gene PCR-DGGE of three WL subcultures. Dehalobacter (DHB) and Dehalococcoides (DHC) bands are indicated. The left lane is a reference ladder composed of bands amplified from a mixture of 16S rRNA gene clones derived from the WL subcultures. The image is a negative of a 1% ethidium bromide-stained DGGE gel.
Species-specific qPCR was used to further explore community composition differences revealed by DGGE. Total genomic DNA extracted from each culture was assayed for the presence of Dehalobacter and Dehalococcoides 16S rRNA gene copies, normalized to total DNA extracted from the culture sample. WL/1,2-DCA contained more Dehalococcoides than Dehalobacter (Table 1), but in WL/1,1,2-TCA these values were very similar. Most notably, Dehalococcoides 16S rRNA gene copies were 2 orders of magnitude lower than Dehalobacter in the WL/Mix subculture. Given that it is thought that only organisms that make up >1% of the total community can be detected by DGGE (43), the qPCR results agreed well with the presence or absence of Dehalobacter and Dehalococcoides bands in Fig. 1.
TABLE 1.
Proportions of Dehalobacter and Dehalococcoides in WL subculture DNAa
| Subculture | 16S rRNA gene copies/ng of DNA
|
|
|---|---|---|
| Dehalobacter | Dehalococcoides | |
| WL/1,2-DCA | 4.5 (± 0.3) × 105 | 3.0 (± 0.1) × 106 |
| WL/1,1,2-TCA | 3.2 (± 0.2) × 106 | 2.6 (± 0.1) × 106 |
| WL/Mix | 2.6 (± 0.1) × 106 | 3.0 (± 0.1) × 104 |
Error (in parentheses) is the range of duplicate qPCR reactions.
DGGE evidence for 1,2-DCA-dependent growth.
The presence of two putative dechlorinating organisms in WL subcultures raised the question of whether both were directly involved in degradation of the chlorinated ethanes on which the cultures had been enriched. To examine this issue, WL/1,2-DCA culture was first starved (denied exogenous electron donor or acceptor) for >5 months. Aliquots of this starved culture were then amended with either just H2-CO2 or both H2-CO2 and 1,2-DCA, so that organisms involved in dechlorination could be distinguished from those growing due to nondechlorination processes (e.g., acetogenesis). In the 1,2-DCA-amended bottles, dechlorination to ethene commenced within 4 h of amendment (data not shown) despite the starvation period and was 80% complete in 12 days.
DGGE analysis (Fig. 2) revealed that the banding pattern after the culture was extensively starved was similar to that of the parent WL/1,2-DCA subculture (Fig. 1). Despite no carbon source or electron donor or acceptor, the Dehalococcoides band persisted. Upon amendment with H2 and CO2 in one treatment bottle, an unknown band (band 1) appeared that was present in the parent subculture; this was probably an organism that can utilize H2 directly, perhaps for acetogenesis, since there was no competition for H2 by reductive dechlorination processes. With 1,2-DCA amendment, however, a unique band appeared: that of Dehalobacter. This indicated that this organism benefited from the presence of 1,2-DCA and thus was likely involved in dechlorination. qPCR for Dehalococcoides and Dehalobacter during starvation and reamendment with 1,2-DCA agreed with the DGGE data: during starvation, Dehalococcoides and Dehalobacter 16S rRNA gene copies decreased 3- and 108-fold, respectively; during reamendment, Dehalococcoides and Dehalobacter 16S rRNA gene copies increased 3- and 150-fold, respectively (data not shown).
FIG. 2.
