Abstract
Free-living protists are ubiquitous in the environment and form a potential reservoir for the persistence of animal and human pathogens. Mycobacterium avium subsp. paratuberculosis is the cause of Johne's disease, a systemic infection accompanied by chronic inflammation of the intestine that affects many animals, including primates. Most humans with Crohn's disease are infected with this chronic enteric pathogen. Subclinical infection with M. avium subsp. paratuberculosis is widespread in domestic livestock. Infected animals excrete large numbers of robust organisms into the environment, but little is known about their ability to replicate and persist in protists. In the present study we fed laboratory cultures of Acanthamoeba polyphaga with bovine and human strains of M. avium subsp. paratuberculosis. Real-time PCR showed that the numbers of the pathogens fell over the first 4 to 8 days and recovered by 12 to 16 days. Encystment of the amoebic cultures after 4 weeks resulted in a 2-log reduction in the level of M. avium subsp. paratuberculosis, which returned to the original level by 24 weeks. Extracts of resection samples of human gut from 39 patients undergoing abdominal surgery were fed to cultures of A. polyphaga. M. avium subsp. paratuberculosis detected by nested IS900 PCR with amplicon sequencing and visualized by IS900 in situ hybridization and auramine-rhodamine staining was found in cultures derived from 13 of the patients and was still present in the cultures after almost 4 years of incubation. Control cultures were negative. M. avium subsp. paratuberculosis has the potential for long-term persistence in environmental protists.
Free-living protists, including amoebae, flagellates, and ciliates, occur widely in the biosphere in both temperate and arid regions and are thought to be globally ubiquitous (20). They populate soils and sediments, suspended aggregates in fresh and marine waters, roots and surfaces of plants, and bodies of animals. They are dispersed by the movement of water and wind (1, 19, 20, 36-39). They feed by predation on prokaryotes and have a dynamic role in the maintenance and selection of bacterial populations. The bacterial response to predatory pressure has been the evolution of defenses, such as biofilm formation, multicellular aggregation, the production of cytotoxins, and the ability of selected prokaryotic species to survive within eukaryotic cells by avoiding or neutralizing digestive mechanisms (27, 32). These adaptations have contributed to the evolution of pathogenic phenotypes (5, 29, 41).
Environmental protists form a potential reservoir for animal and human pathogens which can survive in them, such as Listeria monocytogenes, Legionella sp., Escherichia coli O157, Campylobacter jejuni, Rickettsiales, and mycobacteria (4, 6, 21, 25, 31, 44, 45). Passage from the extracellular environment to the intracellular environment within Acanthamoeba sp. results in adoption of a different microbial phenotype that is characterized by an enhanced ability to invade epithelial cells, increased virulence in experimental infections, and resistance to antimicrobial drugs (7-9, 12, 13, 28). Environmental mycobacteria, especially those in water and aerosols, are increasingly implicated in diseases of animals and humans (2, 17, 18, 33, 34).
Mycobacterium avium subsp. paratuberculosis is a pathogen which causes chronic inflammation of the intestine (Johne's disease) in ruminants and other animals, including primates, and is implicated in the causation of Crohn's disease in humans (15, 22-24). The main burden of infection is in domestic livestock. Subclinical infection, especially in dairy cattle, is widespread in Europe and North America, as well as in other regions (26). Infected animals excrete large numbers of M. avium subsp. paratuberculosis cells onto pastures, from which the microorganisms run off into surface waters and rivers (33). These robust pathogens can survive for long periods, and so far no upper limit on their persistence in the environment has been established (33, 35, 46). It is likely that environmental M. avium subsp. paratuberculosis is taken up by protists (24), and transmission is likely to occur where the geographic ranges of M. avium subsp. paratuberculosis, protozoans, animals, and humans overlap. Little information is available on the of ability of this bacterium to replicate and persist in the unicellular eukaryotes.
In the present study we examined the ability of bovine and human strains of M. avium subsp. paratuberculosis to replicate and persist in cultures which included trophozoite and encysted forms of Acanthamoeba polyphaga over periods of up to 24 weeks. We also explored the use of A. polyphaga for selective capture of native M. avium subsp. paratuberculosis directly from lysates of infected human intestinal tissues. We found that this low-abundance pathogen from humans survived in cultures of amoebae in both trophozoite and encysted forms for almost 4 years.
MATERIALS AND METHODS
Culture of M. avium subsp. paratuberculosis.
