Abstract
Genetically altered or tagged Vibrio fischeri strains can be observed in association with their mutualistic host Euprymna scolopes, providing powerful experimental approaches for studying this symbiosis. Two limitations to such in situ analyses are the lack of suitably stable plasmids and the need for a fluorescent tag that can be used in tandem with green fluorescent protein (GFP). Vectors previously used in V. fischeri contain the p15A replication origin; however, we found that this replicon is not stable during growth in the host and is retained by fewer than 20% of symbionts within a day after infection. In contrast, derivatives of V. fischeri plasmid pES213 were retained by ∼99% of symbionts even 3 days after infection. We therefore constructed pES213-derived shuttle vectors with a variety of selectable and visual markers. To include a visual tag that can be used in conjunction with GFP, we compared seven variants of the DsRed2 red fluorescent protein (RFP): mRFP1, tdimer2(12), DsRed.T3, DsRed.T4, DsRed.M1, DsRed.T3_S4T, and DsRed.T3(DNT). The last variant was brightest, displaying >20-fold more fluorescence than DsRed2 in V. fischeri. RFP expression did not detectably affect the fitness of V. fischeri, and cells were readily visualized in combination with GFP-expressing cells in mixed infections. Interestingly, even when inocula were dense enough that most E. scolopes hatchlings were infected by two strains, there was little mixing of the strains in the light organ crypts. We also used constitutive RFP in combination with the luxICDABEG promoter driving expression of GFP to visualize the spatial and temporal induction of this bioluminescence operon during symbiotic infection. Our results demonstrate the utility of pES213-based vectors and RFP for in situ experimental approaches in studies of the V. fischeri-E. scolopes symbiosis.
Vibrio fischeri is a useful model for studies of bioluminescence, quorum sensing, and beneficial animal-bacterium interactions. Genetic tools have advanced each of these fields and have been especially important for studying the light organ symbiosis between V. fischeri and the Hawaiian bobtail squid, Euprymna scolopes. This squid acquires V. fischeri from its surroundings after hatching, allowing reconstitution of this association with wild-type or genetically altered V. fischeri. In such experiments, mutant analyses have revealed colonization mechanisms (1, 8, 11, 14-16, 38, 42, 44) and visualization of wild-type cells tagged with green fluorescent protein (GFP) has elucidated early infection events (28, 30, 31). GFP also holds great promise as an in situ reporter of gene activity, because cells expressing GFP can be visualized in the light organ. However, despite the utility of genetic tools in this model symbiosis, researchers still lack suitably stable plasmids and a useful fluorescent tag that is compatible with GFP.
To date, plasmids used to maintain genes in V. fischeri have contained the Escherichia coli plasmid p15A origin of replication (17, 39, 43). Although p15A-derived plasmids replicate in V. fischeri, they require antibiotic selection for plasmid maintenance, leading to at least two problems for conducting and interpreting symbiotic experiments. First, placing hatchlings in antibiotic-containing seawater likely leads to a gradient of antibiotic from peripheral to deep tissue and therefore uneven selective pressure. Second, antibiotic selection does not increase plasmid stability but rather inhibits the growth of plasmid-free cells, potentially causing an artificial situation where gene expression and/or growth ceases in a subset of symbionts. Perhaps for these reasons, we find that antibiotics alter the kinetics of symbiotic infection even for antibiotic-resistant plasmid-containing symbionts (unpublished data).
To overcome the limitations of p15A vectors, we characterized plasmid pES213 (12), which was isolated from a symbiotic V. fischeri strain (5) and is stably maintained in V. fischeri even in the absence of antibiotic pressure. We found that the pES213 replication origin resembles theta-type replicons and directs replication without requiring any plasmid-encoded proteins (12). Because certain common broad-host-range plasmids (e.g., RK2 and pBBR1) apparently do not replicate in V. fischeri (our unpublished data and K. Visick, personal communication), and considering the limited stability of p15A plasmids, we speculated that pES213-based plasmids would be ideal Vibrio/E. coli shuttle vectors.