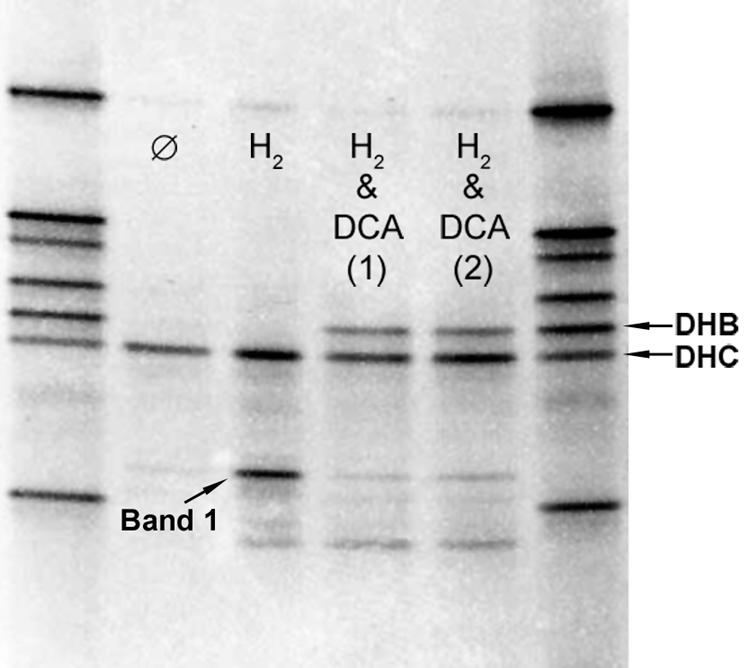
PCR-DGGE of starved and reamended WL/1,2-DCA culture. Dehalobacter (DHB) and Dehalococcoides (DHC) bands are indicated. Lane ⊘, nonamended culture starved for 99 days; lane H2, bottle amended with H2 and CO2 only (no electron acceptor); lanes “H2 & 1,2-DCA (1)” and “H2 & 1,2-DCA (2)”, bottles amended with H2, CO2, and 1,2-DCA; left and right lanes, clone reference ladder. Image is a negative of a 1% ethidium bromide-stained DGGE gel.
1,1,2-TCA and 1,2-DCA degradation time course experiments.
Given the indication by DGGE that Dehalobacter was involved in 1,2-DCA degradation, a more quantitative demonstration of this activity, as well as an explanation of the role of Dehalococcoides in the WL subcultures, was sought. For this purpose, time course experiments, in which the Dehalobacter and Dehalococcoides populations could be monitored during degradation, were performed. For consistency, only the WL/1,1,2-TCA culture was used here. Treatments amended with either just electron donor (ethanol) or electron donor and electron acceptor were prepared. DNA was extracted from both treatments at the beginning of the experiment and at several points during degradation of the chlorinated compounds. The extracted DNA was then analyzed for the presence of Dehalobacter and Dehalococcoides 16S rRNA genes by qPCR. No degradation of the tested chlorinated compounds was observed in uninoculated controls.
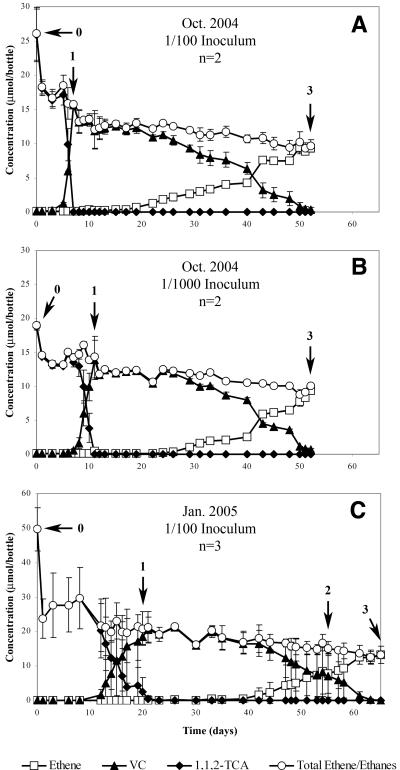
1,1,2-TCA-degradation time course experiments were performed twice over 4 months, with two different inoculum dilutions of WL/1,1,2-TCA tested. The 1,1,2-TCA degradation curves for both the October 2004 (1/100 and 1/1,000 inocula) and January 2005 (1/100 inoculum) experiments are shown in Fig. 3. After a period of several days, 1,1,2-TCA was quickly and completely dichloroeliminated to VC. At this point DNA was extracted from an aliquot from all treatments. VC degradation proceeded very slowly, with a lag of 6 to 11 days between completion of 1,1,2-TCA dichloroelimination and the start of VC reductive dechlorination. Complete conversion of VC to ethene took 25 to 40 days. Methanogenesis was absent during 1,1,2-TCA conversion but generally commenced several days prior to the start of VC degradation. The large drop in mass after T = 0 is due to the removal of 50 ml of culture (one-third of total culture volume) for DNA extraction. The variation in compound concentrations in Fig. 3C was an artifact of the asynchronous degradation of 1,1,2-TCA and VC in the three biological replicate bottles during that experiment.