M. avium subsp. paratuberculosis bovine strains K5 and K10 and human strain SN6 (30) were prepared for inoculation into cultures of A. polyphaga by growth in MGIT medium (Becton Dickinson, Oxford, United Kingdom) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (Sigma, United Kingdom), PANTA (Becton Dickinson) (40 U/ml polymixin B, 4 μg/ml of amphotericin B, 16 μg/ml nalidixic acid, 4 μg/ml of trimethoprim, 4 μg/ml of azlocillin), and 2 μg/ml mycobactin J (Allied Monitor, Fayette, Mo.) (all final concentrations). Sterile 3-mm glass beads were added to the liquid cultures, and the tubes were placed horizontally in a shaking incubator at 37°C. The cultures in which clumps of M. avium subsp. paratuberculosis were partially dispersed were harvested after 4 weeks. The numbers of organisms were estimated by counting dilutions of stained organisms, as well as by real-time PCR.
Culture of amoebae.
A. polyphaga strain CCAP 1501/18 was used throughout this study. All stock cultures were tested and were shown to be negative by IS900 PCR before inoculation. Cultures were prepared in sealed 75-ml flasks containing 25 ml PPG broth (18 g/liter proteose peptone L85 [Oxoid, Basingstoke, United Kingdom], 0.08 M d-glucose, 0.005% saline solution 1 [0.4 M NaCl, 3.2 mM MgSO4 · 7H2O, CaCl2 · 6H2O], 0.005% saline solution 2 [0.2 M Na2HPO4, 0.2 M KH2PO4]). Before inoculation all cultures were allowed to achieve confluence for 2 weeks after passage and were then maintained on a bench at room temperature (RT) with addition of fresh medium at intervals of 4 to 5 days.
Experimental infection of cultures of A. polyphaga.
Each 75-ml culture flask containing confluent A. polyphaga contained about 1 × 106 adherent trophozoites with no visible encystment. The cultures were infected at a multiplicity of 10:1. In the first short-term experiment M. avium subsp. paratuberculosis strains K5 and SN6 were each inoculated into a group of five flasks containing A. polyphaga. One flask was withdrawn and processed on days 1, 4, 8, 12, and 16. During processing each culture was washed twice with 10 ml phosphate-buffered saline (PBS), and the adherent amoebae were harvested into 1 ml PBS using a cell scraper. A second, longer-term experiment was carried out to examine the behavior of M. avium subsp. paratuberculosis strains in amoebae over a 24-week period. In these studies two groups of six 75-ml flasks containing amoebae inoculated with either K10 or SN6 were prepared. At weeks 2 and 4 nonadherent bacteria were removed from all flasks by washing, and the medium was changed. One flask from each group was removed for processing at weeks 2, 4, 6, 8, and 10. Processing at weeks 2 and 4 consisted of harvesting adherent trophozoites as described above. After week 4, processing consisted of harvesting both adherent trophozoites and encysted forms which emerged in the supernatant. The sixth flask was left for 24 weeks, during which addition of fresh medium was withheld for two 4-week periods so that the amoebae underwent two cycles of encystment followed by trophozoite reactivation.
DNA extraction.
Harvested suspensions of amoebae were centrifuged at 10,000 × g, and each pellet was resuspended in 600 μl of mycobacterial lysis buffer (2 mM EDTA, 400 mM NaCl, 10 mM Tris HCl, 0.6% sodium dodecyl sulfate, 33 μg/ml proteinase K) and incubated with shaking in a Ribolyser tube (Lysing matrix B; catalog no. 6911-100; Qbiogene, Cambridge, United Kingdom) overnight at 37°C. The Ribolyser tubes were then chilled on ice for 5 min, and the contents were mechanically disrupted with a FastPrep Ribolyser (Qbiogene) at a setting of 6.5 m s−2 for 45 s and then immediately chilled again on ice for 15 min. Six hundred microliters of phenol (pH 6.7) saturated in 1× TE (10 mM Tris HCl, 1 mM sodium EDTA; pH 8.0) was added, and the mixture was vortexed for 20 s and then centrifuged at 10,000 × g for 1 min. The aqueous layer was transferred to a new screw-cap reaction tube containing an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1; Sigma, Poole, United Kingdom), vortexed for 20 s, and centrifuged (10,000 × g for 1 min). The aqueous layer was transferred to a new screw-cap tube containing 500 μl of chloroform-isoamyl alcohol (24:1), vortexed for 30 s, and centrifuged (10,000 × g for 1 min). The final aqueous layer (450 μl) was transferred to a new tube containing 90 μl of 10 M ammonium acetate and mixed thoroughly. One milliliter of ice-cold 100% ethanol was added to enable DNA precipitation at RT for 1 h. The samples were then centrifuged (10,000 × g for 20 min) at RT, and the pellets were washed in 750 μl of ice-cold 70% ethanol. The pellets were dried at RT for 30 min, resuspended in 50 μl of 1× TE, and allowed to redissolve at 4°C overnight.