To expand the usefulness of the vectors we developed, we also sought a visual tag that could be used alongside GFP. One candidate is red fluorescent protein (RFP) from Discosoma corals, because the relatively long-wavelength excitation and emission maxima of RFP make it compatible with GFP for multicolor labeling. Unfortunately, the original RFP “DsRed” allele (23) has several limitations, including slow or incomplete maturation and obligate tetramerization (2). Both DsRed and the improved DsRed2 yielded low and slow-developing fluorescence in V. fischeri, and we were unable to use them effectively in symbiotic studies. New DsRed variants have been developed to address these limitations in eukaryotes (3, 9, 36, 41), and these variants have the potential to become widely used in bacteria. However, they have not been tested in prokaryotes under isogenic conditions to identify the most useful version.
Our goal in this study was to explore the utility of pES213 derivatives and new RFP variants in V. fischeri. In this paper we report the efficacy of pES213-derived plasmids for symbiotic experiments with V. fischeri, we present a series of useful pES213-derived vectors, and we demonstrate that an improved RFP allele can be used in combination with GFP for strain-tagging or gene expression studies in the V. fischeri-E. scolopes symbiosis.
MATERIALS AND METHODS
Bacteria, media, and enzymes.
Except where noted, we used V. fischeri strain ES114, an isolate from E. scolopes (4), and cloning was performed in E. coli DH5α or DH5αλpir. Plasmids were transferred from E. coli to V. fischeri by triparental mating using conjugative helper strain CC118λpir(pEVS104) (39). V. fischeri was grown at 28°C either in LBS (37), which contained, per liter of water, 10 g of tryptone, 5 g of yeast extract, 20 g of NaCl, and 20 mM Tris-hydrochloride (Tris pH 7.5), or in SWT (4), which contained 5 g of tryptone, 3 g of yeast extract, 3 ml of glycerol, and 700 ml of Instant Ocean (Aquarium Systems, Mentor, Ohio) per liter. Agar (15 mg ml−1) was added to solidify media for plating. E. coli was grown in LB medium (26) at 37°C. Trimethoprim, chloramphenicol, kanamycin, and tetracycline were used at concentrations of 10, 20, 40, and 5 μg ml−1, respectively, for selection of E. coli and added to LBS at 10, 2, 100, and 5 μg ml−1, respectively, for selection of V. fischeri. To screen for lacZ expression in V. fischeri, LBS agar was supplemented with 50 μg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal). Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Beverly, Mass.). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, Iowa) or the University of Georgia Molecular Genetics Instrumentation Facility.
Molecular techniques.
PCR was performed on an iCycler (Bio-Rad Laboratories, Hercules, Calif.), using KOD HiFi DNA polymerase (Novagen, Madison, Wis.) except where noted. PCR products were purified using the DNA Clean and Concentrator-5 kit (Zymo Research, Orange, Calif.). Plasmids were isolated using QIAGEN (Valencia, Calif.) miniprep kits with optional washes in the manufacturer's protocol when V. fischeri was the host. Plasmid copy number per genome was determined with real-time PCR by comparing the abundances of aph, which encodes kanamycin resistance, on plasmids or on chromosome 2 of strain EVS401 (38) as previously described (12). DNA concentration was measured with a Brinkmann Instruments (Westbury, NY) BioPhotometer.
Construction of pES213-derived plasmids.