FIG. 3.
Degradation profiles for 1,1,2-TCA timecourse experiments. (A) Degradation during the October 2004 experiment with a 1/100 inoculum of WL/1,1,2-TCA. (B) Degradation during the October 2004 experiment with a 1/1,000 inoculum. (C) Degradation during January 2005 experiment with a 1/100 inoculum of WL/1,1,2-TCA. Numbers indicate the approximate time points for DNA extraction in the respective experiments. Each curve shows the mean values of replicates. Error bars indicate the range of duplicates (A and B) or the standard deviation of three replicates (C).
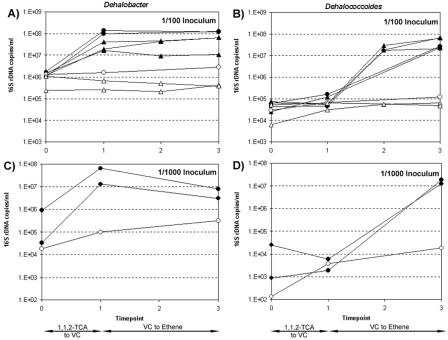
The change in concentration of Dehalobacter and Dehalococcoides 16S rRNA genes during the conversion of 1,1,2-TCA to VC and then to ethene for individual experimental bottles is shown in Fig. 4. During the initial dichloroelimination of 1,1,2-TCA to VC, Dehalobacter 16S rRNA gene copies increased by over an order of magnitude, whereas Dehalococcoides did not change. During subsequent VC dechlorination there was no significant change in the Dehalobacter population, whereas Dehalococcoides increased by over 2 orders of magnitude. After 1,1,2-TCA was fully degraded, Dehalobacter reached a concentration of 107 to 108 16S rRNA gene copies/ml; after VC was fully degraded, Dehalococcoides consistently reached a concentration of 107 16S rRNA gene copies/ml. Relative to the 1,1,2-TCA-amended bottles, no significant growth of either organism was observed in the bottles amended with ethanol only, verifying that indeed a chlorinated compound is required by both organisms as an electron acceptor for growth.
FIG. 4.
Dehalobacter and Dehalococcoides growth in individual bottles during 1,1,2-TCA degradation. (A and C) Dehalobacter; (B and D) Dehalococcoides. Circles represent bottles from the October 2004 experiments, and triangles represent bottles from the January 2005 experiments. Open symbols represent controls (amended with ethanol only); closed symbols represent bottles amended with ethanol and 1,1,2-TCA. On the x axis four time points are shown for consistency, although T = 2 values were determined for January 2005 replicates only. 16S rRNA gene copy values for each bottle are averages of duplicate qPCR reactions and are expressed as copies per ml of culture, assuming 100% DNA extraction efficiency.
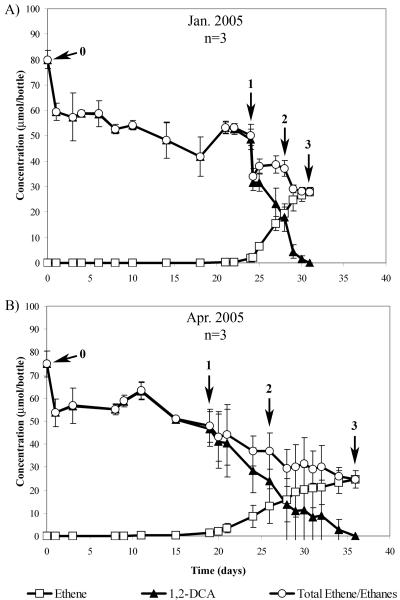
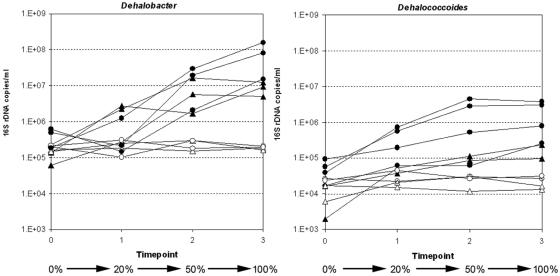
In the 1,2-DCA time course experiments, there was a substantial delay (∼18 days) before dichloroelimination commenced (Fig. 5). It should be noted that, prior to these experiments, the WL/1,1,2-TCA subculture used for inoculation had not been exposed to 1,2-DCA since the subculture was established 2.5 years ago. Once degradation commenced, however, it proceeded completely, with 0.4 mM 1,2-DCA being converted to ethene within 20 days. In addition, methanogenesis was absent throughout 1,2-DCA conversion, and no CA was detected. The variation in compound concentrations in Fig. 5B was an artifact of the asynchronous degradation of 1,2-DCA in the three replicate bottles during that experiment. During dechlorination, both Dehalobacter and Dehalococcoides 16S rRNA gene copies increased 1 to 2 orders of magnitude, although there was considerable variability between replicate bottles (Fig. 6). As in the 1,1,2-TCA degradation experiments, no significant growth of either organism was observed in bottles amended with only ethanol.