Real-time PCR.
The numbers of M. avium subsp. paratuberculosis cells in amoebae harvested in the short- and long-term experimental infection experiments were determined by real-time PCR by using a Stratagene MX4000 cycler (Stratagene, United Kingdom) with primers TJ60 (5′-CGACGTTCCCGATCTCTGGTG-3′) (0.4 μM) and TJ61 (5′-AGCGAGTAAGCAGGATCAGC-3′) (0.4 μM) and internal probe TJ62 (5′-6-carboxyfluorescein-GACTACAACAAGAGCGTGCC) (0.2 μM) in a 50-μl reaction mixture each containing 25 μl 1× ABsolute QPCR-dUTP master mixture plus 100 mM ROX reference dye (catalogue no. AB-1140/a; ABgene, Surrey, United Kingdom). The reaction conditions included activation at 95°C for 15 min and 40 cycles of 95°C for 30 s, 58°C for 30 s, and 60°C for 1 s. The standard curve was derived from serial dilutions of a standardized concentration of plasmid pIDL60 containing a single copy of IS900. Bacterial counts were derived by assuming that there were 15 copies of IS900 per genome.
Direct infection of A. polyphaga from surgical samples of human gut tissue.
Between April 2001 and August 2002 full-thickness samples of fresh human gut tissue removed during abdominal surgery from 39 patients were obtained from hospitals in Carmarthen, Llanelli, Swansea, and Cardiff (Wales, United Kingdom). Ethical approval for the study was given by the relevant health authorities. Tissues were obtained from 13 patients with Crohn's disease, 18 patients with colon cancer, 2 patients with ulcerative colitis, 1 patient with indeterminate colitis, 1 patient with diverticulitis, 1 patient with radiation colitis, 1 patient with volvulus, 1 patient with a mesenteric vascular occlusion, and 1 patient with carcinoma of the bladder infiltrating the colon. The unfrozen tissues in sealed sterile containers were mailed to St. George's University of London.
Preparation of tissue extracts and feeding to cultures of A. polyphaga.
In a laminar flow hood about 0.5 g of tissue from each sample was diced between crossed scalpels, ground in a disposable tissue grinder (Kendall 3500-SA; Tyco Healthcare) with homogenization buffer containing 0.1% trypsin-collagenase, and then incubated with shaking at 37°C for 3 h. The digest was again ground and incubated for a further 30 min. An equal volume of NALC buffer (BBL Mycoprep; Becton Dickinson) was added, and the solution was incubated at room temperature for 20 min and then neutralized with sterile 0.5 M Tris-HCl, pH 4.0. After centrifugation (10,000 × g for 20 min) the pellet was divided between duplicate cultures of A. polyphaga. Control cultures inoculated with only sterile culture medium were established at the same time. The amoebic cultures fed with the tissue extracts were incubated at RT for between 34 and 47 months with occasional additions to the culture medium. DNA extracted from cultures were then examined using nested IS900 PCR, PCR for the MAP2204c gene, in situ hybridization using an IS900 probe, and auramine-rhodamine staining.
Nested IS900 PCR.
DNA extraction was performed as described previously (11). Briefly, the first round was performed in a 50-μl reaction mixture containing 5 μl of sample DNA, 2 μM (final concentration) of each primer (primers L1 [5′-CTTTCTTGAAGGGTGTTCGG-3′] and L2 [5′-ACGTGACCTCGCCTCCAT-3′], 1× Expand High Fidelity reaction buffer containing 1.5 mM MgCl2, 10% dimethyl sulfoxide, 200 μM (each) dATP, dCTP, dGTP, and dTTP, and 3.5 U of Expand High Fidelity Taq polymerase (Expand High Fidelity PCR system; Roche, Lewes, United Kingdom). The cycling conditions were as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 3 min and then 72°C for 7 min. In order to avoid cross-contamination, the DNA amplified during the first step was treated with 1 U of uracil-DNA glycosylase (Roche) at RT for 10 min. For the nested PCR, 2 μl of the first reaction mixture was added to a reaction mixture that was identical to the first reaction mixture except that it contained primers AV1 (5′-ATGTGGTTGCTGTGTTGGATGG-3′) and AV2 (5′-CCGCCGCAATCAACTCCAG-3′) and 400 μM dTUP instead of dTTP. The 298-bp amplicons were visualized after electrophoresis in a 1.5% agarose gel, followed by ethidium bromide staining. All IS900 PCR amplicons were sequenced to verify their identities. After agarose gel electrophoresis the band was cut out, purified with a Minielute PCR purification kit (QIAGEN, Sussex, United Kingdom), and then eluted in 25 μl of TE. Amplicons were sequenced in both directions using the primers AV1 and AV2 and then aligned with the IS900 sequence (GenBank accession no. AEO16958).