Each of the pES213-derived vectors described below contains a similar pES213 fragment. Specific cloning details have been included in Table S1 in the supplemental material. pVSV2 was constructed from the aph (kanamycin resistance)-carrying pES213-based vector pES213Kn (12) by adding gfp from pQBI63 (Quantum Biotechnologies, Montreal, Canada) downstream of the lac promoter from pGEM3Z (Promega, Madison, Wis.). pVSV3 was generated by adding the full-length E. coli lacZ to pES213Kn. pVSV4 was generated by cloning lacZα from pGEM3Z into pES213Kn and cloning a small linker to add restriction sites in lacZα. pVSV102, pVSV103, and pVSV104 were made from pVSV2, pVSV3, and pVSV4, respectively, by adding the R6Kγ replication origin. pVSV105 was made by deleting aph and some pES213 sequences from pES213C (12) and inserting lacZα from pGEM3Z. In pEVS106, the cat gene in pEVS105 was replaced with tetM, and lacZα from pBluescript SK+ (Stratagene, La Jolla, Calif.) was added. pVSV107 was made from pES213Tp (12) by adding the R6Kγ origin and lacZα from pBluescript SK+ (Stratagene). pVSV33 was constructed such that it contains a pES213Kn backbone and a reporter with three upstream transcriptional terminators, a multiple cloning site, promoterless cat and gfp genes as a reporter operon, and a downstream terminator. As part of another study, we identified a copy-up derivative of the replication origin, which replaced the origin in pVSV104 to generate pVSV104H. pVSV209 is composed of the rfp cassette from pVSV208 (described below) in pVSV33, and pVSV210 is identical to pVSV209 but has the V. fischeri luxIR intergenic region cloned so that the luxICDABEG promoter is driving cat and gfp expression.
Allelic variants of RFP were obtained or generated by PCR and cloned with isogenic upstream sequences into pVSV105. pDsRed2 was obtained from Clontech (Mountain View, Calif.). pDSRed.T3-N1, pDsRed.T4-N1, and pDsRed.M1-N1 were obtained from B.S. Glick (3, 41), although the DSRed.M1 allele is now also available from Clontech. pRSETb containing genes encoding mRFP1 or tdimer2(12) were obtained from R.Y. Tsien (9). pDsRed.T3-N1 was used as a template for PCR to generate the DsRed.T3_S4T and DsRed.T3(DNT) alleles. A two-step PCR protocol was used to amplify each DsRed gene and to change the upstream region between the lac promoter and ATG start codon (in boldface) to 5′-GCA TGC TAA GAA GGA GAT ATA CAT ATG-3′. Primers (Table 1) used for the first PCR step were F1a and R1 (DsRed2, DsRed.T3, and DsRed.T4), F1a and R2 [mRFP1 and tdimer2(12)], F1b and R1 (DsRed.T3_S4T), F1c and R3 (DsRed.M1), and F1d and R1 [DsRed.T3(DNT)]. The PCR products were purified, diluted 1,000-fold, and used as templates for a second PCR step using the primer sets F2a and R1 (DsRed2, DsRed.T3, DsRed.T4, and DsRed.T3_S4T), F2a and R2 [mRFP1 and tdimer2(12)], F2b and R3 (DsRed.M1), and F2c and R1 [DsRed.T3(DNT)]. These PCR products were purified, SphI digested, and directionally cloned into HincII- and SphI-digested pVSV105. The plasmid expressing DsRed.T3(DNT) was designated pVSV207, and a spontaneous mutant of pVSV207 with higher fluorescence was designated pVSV208. Each insert was sequenced at the University of Michigan DNA Sequencing Core Facility to verify upstream and coding sequences.
TABLE 1.
Oligonucleotides used to generate and clone variant rfp alleles
| Primer | Sequence (5′→3′) |
|---|---|
| F1a | AGGAGATATACATATGGCCTCCTCCGAGGA |
| F1b | AGGAGATATACATATGGCCTCCACCGAGGA |
| F1c | AGGAGATATACATATGGACAACACCGAGGA |
| F1d | AGGAGATATACATATGGACAACACCGAGGA |
| F2a | GGGTGCATGCTAAGAAGGAGATATACATAT GGC |
| F2b | GGGTGCATGCTAAGAAGGAGATATACATAT GGA |
| F2c | GGGTGCATGCTAAGAAGGAGATATACATAT GGA |
| R1 | AAAAGGATCCCCGCTACAGGAACAGGTG |
| R2 | GGTTGGTACCGGATCAAGCTTCGAATTC |
| R3 | AAAAGGATCCCCGCTACTGGGAGCCGGA |
Plasmid stability and in-culture competition.
