Abstract
To investigate the effects of administration of raffinose and encapsulated Bifidobacterium breve JCM 1192T cells on the rat cecal microbiota, in a preclinical synbiotic study groups of male WKAH/Hkm Slc rats were fed for 3 weeks with four different test diets: basal diet (group BD), basal diet supplemented with raffinose (group RAF), basal diet supplemented with encapsulated B. breve (group CB), and basal diet supplemented with both raffinose and encapsulated B. breve (group RCB). The bacterial populations in cecal samples were determined by fluorescence in situ hybridization (FISH) and terminal restriction fragment length polymorphism (T-RFLP). B. breve cells were detected only in the RCB group and accounted for about 6.3% of the total cells as determined by FISH analysis. B. breve was also detected only in the RCB group by T-RFLP analysis. This was in contrast to the CB group, in which no B. breve signals were detected by either FISH or T-RFLP. Increases in the sizes of the populations of Bifidobacterium animalis, a Bifidobacterium indigenous to the rat, were observed in the RAF and RCB groups. Principal-component analysis of T-RFLP results revealed significant alterations in the bacterial populations of rats in the RAF and RCB groups; the population in the CB group was similar to that in the control group (group BD). To the best of our knowledge, these results provide the first clear picture of the changes in the rat cecal microbiota in response to synbiotic administration.
The human gastrointestinal tract harbors various kinds of bacteria that may have positive or negative effects on the health of the host (9). There is increasing interest in the idea that diet can promote or maintain beneficial colonic bacteria that improve immunopotentiation (25), prevent the invasion of pathogenic bacteria (45), and provide metabolic energy for the host (17). This concept has led to the terms “probiotic” (15), “prebiotic” (17), and “synbiotic” (17).
Synbiotic was first defined as a mixture of probiotics and prebiotics that beneficially affects the host by improving the survival and implantation of live microbial dietary supplements in the gastrointestinal tract by selectively stimulating the growth and/or by activating the metabolism of one or a limited number of health-promoting bacteria and thus improving host welfare (17). Synbiotics are believed to increase the persistence of the probiotic bacteria in the gastrointestinal tract. Although the concept of synbiotics has been introduced, we believe that there has not been a clear demonstration of synbiotic effects due to unclear experimental design or unreliable methods for microbiota analysis. Molecular ecological methods, which reveal the entire range of bacterial diversity and more accurately detect population changes, are now considered more appropriate for evaluating synbiotic effects.
In this study, alteration of the rat cecal microbiota upon administration of raffinose in combination with Bifidobacterium breve JCM 1192T was analyzed. B. breve JCM 1192T was employed in a gelatin-encapsulated form to protect the bacterial cells from the acidic environment of the stomach and to increase bacterial access to the intestine. This strain has recently been reported to accumulate large amounts of cholic acid, a primary bile acid in humans, in an energy-dependent manner (26). Thus, we intended to amplify this strain in the rat cecum in a preclinical study in order to design a rational human trial to determine the effects on lipid metabolism and on intestinal bile acid composition. In in vitro growth experiments, B. breve JCM 1192T grew better on raffinose than on the other fermentable sugars tested. Therefore, raffinose was chosen as a suitable carbon source for use in combination with encapsulated B. breve JCM 1192T. Although this mixture (i.e., raffinose and encapsulated B. breve JCM 1192T) has not yet been demonstrated to be a synbiotic in accord with the definition of Gibson and Roberfroid (17), below we sometimes refer to this mixture as “synbiotic” for simplicity and clarity. Similarly, encapsulated B. breve JCM 1192T is tentatively referred to as a “probiotic.” The rat experiment was designed with four groups (control, prebiotic, probiotic, and synbiotic). Two molecular techniques, fluorescence in situ hybridization (FISH) (4) and terminal restriction fragment length polymorphism (T-RFLP) analysis (29, 31), were used for the first time to monitor alteration of the rat microbiota in the synbiotic experiment. As a result, proliferation of B. breve JCM 1192T was successfully observed only in the synbiotic group. Furthermore, we analyzed other members of the rat microbiota and determined the organic acid content in the cecum to obtain a more comprehensive picture of the changes in the rat intestinal microbiota. We believe that our results provide the first clear picture of changes in the microbiota in the rat cecum after administration of a synbiotic and also provide fundamental information concerning rat intestinal microbiology.
MATERIALS AND METHODS
Bacterial strain and media.
B. breve JCM 1192T was obtained from the Japan Collection of Microorganisms (Wako, Japan). Cultures were grown in half-strength MRS medium (10) (0.5× MRS) containing filter-sterilized l-cysteine-HCl at a concentration of 0.25 g/liter unless indicated otherwise. TOS propionate agar (Yakult Pharmaceutical Ind. Co., Ltd., Tokyo, Japan) was used for isolation of bifidobacteria from rat cecal samples.
Measurement of the growth rate.
A seed culture for B. breve JCM 1192T growth experiments was grown overnight in 0.5× MRS. At an optical density at 660 nm (OD660) of 0.05 the seed culture was transferred to the same medium containing one of the following carbohydrates (10 g/liter): glucose, sucrose, raffinose (Sigma-Aldrich, St. Louis, Mo.), kestose, or nystose (kestose and nystose were donated by the Hokuren Federation of Agricultural Cooperatives, Sapporo, Japan). Bacterial growth was periodically monitored by determining the OD660 of the culture broth. All the cultures were incubated at 37°C under anaerobic conditions using mixed gas (N2-CO2-H2, 8:1:1). Specific growth rates (μ) (in h−1) were calculated during the logarithmic growth phase using the following equation: μ = (lnxt2 − lnxt1)/(t2 − t1), where xt2 and xt1 are the OD660 values at times t2 and t1, respectively.
Animals and diets.