MAP2204c PCR.
Two cultures of A. polyphaga which were strongly positive as determined by IS900 PCR were also analyzed by PCR for the integrity of the MAP2204c gene in M. avium subsp. paratuberculosis at IS900 locus 5. PCR was performed as previously described (10) using flanking primers Loc5R (5′-GACAATCTGCCGTCGTATCA-3′) and Loc5L (5′-TCCCGGTAGAAGATCATGTG3-3′), each at a final concentration of 2 μM. The 50-μl reaction mixture contained 5 μl of sample DNA, 1× Expand High Fidelity reaction buffer containing 1.5 mM MgCl2, 10% dimethyl sulfoxide, 200 μM (each) dATP, dGTP, dTTP, and dCTP, and 3.5 U of Expand High Fidelity Taq polymerase (Roche, United Kingdom). The reaction conditions were as follows: 94°C for 5 min, followed by 40 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min and then 72°C for 7 min.
In situ hybridization.
After the flasks were tapped to release some trophozoites, 1 ml of culture supernatant was removed from each flask, centrifuged (5,000 × g for 40 min), washed with PBS, and then applied to a positively charged slide (SuperFrost Plus; Menzel-Glaser, Braunschweig, Germany) and dried. The adherent amoebae were washed in PBS and air dried, and the slides were heated at 56°C for 1 min and then fixed with 10% formalin in PBS. After washing again in PBS, the slides with mostly trophozoite forms were treated with 40 μg/ml of proteinase K in PBS for 20 min at 37°C, and those with mostly encysted forms were treated with 50 μg/ml of proteinase K for 1 h at 37°C. The slides were then dehydrated by using a gradient up to absolute ethanol.
The IS900 probe was prepared by amplification of M. avium subsp. paratuberculosis DNA with 2 μM primer TJ3 (5′-CAGCGGCTGCTTTATTATATTCC-3′) and 2 μM primer TJ4 (5′ GGCACGGCTCTTCTTGTAGT) by using the reaction conditions described above for first round of the nested PCR, except that the nucleotides were 200 μM dATP, 200 μM dGTP, 200 μM dTTP, 100 μM dCTP, and 100 μM biotin-labeled dCTP. After electrophoresis in 1.5% agarose gels, the 300-bp PCR product was cut out and purified with a Minielute PCR purification kit (QIAGEN). For hybridization the probe was suspended in hybridization buffer (4× SSC, 0.2 M sodium phosphate [pH 6.5], 2× Denhardt's solution, 0.1 mg/ml sodium azide [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at twice the desired concentration (0.5 to 1 μg/μl), an equal volume of 20% dextran sulfate in formamide was added, and 20 μl of the resulting solution was applied to the adherent amoebae and covered with a coverslip. To denature the DNA, slides were preheated at 94°C for 7 min, and hybridization was performed overnight at 42°C in a humidified chamber. The slides were then immersed in 0.2× SSC for 15 min to detach the coverslips and washed twice with 0.2× SSC for 15 min. The slides were then incubated for 15 min in blocking solution (50 mg/ml bovine serum albumin in Tris-HCl [pH 7.8], 150 mM NaCl, 0.2 mg/ml sodium azide) at room temperature in a humidified chamber. The blocking solution was replaced with 100 μl fluorescein isothiocyanate-streptavidin (40 μg/ml) in conjugate dilution buffer (100 mM Tris-HCl, 150 mM MgCl2, 10 mg/ml bovine serum albumin, 0.2 mg/ml sodium azide) for 1 h at room temperature in a humidified chamber. After washing in distilled water, the sample was examined with a fluorescence microscope. For each reaction, a negative control consisting of uninfected amoebae and a positive control consisting of amoebae that were experimentally infected with the K10 strain were used.
Auramine-rhodamine staining.
The amoebae were detached from the bottom of a flask mechanically, and 1 ml of the culture was centrifuged (5,000 × g for 40 min), washed in PBS, and resuspended in 20 μl of distilled water. The sample was then applied to a slide as described above, fixed with 10% formalin, stained with auramine-rhodamine for 15 min, washed with distilled water, destained with alcohol-acid for 10 min, washed with distilled water, and counterstained with potassium permanganate. The slide was then washed, dried, and examined by fluorescence microscopy.
RESULTS
Experimental infection of cultures.