To assess the stability of pEVS126, an aph (kanamycin resistance)-carrying p15A-based vector, and pES213Kn (12), overnight cultures of plasmid-carrying cells were diluted 100-fold in LBS containing kanamycin and grown to an optical density at 595 nm (OD595) of approximately 0.5, which was designated “generation 0.” The cells were washed, diluted 210-fold in nonselective LBS medium, and regrown to the starting OD595 of 0.5, which requires 10 generations. This process was repeated, and after 30 and 60 generations cells were plated onto LBS agar and incubated overnight. Isolated colonies were patched onto LBS agar with and without kanamycin to determine the percentage of cells retaining the plasmid of interest. Plasmid stability in the symbiosis was determined by patch plating colonies recovered from homogenized squid. The competitiveness of two strains in mixed culture was similarly assessed, with the number of generations in coculture determined by starting “generation 0” of a mixed culture at a set OD595, diluting 25-fold, regrowing through five generations to reachieve the initial OD595, plating “generation 5,” and repeating the dilution, regrowth, and plating process for subsequent generations. Strain ratios were determined by screening colonies for blue color, which is indicative of the plasmid-borne lacZ tag, after plating on LBS medium containing X-Gal.
Determination of fluorescence intensities of DsRed variants.
To prepare log-phase cells carrying DsRed variants, overnight cultures were diluted 100-fold in fresh LBS, grown to an OD595 of 1.0, pelleted, and resuspended in filter-sterilizedInstant Ocean. To prepare stationary-phase cells, fresh overnight cultures grown in LBS were pelleted, resuspended, and diluted to an OD595 of 1.0 in filter-sterilized Instant Ocean. Resuspended cultures were placed on ice, and the relative fluorescence intensity for 1 ml of cells was determined using a TD-700 fluorometer (Turner Designs, Sunnyvale, Calif.) equipped with a 550-nm excitation filter (550FS10-25) and a >570-nm band pass emission filter. The optimal range and sensitivity of the fluorometer were set using multioptional mode raw-fluorescence calibration, with stationary-phase cells expressing DsRed.T3_S4T used as the 900-unit standard and cells harboring pVSV105 used as the zero-unit blank. To examine cells expressing the brightly fluorescent DsRed.T3(DNT) in pVSV207 and pVSV208, the fluorometer was calibrated as described above except that the standard was calibrated to 100 units and the readings were converted to the same scale as the other data by multiplying them by nine. Fluorescence readings of cells expressing DsRed2 and DsRedT3_S4T taken with the standard set to 100 units and converted were identical to readings taken with the standard set to 900 units, validating this conversion method. Each experiment was repeated three times using independent cultures.
Symbiosis assays.
V. fischeri were grown in SWT to an OD595 of between 0.4 and 0.8 and diluted to between 1,000 and 7,000 CFU ml−1 in inocula. To generate infections with both GFP- and RFP-labeled bacteria, juvenile E. scolopes hatchlings were exposed to mixed inocula with roughly equal numbers of pVSV102- and pVSV208-containing cells for 12 to 14 h before being rinsed in inoculum-free seawater. Ratios of RFP- to GFP-expressing V. fischeri were determined, after dilution plating, either by patching on differentially selective media or by scoring the number of red and green colonies. Relative competitive indices (RCIs) were calculated by dividing the ratio of pVSV102- to pVSV208-containing V. fischeri in the squid after 48 h by the ratio in the inoculum. A one-sample t test was applied to log-transformed RCIs to determine if RFP-expressing cells were significantly outcompeted. Five independent experiments yielded similar results, and the data were combined. In separate experiments, GFP- and RFP-marked symbionts were visualized using a Zeiss (Thornwood, NY) LSM510 laser-scanning confocal microscope or a Nikon (Melville, NY) Eclipse E600 epifluorescence microscope. The latter was equipped with a Nikon 96165 tricolor filter cube and a Nikon Coolpix 5000 camera to obtain images of RFP- and GFP-tagged bacteria in animals stained with CellTracker Blue CMF2HC (Molecular Probes, Eugene, Oreg.).