FIG. 5.
Degradation profile for 1,2-DCA time course experiments. (A) Degradation during the January 2004 experiment. (B) Degradation during the March 2005 experiment. Numbers indicate the approximate time point for DNA extraction in the respective experiments. Each curve shows the mean values of replicates. Error bars represent standard deviations.
FIG. 6.
Dehalobacter and Dehalococcoides growth in individual bottles during 1,2-DCA degradation. Triangles represent bottles from the January 2005 experiments, while circles represent bottles from the March 2005 experiment. Open symbols represent controls (amended with ethanol only); closed symbols represent bottles amended with ethanol and 1,2-DCA. 16S rRNA gene copy values for each bottle are averages of duplicate qPCR reactions and are expressed as copies per milliliter of culture assuming a 100% DNA extraction efficiency. Percentages represent the approximate proportion of 1,2-DCA (concentration) degraded at each time point.
Given the lack of growth of Dehalococcoides during the dichloroelimination of 1,1,2-TCA and the lack of growth of Dehalobacter during VC reductive dechlorination, yields on each compound could be determined by calculating the actual amount of dechlorination that occurred between the extraction time points, taking into account substrate mass loss due to culture volume removed for DNA extraction. The Dehalobacter yield was 3.07 (±2.88) × 108 16S rRNA gene copies/μmol 1,1,2-TCA degraded (n = 7), while the Dehalococcoides yield was 8.68 (±2.62) × 107 16S rRNA gene copies/μmol VC degraded (n = 7). The latter value agrees closely with the yield reported by He et al. (25) for strain BAV1, but it is 50% lower than that reported for strains VS (8) and KB-1/VC (14). Due to the simultaneous growth of Dehalobacter and Dehalococcoides during 1,2-DCA degradation, it was not possible to determine individual yields on 1,2-DCA, since it was not known how much 1,2-DCA was transformed by each organism. However, based on the overall yield of the two putative dechlorinating organism, it was determined that Dehalobacter accounted for >95% of 16S rRNA gene copy yield.
DISCUSSION
For the last decade the study of the stepwise degradation of chlorinated ethanes and ethenes has concentrated on isolates, with particular focus on members of the genus Dehalococcoides that are capable of degrading chlorinated ethenes. Dehalococcoides sp. strains 195 (39), BAV1 (25), FL2 (26), VS (8), and KB-1/VC (14) can all grow through the stepwise reductive dechlorination of PCE or its by-products, but none can fully degrade PCE to ethene. Therefore, in mixed cultures that can fully degrade PCE to ethene, two or more organisms are likely involved. Isolates of the genera Dehalobacter (29, 56), Desulfitobacterium (13, 21, 40, 51), Desulfuromonas (50), and Sulfospirillum (37) can grow through the dechlorination of PCE or TCE to cDCE, but dechlorination does not proceed further. Here it has been found that in the WL cultures dechlorination of 1,1,2-TCA proceeds through an analogous situation: 1,1,2-TCA was initially transformed to VC by a Dehalobacter sp., but VC was only degraded by the Dehalococcoides sp., as each organism only grew significantly during the respective dechlorination steps. To our knowledge, this is the first demonstration by molecular techniques that the complementary action of two dechlorinating organisms is required for the complete detoxification of a chlorinated ethane or ethene. Conversely, the growth of both Dehalobacter and Dehalococcoides during the degradation of 1,2-DCA points to a very different situation, where two organisms are competing for the same growth substrate. It is interesting that after years of enrichment two “competing” strains are both abundant, suggesting slightly different niches exist for each and not direct competition. One possible difference between these organisms is their respective decay rates. Upon starvation, Dehalobacter numbers decreased more significantly than did Dehalococcoides, indicating that Dehalococcoides may be more resistant to periods of adverse growth conditions. Alternatively, the two organisms may have optimal growth rates at different concentrations, i.e., one grows faster at a high substrate concentration and the other at a low substrate concentration.