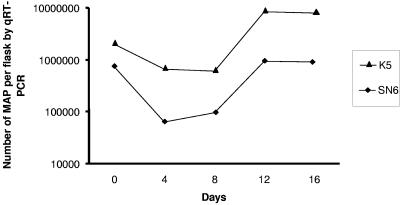
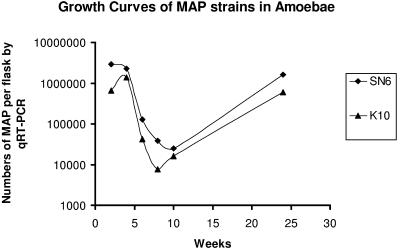
Initial studies using quantitative real-time PCR showed that only 2.5 to 11% of the M. avium subsp. paratuberculosis cells added to the cultures were taken up by A. polyphaga trophozoites. The numbers of organisms per flask after inoculation in the short-term study are shown in Fig. 1. Care was taken to ensure that any residual extracellular M. avium subsp. paratuberculosis cells were removed by washing the cultures. This was done at 24 h after infection and prior to harvesting and lysis. The numbers of cells of human strain SN6 declined about 10-fold and the numbers of cells of strain K5 declined by about fivefold over the first 4 days. The numbers of cells of both strains then stabilized, and the number of cells of each strain increased substantially from day 8 to day 16. The concentration of strain SN6 returned to the starting level, while number of the faster-growing bovine strain K10 cells was about fivefold greater by day 12. The results of the longer-term study are shown in Fig. 2. During the first 4 weeks the numbers of M. avium subsp. paratuberculosis K10 cells increased, whereas the numbers of SN6 cells remained unchanged. Then approximately 100-fold decreases in the numbers of mycobacterial cells occurred between weeks 4 and 8, and the numbers stabilized by week 10. The numbers then increased, returning to the original starting level by week 24.
FIG. 1.
Short-term growth over 16 days of M. avium subsp. paratuberculosis bovine strain K5 and human strain SN6 in A. polyphaga trophozoites. MAP, M. avium subsp. paratuberculosis; qRT-PCR, quantitative real-time PCR.
FIG. 2.
Long-term growth over 24 weeks of M. avium subsp. paratuberculosis bovine strain K10 and human strain SN6 in dynamic long-term cultures of A. polyphaga undergoing encystment and reactivation to trophozoite forms. MAP, M. avium subsp. paratuberculosis; qRT-PCR, quantitative real-time PCR.
M. avium subsp. paratuberculosis from human gut samples fed to A. polyphaga.
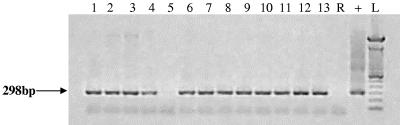
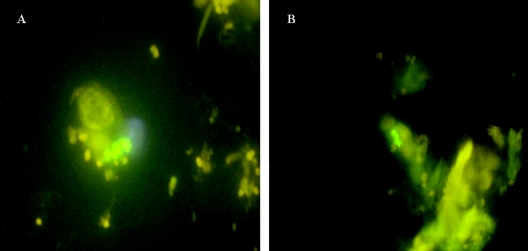
The mean time interval between sampling of tissues and processing in the laboratory was 2.2 days (range, 1 to 10 days). The initial examination of the duplicate culture flasks began in June 2003, so the minimum incubation time was about 1 year. By this time the organisms in the flasks from only 4 of the 39 patients were still in the trophozoite form. The rest of the organisms consisted of variable numbers of mature cysts. From this time all flasks were replenished at intervals with fresh medium. Thirteen of the 39 patient flasks (33%) (five flasks for patients with Crohn's disease, one flask for a patient with ulcerative colitis, one flask for a patient with diverticulitis, and six flasks for patients with colorectal cancers) were positive as determined by nested IS900 PCR. Control flasks were negative. Amplicon sequences confirmed the presence of IS900. Nested IS900 PCR was repeated for all cultures in May 2005 after total incubation periods ranging from 35 to 47 months. Identical results were obtained (Fig. 3). All 13 IS900 PCR-positive flasks were also positive for M. avium subsp. paratuberculosis as determined by IS900 in situ hybridization with control flasks reporting correctly (Fig. 4). Of the 13 cultures which were positive for M. avium subsp. paratuberculosis as determined by IS900 PCR and in situ hybridization, 6 contained bacilli which were auramine-rhodamine acid-fast stain positive (Fig. 5). Control cultures of A. polyphaga were negative. DNA extracts from two A. polyphaga cultures which were strongly positive as determined by IS900 PCR and contained the greatest numbers of M. avium subsp. paratuberculosis amplified with PCR primers specific for the mycobacterial MAP2204c gene with a product of the predicted 387 bp characteristic of human strains.
FIG. 3.