To examine gfp expression driven by the lux promoter, squid were inoculated with ES114 harboring pVSV210 within 3 h of hatching. At each time point, between three and eight animals were removed and anesthetized with 2% ethanol in mPBS (500 mM NaCl, 50 mM NaH2PO4, pH 7.4). A subset of the animals was fixed in mPBS supplemented with 4% formaldehyde, and the remaining animals were viewed immediately by confocal microscopy prior to fixation. All animals were removed from fixative within 4 h, washed twice in mPBS, and stored in mPBS at 4°C for up to 2 weeks. Light organs were then dissected, washed in mPBS twice for 10 min, permeabilized in 1% Triton X-100 in mPBS, incubated with 16 μg ml−1 rhodamine phalloidin (Molecular Probes) and 0.5% Triton X-100 in mPBS for 18 h at 4°C, and washed twice in mPBS for 10 min prior to viewing. Stained light organs were viewed by laser-scanning confocal microscopy within 2 days of mounting to locate symbionts with green or red fluorescence.
RESULTS
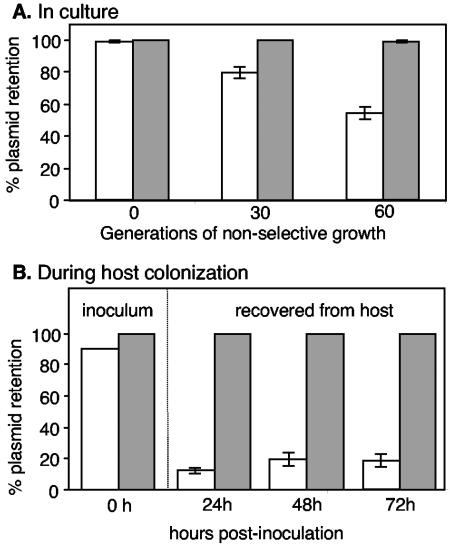
The goal of this study was to improve upon the p15A-based plasmids currently used to introduce genes into V. fischeri. We recently described V. fischeri plasmid pES213 and reported that it was stable in culture-grown V. fischeri (12). We therefore compared the stabilities of pES213Kn and pEVS126, which are derived from pES213 and p15A, respectively, during nonselective growth in culture and during colonization of the E. scolopes light organ. pES213Kn was more stable than pEVS126 under both conditions (Fig. 1), and this was particularly evident during the first 24 h of symbiosis (Fig. 1B), a period when a small number of bacteria from the inoculum colonize the host and grow through many generations to establish the infection (24, 34, 42). Moreover, pES213Kn was rarely lost from cells either in culture or in the light organ and was retained by ≥99% of V. fischeri cells at the end of each experiment. pES213Kn and pEVS126 are both small plasmids that encode kanamycin resistance, and they differ primarily in their replication origins. Other pES213- and p15A-derived vectors also exhibited stabilities similar to those of pES213Kn and pEVS126, respectively (data not shown). These findings suggested that the pES213 origin is superior to the p15A origin for use in V. fischeri and that pES213-derived plasmids would be good candidates to replace or augment existing p15A-derived vectors.
FIG. 1.
Stability of pEVS126 (unshaded bars) and pES213Kn (shaded bars) in V. fischeri after growth in nonselective LBS medium (A) or during squid colonization without kanamycin selection (B). Panel B includes the percent plasmid retention in cells from the inoculum that squid were exposed to and in cells recovered from colonized squid. Error bars (some too small to visualize) indicate standard errors; n = 3 (cultures) in panel A, and n = 8 (squid) at the 24-h, 48-h, and 72-h time points in panel B.