Although 16S rRNA gene PCR-DGGE is often used to explore the spatial and temporal variation in microbial populations, complex samples can make it difficult to draw clear conclusions from these analyses. Enriched cultures, with fewer microbial species, allow for more manageable analyses, and thus true differences in populations can be more easily observed. Extensively starving the enrichment cultures helps to decrease the background organisms, such that, upon amendment of the targeted compound, only organisms directly benefiting from the compound should grow initially and thus be detectable as a band difference by DGGE. A limitation of this method is that if an organism can persist despite starvation, its growth upon reamendment will not be detectable through the differential appearance of a band, as was seen here for Dehalococcoides (Fig. 2). An alternative may be to use reverse-transcribed RNA as a template for PCR-DGGE instead of DNA, with the assumption that metabolically active organisms will have more ribosomes than nonactive ones (19). Other authors have found various results with this method (22, 35, 41); preliminary studies in our lab have shown no distinct differences between using DNA or RNA as a template for PCR-DGGE.
Although DGGE was useful for demonstrating the dechlorination-linked growth of Dehalobacter in the WL/1,2-DCA culture, qPCR gave a much clearer result, since it revealed that both the Dehalobacter and the Dehalococcoides spp. grew during 1,2-DCA degradation and that growth of both organisms was dependent on the presence of 1,2-DCA. The usefulness of qPCR was also demonstrated by being able to distinguish differential growth of the two organisms during the two-step degradation of 1,1,2-TCA. Furthermore, this was done in the context of an undefined mixed culture, which highlights that one of the biggest advantages of qPCR is the ability to quantify single templates in a mixture. The technique can be expanded to monitor multiple organisms over time and across treatments to indicate functional roles in the culture, as long as the function is linked to growth. The technique would not detect cometabolic dechlorination by methanogens or acetogens.
A caution involved in the use of qPCR is that relatively similar copy concentrations may be indistinguishable due to error inherent in the technique. Although an increase in copy concentration from 104 16S rRNA gene copies/ml to 107 copies/ml due to dehalorespiration represents virtually the same increase in biomass as an increase from 1 × 107 copies copies/ml to 2 × 107 copies/ml, far more confidence can be placed in the former result because of the change of 3 orders of magnitude. Therefore, a relatively deep starting dilution is required to observe unequivocal increases in gene copy number and thus evidence of growth.
The 16S rRNA gene sequence of the Dehalococcoides WL strain was indistinguishable from that of Dehalococcoides strain 195. Interestingly, Dehalococcoides WL grew during dechlorination of VC to ethene, while this step is cometabolic in strain 195, illustrating, as others have, the lack of discrimination provided by the 16S rRNA gene sequence. Dehalococcoides sp. strain WL is similar to other Dehalococcoides tested in its ability to dichloroeliminate 1,2-DCA to ethene but not 1.1,2-TCA to VC. The underlying reason for this substrate specificity is yet to be determined but may be related to the complement of dehalogenase genes acquired by each specific strain (31, 34, 42; A. Waller et al., in press).
The demonstration of growth of the Dehalobacter sp. WL strain during dichloroelimination of 1,2-DCA and 1,1,2-TCA has not been previously reported for this genus. The three known Dehalobacter isolates described to date have been reported only to carry out reductive dechlorination of PCE or TCE to cDCE (29, 56) or 1,1,1-TCA to chloroethane (49). In addition, a Dehalobacter sp. in coculture was recently shown to dechlorinate hexachlorocyclohexane (HCH) (53), and Dehalobacter was associated with trichlorobenzene (54) and 1,2-dichloropropane (46) transformation. Dehalobacter strains TEA and TCA1 and the HCH-degrading strain have not been characterized beyond the 16S rRNA gene. However, the sequences of several putative dehalogenase genes from the chloroethene-dehalorespiring D. restrictus have been determined using degenerate primers (45). We have begun to identify putative dehalogenase genes using these and other degenerate primers (34) in order to elucidate the unique dichloroelimination activity of this Dehalobacter sp. WL strain, and isolation efforts are continuing to obtain a pure culture.