IS900 PCR analysis of DNA extracts from amoebic cultures showing amplification of the predicted 298-bp product, which was also verified by amplicon sequencing. The amoebic cultures had been fed extracts of human gut tissues and then incubated at room temperature for between 35 and 47 months with long periods of encystment. Lane R, reagent control; lane +, positive control; lane L, size ladder.
FIG. 4.
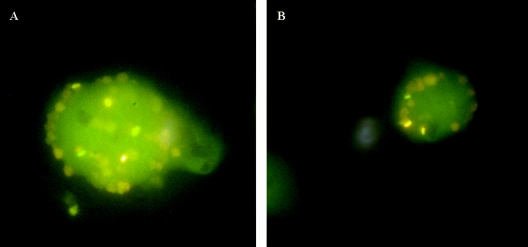
Fluorescent in situ hybridization using an IS900 probe (see text), showing clumps of M. avium subsp. paratuberculosis cells within the cytoplasm of A. polyphaga in cultures maintained for 24 months after feeding with extracts of human gut tissue. (A) Culture fed to maintain the amoebae in the trophozoite form. (B) Culture undergoing long periods of encystment of the amoebae, followed by trophozoite reactivation.
FIG. 5.
Auramine-rhodamine staining of amoebic cultures previously shown to be positive for M. avium subsp. paratuberculosis by PCR and in situ hybridization, showing fluorescent mycobacteria 24 months after feeding with extracts of human gut tissues. (A) Mycobacteria dispersed in the cytoplasm of a trophozoite. (B) Peripheral distribution of mycobacteria in an encysted A. polyphaga cell.
DISCUSSION
The present study showed that, as observed for some other M. avium complex isolates, M. avium subsp. paratuberculosis can survive and replicate within A. polyphaga (44). When cultures were inoculated with M. avium subsp. paratuberculosis, the freshly fed amoebae were all in the trophozoite form. Following experimental infection of cultures of A. polyphaga only a small proportion of the M. avium subsp. paratuberculosis cells, which had been added at a multiplicity of infection of about 10:1, were taken up by the amoebae. This suggests that the amoebae were able to take up the mycobacteria only as single organisms and not as residual clumps present in the suspensions. The numbers of both the bovine and human strains of M. avium subsp. paratuberculosis fell sharply shortly after addition to the cultures, suggesting that only a portion of the M. avium subsp. paratuberculosis cells were able to adapt and survive intracellularly (Fig. 1). The cells that did survive, however, subsequently went through an early period of replication in the trophozoites between days 8 and 12. After 28 days (Fig. 2) the amoebic cultures underwent encystment, and the period from week 4 to week 8 was associated with a dramatic reduction in the numbers of M. avium subsp. paratuberculosis cells in the cultures. This could have been due to death of a portion of the M. avium subsp. paratuberculosis cells during encystment, extrusion of M. avium subsp. paratuberculosis into the medium during encystment, or a reduction in the total numbers of available host cells in the maturing culture. After the 10th week substantial increases in the numbers of M. avium subsp. paratuberculosis cells in the cultures occurred despite two cycles of trophozoite formation and encystment. Much additional work is needed to define the cellular compartments occupied by M. avium subsp. paratuberculosis within amoebae and the effect of internalization on both the host and the microbial phenotype.
The experimental conditions surrounding the transport of human tissues in the present study were not ideal because the tissues had undergone autolysis at the ambient temperature, sometimes for several days before processing. Studies by several research groups using fresh tissues and optimized processing procedures have shown that a majority of Crohn's disease patients are infected with M. avium subsp. paratuberculosis (3, 11, 40, 42, 43). The proportion of autolyzed Crohn's disease tissues from which M. avium subsp. paratuberculosis could be recovered by amoebae in the present study (5/13, 33%) was lower than expected. Despite this, IS900 PCR with amplicon sequencing showed that more than one-third of 13 A. polyphaga cultures (most of which had encysted) which had been fed extracts of surgical samples of human intestinal tissues contained M. avium subsp. paratuberculosis 1 year later. These cultures were then reactivated and maintained with addition of fresh medium for a total of up to 47 months. At the end of this period IS900 PCR with amplicon sequencing showed that M. avium subsp. paratuberculosis was still present in the same amoebic cultures. This was confirmed by in situ hybridization, which also demonstrated that the M. avium subsp. paratuberculosis cells appeared to be intact and harbored within A. polyphaga as single organisms or as small clumps.