Male WKAH/Hkm Slc rats (n = 24; Japan SLC, Hamamatsu, Japan) that were 4 weeks old were acclimatized with the basal diet for 7 days in individual rat cages. Twenty-four rats were divided equally into four groups and fed test diets for 3 weeks. The control group (group BD) received the basal diet, which contained (per kg diet) 250 g casein, 50 g corn oil, 35 g AIN-93G mineral mixture, 10 g AIN-93 vitamin mixture, 2.5 g choline bitartrate, 602.5 g of sucrose, and 50 g/kg crystalline cellulose (Avicel; Asahi Kasei Corporation, Osaka, Japan) (35). Group RAF received the basal diet supplemented with 30 g/kg raffinose (Nippon Beet Sugar Manufacturing Co., Ltd., Tokyo, Japan). Group CB was fed the basal diet supplemented with 30 g/kg gelatin-encapsulated B. breve JCM 1192T (viable cell count, about 5.7 × 107 CFU/g of capsule; provided by Morishita Jintan Co., Ltd., Osaka, Japan). The gelatin content of the capsule was 17.5% (wt/wt). Thus, the CB group received 5.25 g of gelatin/kg of diet. For preparation of the capsule, B. breve JCM 1192T was cultured in a jar fermentor until the late exponential growth phase using 0.5× MRS medium containing both 40 g/liter raffinose as the carbon source and 0.5 g/liter l-cysteine-HCl. The culture was incubated at 37°C with stirring at 200 rpm under anaerobic conditions with CO2 gas in the headspace of the fermentor. The pH of the medium was controlled at 6.5 with NaOH. The cells were harvested by centrifugation, washed twice with 150 mM NaCl, resuspended in 10% (wt/vol) skim milk, and then freeze-dried before encapsulation. Group RCB received the basal diet supplemented with the same amounts of raffinose and encapsulated B. breve JCM 1192T that were in the group RAF and CB diets, respectively. Gelatin-encapsulated B. breve JCM 1192T and raffinose were added at the expense of sucrose in the basal diet. Rats were maintained and handled according to the recommendations of our institutional ethics committee.
Sample preparation.
Individual rat cecal contents were weighed immediately after rats were sacrificed. One portion of the contents was used for pH and organic acid measurements (see below). The remainder (about 0.5 g) was washed three times with ice-cold phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM sodium phosphate buffer; pH 7.2) with low-speed centrifugation at 200 × g for 5 min. Bacteria were pelleted from the pooled supernatant by high-speed centrifugation three times at 9,000 × g for 2 min each time. The bacterial cell pellet was resuspended in PBS, and each preparation was divided into two parts. One part was used for bacterial cell fixation for FISH analysis (see below). As sample amounts were limited, the remaining portions were pooled according to test diet groups for genomic DNA extraction for T-RFLP analysis (see below).
Measurement of organic acids.
The concentrations of organic acids (succinate, lactate, propionate, butyrate, isovalerate, and valerate) in the rat cecal samples were determined by high-performance liquid chromatography (SCL-10AVP; Shimadzu Corporation, Kyoto, Japan) by using the method described previously (18). Briefly, the cecal samples were added to an aqueous sodium hydroxide solution containing crotonic acid as an internal standard. After centrifugation, the fat-soluble substances in the supernatant were removed by extraction with chloroform. The aqueous phase was passed through a membrane filter and subjected to high-performance liquid chromatography.
FISH analysis.
Individual washed samples were fixed in 4% (wt/vol) paraformaldehyde in PBS (pH 7.2) for 24 h. Fixed samples were washed once in PBS and stored in a known volume of 50% (vol/vol) ethanol-PBS at −20°C until use. Aliquots (3 μl) of fixed cells were applied to Teflon printed glass slides (ADCELL; 12 wells; diameter, 5 mm; Erie Scientific Company, Portsmouth, N.H.) and air dried. The cells were then dehydrated with a series of solutions containing 50%, 80%, and 99.5% ethanol (3 min for each concentration). The cells fixed on the glass slides were hybridized by addition of 8 μl of hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl, 20% deionized formamide; pH 7.2) with 1 μl of Cy3-labeled oligonucleotide probe (25 ng/μl; Tsukuba Oligo Service Co., Ltd., Tsukuba, Japan). The slides were hybridized at 46°C for 16 h in a moist chamber. After hybridization, the slides were rinsed with warm hybridization buffer at 48°C and washed in prewarmed washing buffer (225 mM NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl; pH 7.2) for 20 min at 48°C. The washed slides were stained with a DAPI (4′,6-diamidino-2-phenylindole dihydrochloride n-hydrate) solution for 5 min at room temperature to stain the chromosomes as a control signal. The slides were washed with distilled water for 5 min at room temperature and air dried in the dark. The dried slides were mounted with Vectashield (Vector Laboratories Inc., Burlingame, Calif.) and examined with an Olympus BX50 epifluorescence microscope (Olympus Corporation, Tokyo, Japan) equipped with a SenSys charge-coupled device camera (Photometrics Ltd., Tucson, Ariz.) operated by the IPLab software (Scanalytics, Inc., Fairfax, Va.). DAPI and Cy3 signals were captured in pairs of 10 to 15 random microscopic fields (about 500 cells per microscopic field). Hybridization images were manually counted and were colorized when necessary using Adobe Photoshop 5.5 (Adobe Systems Incorporated, San Jose, Calif.). Specific signals from the probes were expressed as average percentages of the total cells visualized by DAPI signals in the same microscopic field.
Oligonucleotide probes.
The oligonucleotide probes and the target microorganisms used in this study are shown in Table 1; a target region alignment for PBR2 and Bani449 (created in this study) is shown in Fig. 1. Computer alignment of the 16S rRNA genes from bifidobacteria revealed that Bifidobacterium animalis and some other bifidobacteria are not targets of probe Bif164 because the 16S rRNA sequences of these bacteria showed a single-base mismatch with Bif164 (27). The mismatched nucleotide (C or T) was replaced with Y to create a new probe designated Bif164m. No cross hybridization was observed with nontarget bacterial strains, including Lactobacillus, Clostridium, Bacteroides, Streptococcus, and Enterococcus strains (data not shown). In a preliminary experiment to visualize the secondary structures of the 16S rRNA of B. breve and B. animalis by computer simulation (GENETYX-Win; Software Development, Tokyo, Japan), it was found that the target regions for PBR2 and Bani449 were difficult to access due to RNA-RNA interactions. Therefore, we used unlabeled oligonucleotides (helper probes), which were complementary to the regions up- and downstream of the probe target site on the 16S rRNA, to increase the target accessibility for the probes, as suggested by Fuchs et al. (14). The sequences of these helper probes are also shown in Table 1. Indeed, the hybridization efficiencies of PBR2 and Bani449 were significantly increased in both target strains by use of these helper probes (data not shown).