The WL/Mix culture's ability to only reductively dechlorinate TCE to cDCE starkly contrasts with the ability of the WL/1,2-DCA and WL/1,1,2-TCA subcultures to completely degrade PCE to ethene. As shown by DGGE and qPCR, this difference is likely attributable to a lack of a Dehalococcoides sp. in the WL/Mix subculture and indicates that the Dehalobacter in these cultures can only dechlorinate TCE to cDCE.
In summary, it has been shown that in an anaerobic 1,1,2-TCA-dechlorinating enrichment culture, two distinct organisms are required for the complete conversion of 1,1,2-TCA to ethene and that the presence of multiple dechlorinating organisms in multiple genera significantly enhances the substrate range of the mixed culture. This also emphasizes the advantage of using mixed cultures rather than a pure culture for bioaugmentation for site remediation: the complementary substrate ranges of multiple dechlorinating organisms can improve the potential for remediation of sites that are contaminated with multiple chlorinated compounds that would otherwise be inhibitory or recalcitrant to a single dechlorinating organism.
Acknowledgments
Initial microcosms were prepared by Sandra Dworatzek (SiREM Laboratories, Guelph, Ontario, Canada) with the advice and assistance of Evan Cox (GeoSyntec Consultants, Guelph, Ontario, Canada). We thank Melanie Duhamel for help with DGGE and qPCR techniques; Cristina Chornewich and Ekanki Saxena for conducting the substrate range experiment; and Joyce Dinglasan, YuSan Ong, and Susanna Lam for help with maintaining cultures.
The research was funded by a Natural Sciences and Engineering Research Council (NSERC) Collaborative Research and Development Grant with GeoSyntec Consultants. A. Grostern was supported by a NSERC postgraduate scholarship.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Agency for Toxic Substances and Disease Registry. 2004. ToxFAQs for 1,1,2-trichloroethane. [Online.] http://www.atsdr.cdc.gov/tfacts148.html.
- 3.Agency for Toxic Substances and Disease Registry. 2004. ToxFAQs for 1,2-dichloroethane. [Online.] http://atsdr1.atsdr.cdc.gov/tfacts38.html.
- 4.Bunge, M., L. Adrina, A. Kraus, M. Opel, W. Lorenz, J. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 5.Castro, C. E., and N. O. Belser. 1990. Biodehalogenation-oxidative and reductive metabolism of 1,1,2-trichloroethane by Pseudomonas putida-biogeneration of vinyl chloride. Environ. Toxicol. Chem. 9:707-714. [Google Scholar]
- 6.Castro, C. E., M. C. Helvenston, and N. O. Belser. 1994. Biodehalogenation, reductive dehalogenation by Methanobacterium thermoautotrophicu-comparison with nickel(I)octaethylisobacteriochlorin anion-an F-430 model. Environ. Toxicol. Chem. 13:429-433. [Google Scholar]
- 7.Cox, E., D. Major, and E. A. Edwards. 2000. Natural attenuation of 1,2-dichloroethane in groundwater at a chemical manufacturing facility, p. 217-224. In G. B. Wickramanayake, A. R. Gavaskar, and M. E. Kelley (ed.), Remediation of chlorinated and recalcitrant compounds, natural attenuation considerations and case studies, vol. C2-3. Proceedings of the Second International Conference on Remediation of Chlorinated and Recalcitrant Compounds. Battelle Press, Columbus, Ohio. [Google Scholar]
- 8.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2004. Vinyl chloride and cis-dichloroethene dechlorination kinetics and microorganism growth under substrate limiting conditions. Environ. Sci. Technol. 38:1102-1107. [DOI] [PubMed] [Google Scholar]
- 9.De Wildeman, S., G. Diekert, H. Van Langenhove, and W. Verstraete. 2003. Stereoselective microbial dehalorespiration with vicinal dichlorinated alkanes. Appl. Environ. Microbiol. 69:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Wildeman, S., G. Linthout, H. Van Langenhove, and W. Verstraete. 2004. Complete lab-scale detoxification of groundwater containing 1,2-dichloroethane. Appl. Microbiol. Biotechnol. 63:609-612. [DOI] [PubMed] [Google Scholar]