To capitalize on pES213's stability in V. fischeri, we generated a series of pES213-based pVSV “vibrio shuttle vectors” (Table 2). Each of these plasmids contains the RP4 origin of transfer, oriT, enabling conjugative transfer from E. coli to V. fischeri by using a RP4-derived helper plasmid (39). Unfortunately, the pES213 replication origin functions poorly in E. coli, resulting in plasmid instability and selective pressure for alterations to the origin. We therefore engineered the vectors in Table 2 to contain the gamma replication origin from plasmid R6K, which directs their stable replication in E. coli strains that contain the pir gene (18). Plasmid stability in V. fischeri was unaltered by the R6Kγ replication origin or by the antibiotic resistance and marker genes that we added, and the stability of pES213Kn is representative of those of all the plasmids in Table 2 (data not shown).
TABLE 2.
Selected plasmids generated in this studya
| Use and plasmid | Descriptionb |
|---|---|
| Complementation and gene introduction | |
| pVSV104 | Knr, lacZα-(SphI, AvrII, HpaI, SalI, NcoI, KpnI, SacI) |
| pVSV105 | Cmr, lacZα-(SphI, SalI/HincII, XbaI, SmaI/XmaI, KpnI, SacI) |
| pVSV106 | Tcr, lacZα-(ApaI, XhoI, SalI/HincII, SmaI/XmaI, XbaI) |
| pVSV107 | Tpr, lacZα-(KpnI, ApaI, XhoI, SalI, SpeI, XbaI, NotI, SacI) |
| Copy number variant | |
| pVSV104H | pVSV104 with single-base-pair change in origin and increased copy number |
| Strain tagging | |
| pVSV102 | Knr, gfp |
| pVSV103 | Knr, lacZ |
| pVSV208 | Cmr, rfp |
| Gene expression analyses | |
| pVSV33 | Knr, transcriptional terminators-(SphI, AvrII, KpnI, XbaI, SalI, StuI)-promoterless Cmr and gfp |
| pVSV209 | Knr, rfp, transcriptional terminators-(AvrII, SalI, StuI)-promoterless Cmr and gfp |
| pVSV210 | Knr, rfp, Plux-cat-gfp |
Other plasmids are described in Materials and Methods.
All plasmids in this table contain the RP4 oriT and the pES213 and R6Kγ replication origins. Knr, kanamycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance. Parentheses bracket unique restriction sites present in the multiple cloning site.
We generated four vectors that are broadly useful for cloning genes and introducing them into V. fischeri; pVSV104, pVSV105, pVSV106, and pVSV107 (Table 2). These plasmids each contain a multiple cloning site within a lacZα fragment, which allows blue/white screening for the presence of a cloned insert upon transformation of an E. coli strain with the complementary ΔlacZM15 fragment. These plasmids yielded only a pale blue color in DH5αλpir, a limitation offset somewhat by high X-Gal concentrations (≥100 μg ml−1). At least for transformants with pVSV107, blue color was also enhanced in E. coli strains BW20338 and BW22245 (25), which contain the pir-116 allele and maintain R6K-based plasmids in higher copy. To provide a choice of selectable markers, these pVSV vectors contain aph, cat, tetM, or dfr, which encode resistance to kanamycin, chloramphenicol, tetracycline, or trimethoprim, respectively (Table 2). To provide flexibility with respect to gene dosage, we constructed pVSV104H (Table 2), which has a mutation in the “inverted repeat 2” region of the pES213 replication origin (12) and is maintained at higher copy number than parental plasmid pVSV104 in V. fischeri (Fig. 2).
FIG. 2.
High-copy pES213-derivative pVSV104H. (A) Single-base-pair difference (highlighted) between pVSV104 and pVSV104H, which resides within “inverted repeat 2” of the replication origin (12). (B and C) Abundances of pVSV104 and pVSV104 determined by yields from plasmid preparations (B) or real-time PCR (C). Error bars in panels B and C indicate standard errors; n = 5 and n = 6, respectively.