TABLE 1.
16S rRNA-targeted oligonucleotide probes used for molecular analysis of rat cecal samples
| Probe (systematic name) | Probe sequence (5′ to 3′) | Target organism(s) | Target sitea | Reference |
|---|---|---|---|---|
| Eub338 (S-D-Bact-0338-a-A-18) | GCTGCCTCCCGTAGGAGT | Bacteria | 338 | 3 |
| Genus- or group-specific probes | ||||
| Erec482 (S-*-Erec-0482-a-A-19) | GCTTCTTAGTCARGTACCG | Clostridium coccoides-Eubacterium rectale group | 482 | 13 |
| Chis150(S-*-Chis-0150-a-A-23) | TTATGCGGTATTAATCTYCCTTT | Clostridium histolyticum group | 150 | 13 |
| Bac303 (S-G-Bac-0303-a-A-17) | CCAATGTGGGGGACCTT | Bacteroides group | 303 | 30 |
| Lacb722 (S-G-Lacb-0722-a-A-25) | YCACCGCTACACATGRAGTTCCACT | Lactobacillus group | 722 | 41 |
| Strc493 (S-*-Strc-0493-a-A-19) | GTTAGCCGTCCCTTTCTGG | Streptococci and lactococci | 493 | 13 |
| Bif164m (S-G-Bif-0164-b-A-18)b | CATCCGGYATTACCACCC | Bifidobacterium spp. | 164 | This studyc |
| Species- or strain-specific probes | ||||
| PBR2 (S-S-Pbr2-0182-a-A-19) | CCATGCGGTGTGATGGAGC | Bifidobacterium breve | 182 | 43 |
| Bani433 (S-S-Bani-0449-a-A-21) | CACTCAACACGGCCCGAAGCC | Bifidobacterium animalis JCM 1190T | 449 | This study |
| Nonlabeled oligonucleotides (helpers) | ||||
| PBR2-1st-Upper helper | ATCCGGCATTACCACCCGT | Bifidobacterium breve | 163 | This study |
| PBR2-1st-Lower helper | CAAAGGCTTTCCCAACACA | Bifidobacterium breve | 201 | This study |
| PBR2-2nd-Upper helper | TTCCAGGAGCTATTCCGGT | Bifidobacterium breve | 144 | This study |
| PBR2-2nd-Lower helper | GCGACCCCATCCCATGCCG | Bifidobacterium breve | 220 | This study |
| Bani433-1st-Upper helper | GTGCCTTGCCCTTGAACAAAA | Bifidobacterium animalis JCM 1190T | 428 | This study |
| Bani433-1st-Lower helper | CCGGTGCTTATTCGAACAATC | Bifidobacterium animalis JCM 1190T | 470 | This study |
Position in Escherichia coli 16S rRNA.
Bif164m was modified from Bif164 described by Langendijk et al. (27) by changing the eighth 5′-nucleotide from C to Y.
The probe was designed in this investigation.
FIG. 1.
Alignment of probe sequences, their target sites, and the sequences of the corresponding sites in reference microorganisms of the large intestine (type strains). The probe names are in accordance with the Oligonucleotide Probe Database nomenclature (2). N indicates an A/T/C/G wobble nucleotide.
T-RFLP analysis.
To establish standard terminal restriction fragment (T-RF) peak sets for identification of bacterial strains present in the cecal samples by T-RFLP analysis, T-RF lengths were determined for B. breve JCM 1192T and an indigenous rat strain of B. animalis isolated from rat cecal samples using TOS propionate agar. Bacterial genomic DNA was extracted from pure cultures of these microorganisms and from pooled cecal samples from the four experimental groups (groups BD, RAF, CB, and RCB) using an Isoplant DNA extraction kit (Nippon Gene, Co., Ltd., Tokyo, Japan) according to the manufacturer's instructions. Two replicates of each sample were separately amplified by PCR and digested with restriction enzymes as described below. A PCR mixture was prepared from each sample using a GeneAmp PCR System 2400 (Applied Biosystems, Foster City, Calif.) with the 6-carboxyfluorescein-labeled primer 46F (5′-GCYTAACACATGCAAGTCGA-3′) (23), which was synthesized by Applied Biosystems Japan, and unlabeled primer 1080R (5′-CCCAACATCTCACGAC-3′) (33). The PCR conditions were based on the method described by Kaplan et al. (23), with the following modifications: reaction mixture volume, 100 μl; amount of template DNA, 200 ng; 1× Gold buffer (Applied Biosystems); concentration of deoxynucleoside triphosphates, 0.6 mM; concentration of bovine serum albumin, 0.8 μg/liter; concentration of MgCl2, 3.5 mM; concentration of 6-carboxyfluorescein-labeled primer 46F, 0.2 μM; concentration of primer 1080R, 0.2 μM; and amount of AmpliTaq Gold DNA polymerase, 4 U (Applied Biosystems). The thermal cycling program for the fecal samples was as follows: 94°C for 2 min; 35 cycles of 94°C for 2 min, 48.5°C for 1 min, and 72°C for 1 min; and a final cycle of 72°C for 10 min. Amplified fragments were purified by SUPREC PCR (Takara Bio Inc., Otsu, Japan), and the purified fragments from three replications were pooled. Aliquots (about 1 μg) of DNA in purified amplicons were digested with 10 U (each) of HaeIII, HhaI, and MspI (Takara Bio Inc.) at 37°C for 24 h. The digestion reactions were stopped by incubation at 65°C in a water bath for 20 min, followed by immediate cooling in an ice bath. The restriction digest products were subjected to ethanol precipitation and vacuum dried. Fifty microliters of distilled water was added to dissolve each dried sample. Aliquots (1 μl) of dissolved samples were then added to 10 μl of a mixture of formamide and a size standard (GS 500 ROX; Applied Biosystems) (100:5, vol/vol). Each sample was denatured at 95°C for 2 min and then immediately placed on ice. The fluorescently labeled T-RFs were analyzed by electrophoresis with an automatic sequence analyzer (ABI PRISM 3100; Applied Biosystems) in gene scan mode, and the lengths of the T-RFs were determined by comparison with size standards using the Genescan 3.7 software (Applied Biosystems).