- 11.De Wildeman, S., A. Neumann, G. Diekert, and W. Verstraete. 2003. Growth-substrate dependent dechlorination of 1,2-dichloroethane by a homoacetogenic bacterium. Biodegradation 14:241-247. [DOI] [PubMed] [Google Scholar]
- 12.De Wildeman, S., H. Nollet, H. Van Langenhove, G. Diekert, and W. Verstraete. 2002. Reductive biodegradation of 1,2-dichloroethane by methanogenic granular sludge: perspectives for in situ remediation. Water Sci. Technol. 45:43-48. [PubMed] [Google Scholar]
- 13.Drzyzga, O., and J. C. Gottschal. 2002. Tetrachloroethene dehalorespiration and growth of Desulfitobacterium frappieri TCE1 in strict dependence on the activity of Desulfovibrio fructosivorans. Appl. Environ. Microbiol. 68:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duhamel, M., K. Mo, and E. A. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duhamel, M., S. Wehr, L. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-1,2-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, E. A., and D. Grbić-Galić. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ely, R. L., K. J. Williamson, M. R. Hyman, and D. J. Arp. 1997. Cometabolism of chlorinated solvents by nitrifying bacteria: kinetics, substrate interactions, toxicity effects, and bacterial response. Biotechnol. Bioeng. 54:520-534. [DOI] [PubMed] [Google Scholar]
- 18.Fathepure, B. Z., and J. M. Tiedje. 1994. Reductive dechlorination of tetrachloroethylene by a chlorobenzoate-enriched biofilm reactor. Environ. Sci. Technol. 28:746-752. [DOI] [PubMed] [Google Scholar]
- 19.Fennell, D. E., S. Rhee, Y. Ahn, M. M. Haggblom, and L. J. Kerkhof. 2004. Detection and characterization of a dehalogenating microorganism by terminal restriction fragment length polymorphism fingerprinting of 16S rRNA in a sulfidogenic, 2-bromophenol-utilizing enrichment. Appl. Environ. Microbiol. 70:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frascari, D., A. Zannoni, S. Fedi, Y. Pii, D. Zannoni, D. Pinelli, and M. Nocentini. 2005. Aerobic cometabolism of chloroform by butane-grown microorganisms: long-term monitoring of depletion rates and isolation of a high-performing strain. Biodegradation 16:147-158. [DOI] [PubMed] [Google Scholar]
- 21.Gerritse, J., V. Renard, T. M. P. Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hage, J. C., R. T. van Houten, J. Tramper, and S. Hartmans. 2004. Membrane-aerated biofilm reactor for the removal of 1,2-dichloroethane by Pseudomonas sp. strain DCA1. Appl. Microbiol. Biotechnol. 64:718-725. [DOI] [PubMed] [Google Scholar]
- 24.Hamamura, N., C. Page, T. Long, L. Semprini, and D. J. Arp. 1997. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 63:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, J., K. M. Ritalahti, K. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 26.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson, E. R., J. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst, B., and U. Wiesmann. 1996. Kinetics and reaction engineering aspects of the biodegradation of dichloromethane and dichloroethane. Water Res. 30:1069-1076. [Google Scholar]
- 29.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and A. J. B. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 30.Holliger, C., G. Schraa, E. Stupperich, A. J. M. Stams, and A. J. B. Zehnder. 1992. Evidence for the involvement of corrinoids and factor-F430 in the reductive dechlorination of 1,2-dichloroethane by Methanosarcina barkeri. J. Bacteriol. 174:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Gorisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inguva, S., and G. S. Shreve. 1999. Biodegradation kinetics of trichloroethylene and 1,2-dichloroethane by Burkholderia (Pseudomonas) cepacia PR1(31) and Xanthobacter autotrophicus GJ10. Int. Biodeter. Biodegrad. 43:57-61. [Google Scholar]
- 33.Klecka, G. M., C. L. Carpenter, and S. J. Gonsior. 1998. Biological transformations of 1,2-dichloroethane in subsurface soils and groundwater. J. Contam. Hydro. 34:139-154. [Google Scholar]