The MAP2204c gene in M. avium subsp. paratuberculosis encodes a predicted acyl-ACP desaturase with 100% homology to a predicted protein in M. avium subsp. avium (GenBank accession no. NC002943) and 78% identity to the desA1 gene product in M. avium subsp. paratuberculosis. It occurs at IS900 locus 5 and has a consensus site for insertion of IS900 within its coding sequence. The MAP2204c PCR primers flanking the insertion site used in the present study amplified a 1,838-bp product from bovine and ovine strains of M. avium subsp. paratuberculosis, indicating that locus 5 is occupied by the element (confirmed by sequencing), putatively inactivating MAP2204c. In the majority of human isolates of M. avium subsp. paratuberculosis that we have examined so far, these PCR primers produce a 387-bp product, indicating that IS900 locus 5 is stably unoccupied (there is no evidence of reversion in other human strains) and that the MAP2204c gene is probably expressed in these strains and not in bovine or ovine strains. The 387-bp PCR product amplified by MAP2204c PCR from the two amoebic cultures in which M. avium subsp. paratuberculosis was most abundant in the present study is consistent with the human origins of these cultures.
We previously found that a bovine strain of M. avium subsp. paratuberculosis was able to survive in planktonic form in fresh lake water microcosms for at least 2.5 years (33). In the present study native human M. avium subsp. paratuberculosis with the in vivo phenotype, obtained directly from human intestinal tissues, was able to persist within A. polyphaga for almost 4 years. The organisms were still intact and probably viable at the end of this period so that no upper limit on their persistence was established.
A histopathological diagnosis was established for all of the human surgical samples. When Ziehl-Neelsen (ZN) staining was carried out, no ZN-positive organisms were seen in the tissues. ZN staining of amoebic cultures is technically unsatisfactory. Eventual examination of IS900-positive amoebic cultures by auramine-rhodamine staining showed that in 6 the 13 IS900-positive cultures shown to contain M. avium subsp. paratuberculosis by in situ hybridization, the organisms had an auramine-rhodamine acid-fast bacilliary phenotype (43). This suggests that long-term intracellular incubation of M. avium subsp. paratuberculosis from human tissue may be able to induce conversion of the acid-fast phenotype from negative to positive. This phenotypic shift, possibly induced by long periods of latency, has previously been observed during conversion between paucimicrobial and multibacillary forms during M. avium subsp. paratuberculosis infection (Johne's disease) in cattle (14). Humans with Crohn's disease have been found to excrete M. avium subsp. paratuberculosis in their feces (16). If these pathogens are returned to the environment in the treated effluent of domestic sewage plants, they have the potential to persist for a long time.
Acknowledgments
We are most grateful to the surgical and other staffs of the West Wales Hospital Carmarthen, Prince Philip Hospital Llanelli, Singleton Hospital Swansea, and Llandough Hospital Cardiff for their help with tissue sampling. We thank CCAP for the gift of A. polyphaga.
This research was funded by MRC-NERC Initiative for “Environment and Health” grants G9901248 and NERC/A/S/1999/00119, to whom we express our appreciation.
REFERENCES
- 1.Anderson, O. R. 2000. Abundance of terrestrial gymnamoebae at a northeastern U. S. site: a four-year study, including the El Nino winter of 1997-1998. J. Eukaryot. Microbiol. 47:148-155. [DOI] [PubMed] [Google Scholar]
- 2.Angenent, L. T., S. T. Kelley, A. St. Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autschbach, F., S. Eisold, U. Hinz, S. Zinser, M. Linnebacher, T. Giese, T. Loffler, M. W. Buchler, and J. Schmidt. 2005. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn's disease. Gut 54:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axelsson-Olsson, D., J. Waldenstrom, T. Broman, B. Olsen, and M. Holmberg. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, J., and M. R. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 6.Barker, J., T. J. Humphrey, and M. W. Brown. 1999. Survival of Escherichia coli O157 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 7.Barker, J., P. A. Lambert, and M. R. Brown. 1993. Influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect. Immun. 61:3503-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker, J., H. Scaife, and M. R. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brieland, J. K., J. C. Fantone, D. G. Remick, M. LeGendre, M. McClain, and N. C. Engleberg. 1997. The role of Legionella pneumophila-infected Hartmannella vermiformis as an infectious particle in a murine model of Legionnaire's disease. Infect. Immun. 65:5330-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull, T. J., J. Hermon-Taylor, I. Pavlik, I., F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:3285. [DOI] [PubMed] [Google Scholar]
- 11.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke, C. J. 1997. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 116:217-261. [DOI] [PubMed] [Google Scholar]