Data analyses.
The effects of diet on cecal organic acids and microbiota were analyzed separately by two-way analysis of variance (ANOVA) at a P value of <0.05 using the JMP software (SAS Institute Inc., Cary, N.C.). The data obtained in the T-RFLP analysis were processed as described by Kaplan et al. (23). The T-RF peaks with areas less than the threshold value were excluded. The remaining T-RF peaks from three restriction enzymes were combined and examined by principal-component analysis (PCA) using the SPSS 10 software (SPSS Inc., Chicago, Ill.).
RESULTS
Carbon source preference of B. breve JCM 1192T.
Prior to in vivo experiments, we conducted growth experiments with B. breve JCM 1192T to determine the carbon source preference of this bacterium. The results showed that B. breve JCM 1192T grew better on raffinose than on the other carbon sources tested, as assessed by the μ in MRS medium. The μ values (means ± standard deviations; n = 2) on the carbon sources tested were 0.45 ± 0.02 h−1 on glucose, 0.65 ± 0.02 h−1 on raffinose, and 0.64 ± 0.08 h−1 on kestose, while sucrose and nystose were not assimilated at all by this strain. Therefore, raffinose was chosen as a prebiotic for this bacterium.
Animal experiments.
All rats were healthy and alive until the day on which they were sacrificed. The pH values of the cecal contents of rats in the BD and CB groups (pH 8.1 ± 0.1 and 8.0 ± 0.1, respectively) were significantly higher than the pH values of the cecal contents of rats in the RAF and RCB groups (pH 6.8 ± 0.1 and 7.0 ± 0.2, respectively) (Table 2). Cecal samples from group RAF and RCB rats were yellow, and group BD and CB cecal samples were brown-green. In addition, the cecal content weights for rats in the RAF and RCB groups were significantly greater than those for rats in the BD and CB groups (Table 2). To confirm the reproducibility of the results, we conducted a second trial and obtained similar results (data not shown).
TABLE 2.
Characterization of rat cecal samples: organic acids produced, cecal content weight, pH, and two-way ANOVA results
| Characteristic | Group BDa | Group RAF | Group CB | Group RCB | Two-way ANOVAc
|
||
|---|---|---|---|---|---|---|---|
| Raffinose | B. breve | Raffinose × B. breve | |||||
| Lactate concn (μmol/g of cecal contents) | NDb | 16.8 (5.8) | 1.4 (0.9) | 6.0 (2.1) | S | NS | NS |
| Acetate concn (μmol/g of cecal contents) | 42.1 (5.7) | 54.3 (6.7) | 35.7 (2.9) | 53.3 (8.3) | S | NS | NS |
| Propionate concn (μmol/g of cecal contents) | 9.4 (1.0) | 11.1 (0.7) | 12.8 (0.8) | 11.9 (1.3) | NS | S | NS |
| Butyrate concn (μmol/g of cecal contents) | 9.3 (2.2) | 3.4 (1.0) | 8.9 (0.9) | 6.4 (2.3) | S | NS | NS |
| Succinate concn (μmol/g of cecal contents) | 26.7 (2.1) | 21.2 (6.1) | 5.4 (2.0) | 31.1 (7.9) | NS | NS | S |
| Isovalerate concn (μmol/g of cecal contents) | 1.0 (0.3) | 0.1 (0.1) | 1.0 (0.1) | ND | S | NS | NS |
| Valerate concn (μmol/g of cecal contents) | 1.6 (0.1) | 0.4 (0.2) | 1.9 (0.2) | 0.7 (0.2) | S | NS | NS |
| Cecal content wt (g) | 1.21 (0.09) | 2.13 (0.09) | 0.94 (0.05) | 2.29 (0.24) | S | NS | NS |
| Cecal content pH | 8.1 (0.1) | 6.8 (0.1) | 8.0 (0.1) | 7.0 (0.2) | S | NS | NS |
The values are means (standard errors of the means), (n = 6). Group BD, basal diet; group RAF, basal diet supplemented with raffinose (30 g/kg diet); group CB, basal diet supplemented with gelatin-encapsulated B. breve (30 g/kg diet); and group RCB, basal diet supplemented with raffinose (30 g/kg diet) and encapsulated B. breve (30 g/kg diet).
ND, not detected.
S, significantly different (P < 0.05); NS, not significant.
As shown in Table 2, the amounts of lactic and acetic acids in group RAF and RCB cecal samples were significantly higher than those in group BD and CB cecal samples. However, the amounts of butyric, isovaleric, and valeric acids in the cecal samples from the RAF and RCB groups were significantly lower than those in control samples due to the effect of raffinose in the diets. The amount of propionic acid was significantly higher in group CB and RCB samples than in control samples.
FISH analysis of microbiota in the cecal samples.
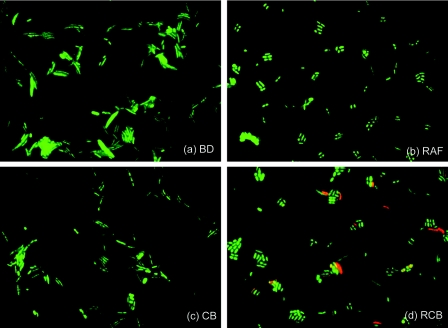
FISH was performed to enumerate the total and target bacterial populations (Table 3). The total bacterial populations of the groups were slightly different (ca. 1.74 × 1010 to 2.19 × 1010 cells/g [wet weight], as determined by the DAPI signal), and the ratio of the total bacteria (probe Eub338) to the total cells (DAPI) decreased from about 90% in the BD and CB groups to about 85% in the RAF and RCB groups. The cell morphologies of the predominant bacteria were different in different samples (Fig. 2). Long curved rods were the major forms in the BD and CB groups, while group RAF and RCB samples contained short rods. The results of the second trial were similar (data not shown).
TABLE 3.