We also generated pES213-derived vectors that carry easily identified tags (Table 2). For example, pVSV103 carries the E. coli lacZ gene, and V. fischeri cells harboring pVSV103 can be identified as blue colonies on media containing X-Gal. Researchers often mix two V. fischeri strains and monitor their relative competitiveness by measuring the ratio of the strains throughout an experiment (13, 19, 21, 24, 28, 38), and the rationale for generating pVSV103 was to provide a rapid method for determining the strain ratios in a mixture. Usually, the strain ratio has been determined by patch plating on differentially selective media; however, by marking one strain with pVSV103, the strain ratio can be determined by counting blue and white colonies.
We tested the efficacy of using pVSV103 as a strain marker in an experiment assessing the competitiveness of recA mutant EVS510 (12) relative to its wild-type parent ES114 in mixed culture (Fig. 3). We found that EVS510 was outcompeted by ES114 at a rate consistent with the 10 to 15% reduced growth rate that we had previously reported for recA mutants in clonal cultures (12). More importantly, results were similar whether EVS510 or ES114 was marked with pVSV103, and ES114 did not outcompete pVSV103-tagged ES114, indicating that loss of pVSV103 or secondary effects of pVSV103 on competitiveness did not skew the results (Fig. 3). We also found that pVSV103-marked colonies developed a deeper blue color and were easier to score if plates were shifted to 37°C, although V. fischeri does not grow at this temperature.
FIG. 3.
Marking strains with pVSV103 to determine strain ratios in mixed cultures. Wild-type strain ES114 and recA mutant EVS510 (12) were marked with pVSV103, which carries lacZ, and mixed in ∼1:1 ratios with unmarked EVS510 or ES114, and the strain ratios were determined during coculturing based on blue/white colony counts on LBS agar containing X-Gal. Relative competitiveness indices represent the strain ratio at each plating divided by the ratio in the inoculum (generation 0) for mixtures of ES114(pVSV103) and ES114 (filled squares), EVS510(pVSV103) and ES114 (filled circles), or EVS510 and ES114(pVSV103) (open circles). The gray line indicates an RCI of 1, which defines no relative competitive defect. The dotted line indicates the calculated RCI values of a strain growing at 90% of the rate of the strain it is competing with.
GFP has been another effective tag for visually identifying V. fischeri cells (28, 30, 31, 40), and we therefore constructed gfp-carrying plasmid pVSV102 (Table 2). The fluorescence intensity of V. fischeri cells harboring pVSV102 was comparable to that of cells harboring the p15A-based gfp delivery vector pKV111 (40), and there appeared to be less cell-to-cell variability in fluorescence using pVSV102 (data not shown). Cells harboring pVSV102 were readily observed in the light organ, but lacking another visual tag, we could not observe two strains simultaneously in the host. We therefore sought to develop RFP as a complementary tag for GFP.