- 15.Cocito, C., P. Gilot, M. Coene, K. M. de, P. Poupart, and P. Vannuffel. 1994. Paratuberculosis. Clin. Microbiol. Rev. 7:328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Prete, R., M. Quaranta, A. Lippolis, V. Giannuzzi, A. Mosca, E. Jirillo, and G. Miragliotta. 1998. Detection of Mycobacterium paratuberculosis in stool samples of patients with inflammatory bowel disease by IS900-based PCR and colorimetric detection of amplified DNA. J. Microbiol. Methods 33:105-114. [Google Scholar]
- 17.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falkinham, J. O., III. 2003. Mycobacterial aerosols and respiratory disease. Emerg. Infect. Dis. 9:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay, B. J., G. F. Esteban, K. J. Clarke, and J. L. Olmo. 2001. Biodiversity of terrestrial protozoa appears homogeneous across local and global spatial scales. Protist 152:355-366. [DOI] [PubMed] [Google Scholar]
- 20.Finlay, B. J., and T. Fenchel. 2004. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 155:237-244. [DOI] [PubMed] [Google Scholar]
- 21.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenstein, R. J., and M. T. Collins. 2004. Emerging pathogens: is Mycobacterium avium subspecies paratuberculosis zoonotic? Lancet 364:396-397. [DOI] [PubMed] [Google Scholar]
- 23.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermon-Taylor, J., T. J. Bull, J. M. Sheridan, J. Cheng, M. L. Stellakis, and N. Sumar. 2000. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521-539. [DOI] [PubMed] [Google Scholar]
- 25.Ly, T. M., and H. E. Muller. 1990. Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33:51-54. [DOI] [PubMed] [Google Scholar]
- 26.Manning, E. J., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev. Sci. Technol. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 27.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 28.Miltner, E. C., and L. E. Bermudez. 2000. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob. Agents Chemother. 44:1990-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molmeret, M., M. Horn, M. Wagner, M. Santic, and K. Y. Abu. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naser, S. A., G. Ghobrial, C. Romero, and J. F. Valentine. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039-1044. [DOI] [PubMed] [Google Scholar]
- 31.Newsome, A. L., T. M. Scott, R. F. Benson, and B. S. Fields. 1998. Isolation of an amoeba naturally harboring a distinctive Legionella species. Appl. Environ. Microbiol. 64:1688-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernthaler, J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537-546. [DOI] [PubMed] [Google Scholar]
- 33.Pickup, R. W., G. Rhodes, S. Arnott, K. Sidi-Boumedine, T. J. Bull, A. Weightman, M. Hurley, and J. Hermon-Taylor. 2005. Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the River Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl. Environ. Microbiol. 71:2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riemann, H. P., and B. Abbas. 1983. Diagnosis and control of bovine paratuberculosis (Johne's disease). Adv. Vet. Sci. Comp. Med. 27:481-506. [PubMed] [Google Scholar]
- 36.Robinson, B. S., S. S. Bamforth, and P. J. Dobson. 2002. Density and diversity of protozoa in some arid Australian soils. J. Eukaryot. Microbiol. 49:449-453. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Zaragoza, S., E. Mayzlish, and Y. Steinberger. 2005. Vertical distribution of the free-living amoeba population in soil under desert shrubs in the Negev desert, Israel. Appl. Environ. Microbiol. 71:2053-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogerson, A., and C. Gwaltney. 2000. High numbers of naked amoebae in the planktonic waters of a mangrove stand in southern Florida, USA J. Eukaryot. Microbiol. 47:235-241. [DOI] [PubMed] [Google Scholar]
- 40.Romero, C., A. Hamdi, J. F. Valentine, and S. A. Naser. 2005. Evaluation of surgical tissue from patients with Crohn's disease for the presence of Mycobacterium avium subspecies paratuberculosis DNA by in situ hybridization and nested polymerase chain reaction. Inflamm. Bowel Dis. 11:116-125. [DOI] [PubMed] [Google Scholar]
- 41.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz, D., I. Shafran, C. Romero, C. Piromalli, J. Biggerstaff, N. Naser, W. Chamberlin, and S. A. Naser. 2000. Use of short-term culture for identification of Mycobacterium avium subsp. paratuberculosis in tissue from Crohn's disease patients. Clin. Microbiol. Infect. 6:303-307. [DOI] [PubMed] [Google Scholar]
- 43.Sechi, L. A., A. M. Scanu, P. Molicotti, S. Cannas, M. Mura, G. Dettori, G. Fadda, and S. Zanetti. 2005. Detection and isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn's disease in Sardinia. Am. J. Gastroenterol. 100:1529-1536. [DOI] [PubMed] [Google Scholar]
- 44.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, S. J., L. J. Ahonen, F. A. de Leij, and J. W. Dale. 2003. Infection of Acanthamoeba castellanii with Mycobacterium bovis and M. bovis BCG and survival of M. bovis within the amoebae. Appl. Environ. Microbiol. 69:4316-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittington, R. J., D. J. Marshall, P. J. Nicholls, I. B. Marsh, and L. A. Reddacliff. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]