Characterization of rat cecal microbiota based on FISH analysis: number of bacterial cells per gram, percentage of total microbiota, and two-way ANOVA results
| Population | Stain or probe | % of microbiota in cecal samplesa
|
Two-way ANOVA
|
|||||
|---|---|---|---|---|---|---|---|---|
| Group BD (control) | Group RAF | Group CB | Group RCB | Raffinose | B. breve | Raffinose × B. breve | ||
| Total cells | DAPI | 100 | 100 | 100 | 100 | |||
| Total bacteria | Eub338 | 89.6 (1.5) | 84.4 (1.4) | 90.4 (0.8) | 86.3 (1.3) | Sc | NSc | NS |
| Clostridium coccoides-Eubacterium rectale group | Erec482 | 23.9 (2.7) | 12.9 (3.3) | 29.7 (4.0) | 23.2 (5.8) | S | S | NS |
| Clostridium histolyticum group | Chis150 | 0.3 (0.1) | 1.2 (0.4) | 0.1 (0.0) | 1.2 (0.4) | S | S | S |
| Bacteroides group | Bac303 | 3.6 (1.4) | 2.1 (0.6) | 0.9 (0.2) | 2.5 (1.1) | NS | NS | S |
| Lactobacillus group | Lacb722 | 0.2 (0.0) | 7.2 (1.1) | 0.8 (0.2) | 2.8 (0.6) | S | S | S |
| Streptococci and lactococci | Strc493 | 0.5 (0.2) | 1.5 (0.9) | 0.8 (0.4) | 0.3 (0.2) | NS | NS | NS |
| Bifidobacterium spp. | Bif164m | 0.4 (0.2) | 19.5 (4.8) | 0.3 (0.1) | 25.4 (4.6) | S | NS | NS |
| Unidentified bacteria | None | 71.1 | 55.6 | 67.4 | 44.6 | |||
| Bifidobacterium breve | PBR2 | NDb | ND | ND | 6.3 (1.9) | S | S | S |
| Bifidobacterium animalis | Bani449 | 0.2 (0.1) | 20.5 (2.2) | 0.1 (0.0) | 18.5 (2.7) | S | NS | NS |
Percentages of the total cells visualized by DAPI signals in the same microscopic field. The values are means (standard errors of the means), (n = 6). The numbers of cells in the cecal contents (means ± standard errors of the means; n = 6) for groups BD, RAF, CB, and RCB were 1.90 × 1010 ± 0.9 × 1010, 1.74 × 1010 ± 0.5× 1010, 2.06 × 1010 ± 0.4 × 1010, and 2.19 × 1010 ± 0.3 × 1010 cells/g (wet weight), respectively.
ND, not detected.
S, significantly different (P < 0.05); NS, not significant.
FIG. 2.
Epifluorescence images of bacterial cells from the four groups of rat cecal samples stained with DAPI (green) and hybridized with a B. breve species-specific oligonucleotide probe (PBR2) (red) in FISH analysis.
As shown in Table 3, at 23.9% of the total cell count, the Clostridium coccoides-Eubacterium rectale group (probe Erec482) accounted for the largest proportion of the bacterial population in group BD rats. The Bacteroides group (probe Bac303) accounted for 3.6% of the total population. Compared with group BD (control) rats, group RAF rats had a significantly lower number of targets for the C. coccoides-E. rectale group (12.9%). The Lactobacillus group (probe Lacb722) and bifidobacteria (probe Bif164m) accounted for 7.2% and 19.5%, respectively, of the total population. These proportions were significantly higher than those in group BD rats (0.2% and 0.4%, respectively). The signals from the Clostridium histolyticum group (probe Chis150) were slightly higher in group RAF samples (1.2%) than in control samples (0.3%). Group RAF rats contained the highest proportion of streptococci and lactococci (probe Strc493) among all the diet groups, although these bacteria did not account for more than 1.5% of the total population. The populations of the group CB rats and the control rats did not differ significantly, although the proportion of the C. coccoides-E. rectale group was slightly higher (29.7%) in the group CB rats. No B. breve (probe PBR2) signals were detected in group CB rats even though the target strain was present in the group CB diet. The group RCB rats contained nearly the same proportions of the C. coccoides- E. rectale group (23.2%) and the Bacteroides group (2.5%) as the group BD rats. The proportion of the Lactobacillus group was higher in the group RCB rats (2.8%) than in the group BD rats (0.2%) but lower than the proportion in the group RAF rats (7.2%). A striking feature of the RCB group was the large proportion of bifidobacteria, which was 25.4% of the total DAPI-stained cells (corresponding to 29.4% of the total cell count using the Eub338 probe) and was the highest value for all the samples. B. breve, the bacterial strain administered, accounted for 6.3% of the total DAPI-stained cells (7.3% of the total cell count using the Eub338 probe), as detected using the PBR2 probe with its helpers. By isolation of bifidobacteria using TOS propionate agar and 16S rRNA gene sequencing (data not shown), the bifidobacteria found in the group RAF rats and the bifidobacteria present in the group RCB rats along with B. breve JCM 1192T were shown to be B. animalis. In addition, a newly designed B. animalis-specific probe (probe Bani449) was used in this study. This species accounted for significant proportions of the populations (20.5% and 18.5% of the total populations in the group RAF and RCB rats, respectively). The second trial revealed similar changes in the populations of total bifidobacteria, B. breve, and B. animalis, confirming the reproducibility of the bifidobacterial responses (data not shown).
The increase in the number of cells of the administered strain was strictly dependent on raffinose (Table 3). These results strongly support the synbiotic concept. Two-way ANOVA suggested that most of the alterations in the bacterial populations, particularly the increases in lactobacilli and bifidobacteria, were significantly influenced by the inclusion of raffinose in the diets.
T-RF analysis of cecal samples.