- 34.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic Identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaPara, T. M., T. Zakharova, C. H. Nakatsu, and A. Konopka. 2002. Functional and structural adaptations of bacterial communities growing on particulate substrates under stringent nutrient limitation. Microb. Ecol. 44:317-326. [DOI] [PubMed] [Google Scholar]
- 36.Lorah, M. M., and M. A. Voytek. 2004. Degradation of 1,1,2,2-tetrachloro ethane and accumulation of vinyl chloride in wetland sediment microcosms and in situ porewater: biogeochemical controls and associations with microbial communities. J. Contam. Hydro. 70:117-145. [DOI] [PubMed] [Google Scholar]
- 37.Luijten, M. L., J. de Weert, H. Smidt, H. T. Boschker, W. M. de Vos, G. Schraa, and A. J. Stams. 2003. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53:787-793. [DOI] [PubMed] [Google Scholar]
- 38.Maymo-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maymó-Gatell, X., Y. Chien, J. Gossett, and S. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetracholoethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 40.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168:513-519. [DOI] [PubMed] [Google Scholar]
- 41.Morgan, C. A., A. Hudson, A. Konopka, and C. H. Nakatsu. 2002. Analyses of microbial activity in biomass-recycle reactors using denaturing gradient gel electrophoresis of 16S rDNA and 16S rRNA PCR products. Can. J. Microbiol. 48:333-341. [DOI] [PubMed] [Google Scholar]
- 42.Muller, A. J., M. B. Rosner, G. von Abendroth, G. Meshulam-Simon, L. P. McCarty, and M. A. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distributions. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nobre, R. C. M., and M. M. M. Nobre. 2004. Natural attenuation of chlorinated organics in a shallow sand aquifer. J. Hazard. Mat. 110:129-137. [DOI] [PubMed] [Google Scholar]
- 45.Regeard, C., J. Maillard, and C. Holliger. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107-118. [DOI] [PubMed] [Google Scholar]
- 46.Schlotelburg, C., C. von Wintzingerode, R. Hauck, F. von Wintzingerode, W. Hegemann, and U. V. Gobel. 2002. Microbial structure of an anaerobic bioreactor population that continuously dechlorinates 1,2-dichloropropane. FEMS Microbiol. Ecol. 39:229-237. [DOI] [PubMed] [Google Scholar]
- 47.Song, J. S., D. H. Lee, K. Lee, and C. K. Kim. 2003. Characteristics of several bacterial isolates capable of degrading chloroaliphatic compounds via hydrolytic dechlorination. J. Microbiol. 41:277-283. [Google Scholar]
- 48.Speitel, G. E., and D. S. McLay. 1993. Biofilm reactors for treatment of gas streams containing chlorinated solvents. J. Environ. Eng. ASCE 119:658-678. [Google Scholar]
- 49.Sun, B., B. N. Griffin, H. L. Ayala-del-Rio, S. A. Hashsham, and J. M. Tiedje. 2002. Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298:1023-1025. [DOI] [PubMed] [Google Scholar]
- 50.Sung, Y., K. M. Ritalahti, R. A. Sanford, J. W. Urbance, S. J. Flynn, J. M. Tiedje, and F. E. Löffler. 2003. Characterization of two tetrachloroethene-reducing, acetate-oxidizing anaerobic bacteria and their description as Desulfuromonas michiganensis sp. nov. Appl. Environ. Microbiol. 69:2964-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suyama, A., R. Iwakiri, K. Kai, T. Tokunaga, N. Sera, and K. Furukawa. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474-1481. [DOI] [PubMed] [Google Scholar]
- 52.Tandoi, V., T. D. Distefano, P. A. Bowser, J. M. Gossett, and S. H. Zinder. 1994. Reductive dehalogenation of chlorinated ethenes and halogenated ethanes by a high-rate anaerobic enrichment culture. Environ. Sci. Technol. 28:973-979. [DOI] [PubMed] [Google Scholar]
- 53.van Doesburg, W., M. H. A. van Eekert, P. J. M. Middeldorp, M. Balk, G. Schraa, and A. J. M. Stams. 2005. Reductive dechlorination of [β]-hexachlorocyclohexane ([β]-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 54:87-95. [DOI] [PubMed] [Google Scholar]
- 54.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Gobel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wild, A., R. Hermann, and T. Leisinger. 1996. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511. [DOI] [PubMed] [Google Scholar]