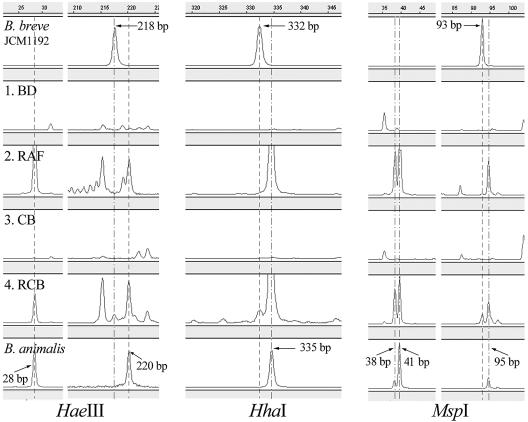
The lengths of T-RFs of B. animalis isolated on TOS propionate agar from rat cecal samples and B. breve JCM 1192T were determined using three restriction enzymes to obtain the standard T-RF peak sets. For the B. animalis isolate the observed T-RF lengths were 28 and 220 bp for HaeIII, 335 bp for HhaI, and 38, 41, and 95 bp for MspI, which differed slightly from the predicted values (36 and 220 bp for HaeIII, 334 and 337 bp for HhaI, and 45 and 98 bp for MspI). The T-RF lengths observed for B. breve JCM 1192T were 218 bp for HaeIII, 332 bp for HhaI, and 93 bp for MspI, whereas the predicted values were 219 bp for HaeIII, 333 bp for HhaI, and 97 bp for MspI. Most of the experimentally determined T-RF lengths were within 4 bp of the predicted lengths, which were close to the smallest size of the DNA standard used (GS 500 ROX). Similar phenomena have been reported previously (23, 36). The T-RF lengths observed for these strains contributed to identification and interpretation of the T-RF patterns from rat cecal samples.
By comparison with the T-RF peak sets for the reference strains, the presence of B. animalis was detected in the RAF and RCB groups (Fig. 3). The T-RFLP electropherogram revealed that B. breve was present only in the RCB group and that it accounted for about 3.3 to 6.0% of the total peak area, as shown in Fig. 3. Although peak areas do not represent the actual proportions of bacteria of interest in populations, these data confirmed the existence of B. breve in the expected cecal samples.
FIG. 3.
Partial electropherogram of HaeIII-, HhaI-, and MspI-derived T-RF profile for four rat cecal samples and two bifidobacteria. The size of each T-RF (in base pairs) is indicated along with the horizontal scale at the top of the GeneScan display.
According to the alignment of the T-RF peak set, seven peak sets were common to the four groups, although the proportions were different. Approximately 29 peak sets present in the control group (group BD) were not found in the RAF or RCB group; two of the indigenous peak sets had smaller areas in the BD group than in the RAF and RCB groups (data not shown). On the other hand, about 16 new peak sets were detected in the RAF and RCB groups, and two indigenous peak sets had areas greater than those of the corresponding peak sets in the BD group (data not shown).
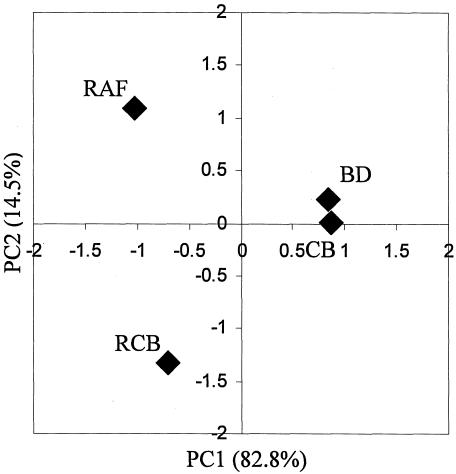
Next we used PCA, an advanced statistical technique, to reveal the variance-covariance structure of the T-RF patterns among all samples (Fig. 4). A total of 209 fragments from three restriction enzyme digests (HaeIII, 54 fragments; HhaI, 82 fragments; and MspI, 73 fragments) were included in the analysis. We found that the correlation of variance was comprised of two principal components, and the yield cumulative sum of squared loading (variance) was 97.3%. The spots representing the RAF and RCB groups were separated from the spots for the BD and CB groups on the PC1 axis (82.8%). This suggested that the first principal component (PC1) separates samples based on the presence of raffinose in the diet and is a major factor in bacterial population alterations (Fig. 4). The spot for the CB group was close to that for the BD group, while the RAF and RCB groups were significantly separated from each other on the PC2 axis (14.5%). The second principal component (PC2) appeared to separate samples based on the presence of B. breve in the RCB group.
FIG. 4.
PCA of T-RFLP profiles from four groups of rat cecal samples. The percent variation accounted for by each principal component is indicated along the corresponding axis, along with the principal-component loading values.
DISCUSSION
The rat and mouse intestinal microbiota consists of complex bacterial communities, which has been confirmed by culture methods and 16S rRNA gene clone library analyses (24, 37, 46). In our FISH analysis, we focused on Bacteroides, Bifidobacterium, Clostridium, Eubacterium, Lactobacillus, and Streptococcus, which have been reported to be the members of the rat and mouse intestinal microbiota (37, 38, 46). Six sets of genus- or group-specific oligonucleotide probes identified about 29 to 55% of the DAPI-stained cells and about 32 to 64% of the active bacterial cells (Eub338) in the rat cecal communities (Table 3). The relatively high proportions of “unidentified bacteria” (about 45 to 71%) in Table 3 seemed to be due to the presence of a large population of an uncharacterized bacterial group called “fusiform-shaped anaerobic bacteria” in the murine microbiota (11, 39). This group of bacteria is morphologically unique and consists of long curved rods with sharp thin ends (39); it was seen in the microbial populations in the BD and CB groups (Fig. 2a and c).
The bifidobacteria were found to dominate the cecal microbial community in the RCB (synbiotic) group (Table 3). The presence of B. breve in this group was clearly demonstrated by FISH analysis (Fig. 2d and Table 3) and was also confirmed by T-RFLP analysis (Fig. 3). The successful proliferation of administered B. breve JCM 1192T observed in the RCB group seems to be attributable to the availability of raffinose for B. breve JCM 1192T cells, because raffinose is not digested in the rat intestine (17) and directly reaches the cecum as the most preferred carbon source for this bacterium. The substantial increase in the number of B. breve JCM 1192T cells in the RCB group strongly demonstrated the effectiveness of the combination of raffinose and B. breve JCM 1192T.
Administration of raffinose alone (group RAF) appeared to induce marked increases in the numbers of indigenous bifidobacteria and lactobacilli (Table 3). The bifidobacteria were identified as B. animalis by FISH and T-RFLP analyses. Although the presence of B. animalis in rat intestine was reported a long time ago (40), the population of this bacterium in the rat intestine has not been reported precisely. This study showed for the first time using molecular ecological methods that B. animalis is a minor member of the population in the rat cecum (0.2%) and that the size of the population can be increased up to about 20% of the microbiota by raffinose administration. The increases in the Bifidobacterium and Lactobacillus populations in the RAF and RCB groups corresponded to the increased production of acetic and lactic acids and the decreased pH values of the cecal contents (Table 2).
Interestingly no proliferation of B. breve JCM 1192T was observed in the CB group, in contrast to the RCB group (Table 3). This may be explained by the fact that this bacterium lacks the ability to utilize sucrose, which is present at high levels in the rat basal diet (602.5g/kg diet). On the other hand, B. breve can utilize glucose and fructose (data not shown), which are the hydrolysis products of sucrose. Thus, we expect that there is a good chance that B. breve is fed by sucrose hydrolysate (glucose and fructose) in the rat intestine. Since we did not observe any proliferation of B. breve cells in the CB group, our results (B. breve proliferation was observed only in the RCB group) illustrate the dependence of this strain on administered prebiotic all the more clearly owing to the inability of this strain to utilize sucrose.
FISH analysis can provide information on bacterial populations at the genus, group, and even species levels in terms of identification and enumeration (4). However, the limited range of the oligonucleotide probe set prevented us from obtaining a complete picture of the microbial population in the rat cecum. Therefore, we performed PCA to distinguish the bacterial communities in the rat cecal contents based on T-RF profiles (Fig. 4). The purpose of PCA is data reduction, which allows interpretation of data through a few linear combinations of the original variables (treatments). PCA confirmed the differences in the rat microbiota in the CB and RCB groups based on T-RFLP profiles. The composition of the microbiota in the CB group was more similar to that in the BD group, indicating that probiotic administration alone was not enough to modulate the bacterial population. When a synbiotic was adminstered, significant changes in the bacterial population were observed along with proliferation of B. breve, as also confirmed by FISH analysis. These analyses led to the conclusion that application of the synbiotic is necessary for the target strain, B. breve JCM 1192T, to proliferate in the rat cecum.
Introduction of the synbiotic concept by Gibson and Roberfroid (17) promoted application of a potential nonindigenous probiotic strain and its preferred carbon source to increase the level of the administered strain in the intestine. Several previously published works on synbiotic application described modulation of the intestinal microbiota (1, 7, 8, 32, 44), reduction of colon carcinogenesis (12, 16, 28), and protection from pathogen infection (6). Recently, synbiotic therapy for improvement of intestinal function (19, 20) or reduction of pathogenic bacteria (21, 22) has also been reported.
However, in many cases, demonstration of synbiotic effects seems to be insufficient from a microbiological point of view. One of the most important criteria for evaluation of synbiotic effects is monitoring the proliferation of the administered probiotic bacteria at the species level, which seems possible only by use of molecular ecological methods for microbiota analysis. In most cases, however, culture-based methods with selective agar medium have been used to count the numbers of bacteria in fecal and cecal samples (6, 8, 28, 32, 44). Selective medium is sometimes not selective or too selective for target bacteria in microbiota analyses (5). Thus, the culture-based methods are not suitable for distinguishing the administered bacteria from similar groups of bacteria and thus allow only group level analysis. In addition, it has been shown that only about 20 to 30% of microorganisms in human intestines are culturable (42). Hence, the reliability of the culture-based methods is thought to be rather low. Besides these technological obstacles, many studies have lacked control experiments, such as probiotic or prebiotic alone (7, 19, 20, 21, 22, 32, 44), making interpretation of the synbiotic results inconclusive.
In contrast to these previous reports, our work may provide the first clear demonstration of the microbiological aspects of synbiotic effects in the rat intestinal microbiota. First, we quantitatively monitored the proliferation of an administered strain at the species level and analyzed the other bacteria using the FISH method. In addition, the T-RFLP technique was used to confirm the FISH results and to compare the whole microbiotas of the experimental groups. These two molecular methods were applied for the first time to microbiota analysis in a synbiotic experiment. As a potential probiotic we used B. breve JCM 1192T, whose carbon source utilization is well characterized. An analysis of a complete set of treatment groups (groups BD, RAF, CB, and RCB) was conducted. Organic acids as indicators of microbial activity were also analyzed. All these parts of the experimental design made precise analysis and interpretation of the data obtained possible.
Although the human diet is complex, the availability of raffinose in the human intestine seems to be relatively low. Raffinose is found in food legumes at relatively high levels (up to about 2%), but the amount is reduced significantly during food processing (34). Therefore, supplementation with raffinose is necessary to support the proliferation of administered B. breve JCM 1192T in the human intestine, and this will be evaluated in human trials.
Acknowledgments
We are grateful for the bacterial strains provided by the Japan Collection of Microorganisms. We also thank Kimiko Minamida for her excellent assistance.
REFERENCES
- 1.Alander, M., J. Mättö, W. Kneifel, M. Johansson, B. Kögler, R. Crittenden, T. Mattila-Sandholm, and M. Saarela. 2001. Effect of galacto-oligosaccharide supplementation on human faecal microflora and on survival and persistence of Bifidobacterium lactis Bb-12 in the gastrointestinal tract. Int. Dairy J. 11:817-825. [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apajalahti, J. H. A., A. Kettunen, P. H. Nurminen, H. Jatila, and W. E. Holben. 2003. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl. Environ. Microbiol. 69:5731-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahara, T., K. Nomoto, K. Shimizu, M. Watanuki, and R. Tanaka. 2001. Increased resistance of mice to Salmonella enterica serovar Typhimurium infection by synbiotic administration of bifidobacteria and transgalactosylated oligosaccharides. J. Appl. Microbiol. 91:985-996. [DOI] [PubMed] [Google Scholar]
- 7.Bartosch, S., E. J. Woodmansey, J. C. M. Paterson, M. E. T. McMurdo, and G. T. Macfarlane. 2005. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 40:28-37. [DOI] [PubMed] [Google Scholar]
- 8.Bielecka, M., E. Biedrzycka, and A. Majkowska. 2002. Selection of probiotics and prebiotics for synbiotics and confirmation of their in vivo effectiveness. Food Res. Int. 35:125-131. [Google Scholar]
- 9.Crittenden, R. G. 1999. Prebiotics, p. 141-156. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, England.
- 10.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 11.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Femia, A. P., C. Luceri, P. Dolara, A. Giannini, A. Biggeri, M. Salvadori, Y. Clune, K. J. Collins, M. Paglierani, and G. Caderni. 2002. Antitumorigenic activity of the prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis on azoxymethane-induced colon carcinogenesis in rats. Carcinogenesis 23:1953-1960. [DOI] [PubMed] [Google Scholar]
- 13.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 16.Gallaher, D. D., and J. Khil. 1999. The effect of synbiotics on colon carcinogenesis in rats. J. Nutr. 129:1483S-1487S. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 18.Hoshi, S., T. Sakata, K. Mikuni, H. Hashimoto, and S. Kimura. 1994. Galactosylsucrose and xylosylfructoside alter digestive tract size and concentrations of cecal organic acids in rats fed diets containing cholesterol and cholic acid. J. Nutr. 124:52-60. [DOI] [PubMed] [Google Scholar]
- 19.Kanamori, Y., K. Hashizume, M. Sugiyama, M. Morotomi, and N. Yuki. 2001. Combination therapy with Bifidobacterium breve, Lactobacillus casei, and galactooligosaccharides dramatically improved the intestinal function in a girl with short bowel syndrome. Dig. Dis. Sci. 46:2010-2016. [DOI] [PubMed] [Google Scholar]
- 20.Kanamori, Y., K. Hashizume, M. Sugiyama, M. Morotomi, N. Yuki, and R. Tanaka. 2002. A novel synbiotic therapy dramatically improved the intestinal function of a pediatric patient with laryngotracheo-esophageal cleft (LTEC) in the intensive care unit. Clin. Nutr. 21:527-530. [DOI] [PubMed] [Google Scholar]
- 21.Kanamori, Y., K. Hashizume, Y. Kitano, Y. Tanaka, M. Morotomi, N. Yuki, and R. Tanaka. 2003. Anaerobic dominant flora was reconstructed by synbiotics in an infant with MRSA enteritis. Pediatr. Int. 45:359-362. [DOI] [PubMed] [Google Scholar]
- 22.Kanamori, Y., M. Sugiyama, K. Hashizume, N. Yuki, M. Morotomi, and R. Tanaka. 2004. Experience of long-term synbiotic therapy in seven short bowel patients with refractory enterocolitis. J. Pediatr. Surg. 39:1686-1692. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, C. W., J. C. Astaire, M. E. Sanders, B. S. Reddy, and C. L. Kitts. 2001. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 67:1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kibe, R., M. Sakamoto, H. Hayashi, H. Yokota, and Y. Benno. 2004. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbiol. Lett. 235:139-146. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl. Environ. Microbiol. 63:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurdi, P., H. Tanaka, H. W. van Veen, K. Asano, F. Tomita, and A. Yokota. 2003. Cholic acid accumulation and its diminution by short-chain fatty acids in bifidobacteria. Microbiology 149:2031-2037. [DOI] [PubMed] [Google Scholar]
- 27.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Leu, R. K., I. L. Brown, Y. Hu, A. R. Bird, M. Jackson, A. Esterman, and G. P. Young. 2005. A synbiotic combination of resistant starch and Bifidobaterium lactis facilitates apoptotic deletion of carcinogen-damaged cells in rat colon. J. Nutr. 135:996-1001. [DOI] [PubMed] [Google Scholar]
- 29.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 31.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 32.Morelli, L., D. Zonenschain, M. L. Callegari, E. Grossi, F. Maisano, and M. Fusillo. 2003. Assessment of a new synbiotic preparation in healthy volunteers: survival, persistence of probiotic strains and its effect on the indigenous flora. Nutr. J. 2:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori, K., K. Yamazaki, T. Ishiyama, M. Katsumata, K. Kobayashi, Y. Kawai, N. Inoue, and H. Shinano. 1997. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int. J. Syst. Bacteriol. 47:54-57. [DOI] [PubMed] [Google Scholar]
- 34.Reddy, N. R., S. K. Sathe, and D. K. Salunkhe. 1989. Carbohydrates, p. 51-74. In D. K. Salunkhe and S. S. Kadam (ed.), CRC handbook of world food legumes: nutritional chemistry, processing technology, and utilization, vol. 1. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 35.Reeves, P. G., F. H. Nielsen, and G. C. Fahey, Jr. 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition Ad Hoc Writing Committee on the Reformulation of the AIN-76A Rodent Diet. J. Nutr. 123:1939-1951. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2003. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J. Med. Microbiol. 52:79-89. [DOI] [PubMed] [Google Scholar]
- 37.Salzman, N. H., H. de Jong, Y. Paterson, H. J. M. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 38.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 39.Savage, D. C., J. S. McAllister, and C. P. Davis. 1971. Anaerobic bacteria on the mucosal epithelium of the murine large bowel. Infect. Immun. 4:492-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scardovi, V., and L. D. Trovatelli. 1974. Bifidobacterium animalis (Mitsuoka) comb. nov. and the “minimum” and “subtile” groups of new bifidobacteria found in sewage. Int. J. Syst. Bacteriol. 24:21-28. [Google Scholar]
- 41.Sghir, A., D. Antonopoulos, and R. I. Mackie. 1998. Design and evaluation of a Lactobacillus group-specific ribosomal RNA-targeted hybridization probe and its application to the study of intestinal microecology in pigs. System. Appl. Microbiol. 21:291-296. [DOI] [PubMed] [Google Scholar]
- 42.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suksomcheep, A., I. N. Sujaya, K. Saito, A. Yokota, K. Asano, and F. Tomita. 2001. Development of 16S rDNA targeted probes for detection of some human health beneficial strains of intestinal microorganisms. Biotechnol. Sust. Util. Biol. Resour. Trop. 15:111-125. [Google Scholar]
- 44.Tortuero, F., E. Fernández, P. Ruperéz, and M. Moreno. 1997. Raffinose and lactic acid bacteria influence caecal fermentation and serum cholesterol in rats. Nutr. Res. 17:41-49. [Google Scholar]
- 45.Vollaard, E. J., and H. A. L. Clasener. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanabe, M., M. Shibuya, T. Gonda, H. Asai, T. Tanaka, K. Sudou, T. Narita, and K. Itoh. 2001. Establishment of specific pathogen-free (SPF) rat colonies using gnotobiotic techniques. Exp. Anim. 50:293-298. [DOI] [PubMed] [Google Scholar]