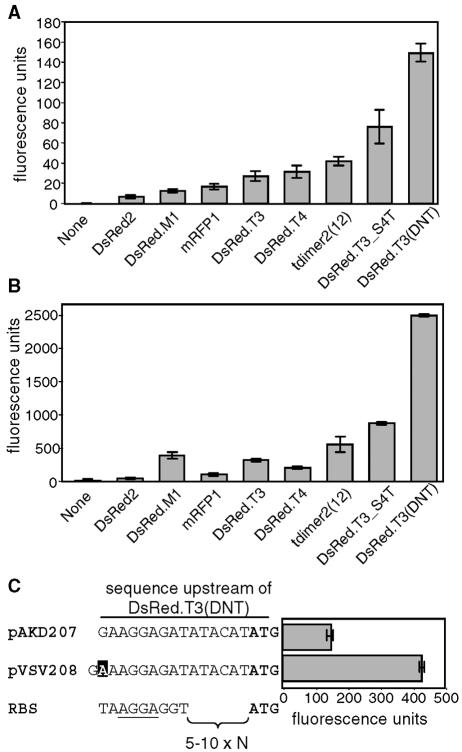
To identify an RFP tag that could be used in conjunction with GFP, we compared the fluorescence intensities of eight variants of DsRed (Table 3) encoded on isogenic plasmids. We found that DsRed.T3(DNT) was the most fluorescent RFP variant during log- or stationary-phase growth in V. fischeri, displaying 21- or 54-fold-higher fluorescence intensity, respectively, than DsRed2 (Fig. 4). While constructing the DsRed.T3(DNT)-encoding plasmid pVSV207, we identified a more fluorescent clone. The corresponding variant plasmid was named pVSV208 and contained a single-nucleotide insertion in the putative ribosome-binding site upstream of the start codon (Fig. 4C). This mutation results in a threefold increase in fluorescence intensity in V. fischeri cells during log-phase growth (Fig. 4C) or in stationary phase (data not shown). Furthermore, the red fluorescence of V. fischeri cells tagged with either pVSV207 or pVSV208 was easily visualized over the autofluorescence of the squid tissue, and we therefore chose the brighter of these, pVSV208 (Table 2), to provide another visual marker for identifying V. fischeri strains in situ.
TABLE 3.
Red fluorescent protein derivatives tested
| Protein | Relevant characteristics | Source or reference |
|---|---|---|
| DsRed2 | Tetramer, 6 amino acid substitutionsa | Clontech |
| DsRed.M1 | Monomer, 46 amino acid substitutionsa (including A2D, S3N, and S4T) | 41 |
| mRFP1 | Monomer, 33 amino acid substitutions,a 25-nm red-shifted excitation and emission wavelengthsa | 9 |
| DsRed.T3 | Tetramer, 8 amino acid substitutions,a enhanced folding,a increased excitation at 488 nm,b brighter fluorophoreb | 3 |
| DsRed.T4 | Tetramer, 9 amino acid substitutions,a approximately two-fold-faster maturationc | 3 |
| tdimer2(12) | Tandem dimer, 17 amino acid substitutionsa | 9 |
| DsRed.T3_S4T | DsRed.T3 with a S4T substitution, increase in fluorescence intensityc | 36 |
| DsRed.T3(DNT) | DsRed.T3_S4T with A2D and S3N substitutions, additional increase in fluorescence intensity | 41 |
FIG. 4.
Fluorescence intensities of DsRed variants in V. fischeri. (A and B) Red fluorescence of V. fischeri carrying DsRed alleles when cells were growing in mid-log phase at an OD595 of 1 (A) or grown overnight to stationary phase and diluted to an OD595 of 1 (B). Data are means with standard errors (n = 3). (C) Sequence upstream of the DsRed.T3(DNT) allele in pVSV207 and pVSV208 and the fluorescence of V. fischeri cells carrying these plasmids during log-phase growth (OD595 of 1). The single-base-pair difference is highlighted, the ATG start codon is in boldface, and a consensus ribosome-binding site (RBS) with the most conserved nucleotides underlined is shown for comparison (35).
Ideally, tagging cells should not decrease their fitness. We found that pVSV102 and pVSV208 did not detectably affect the growth rate or morphology of cultured V. fischeri cells relative to wild-type cells or to cells containing the respective parent vectors (data not shown). Furthermore, 98 to 99% of V. fischeri cells retained pVSV102 or pVSV208 after nonselective growth for 40 generations in culture, or after 48 h of squid colonization, consistent with these plasmids both being stable in V. fischeri and not imposing a selective disadvantage. To further test for effects of these tags during host colonization, we exposed juvenile squid to mixed inocula containing both GFP- and RFP-expressing V. fischeri and determined the ratio of strains present in the squid 48 h postinoculation. GFP- and RFP-expressing V. fischeri competed equally well in these mixed infections (Fig. 5A), indicating that these tags can be used to distinguish between two strains without compromising the relative fitness of either strain.
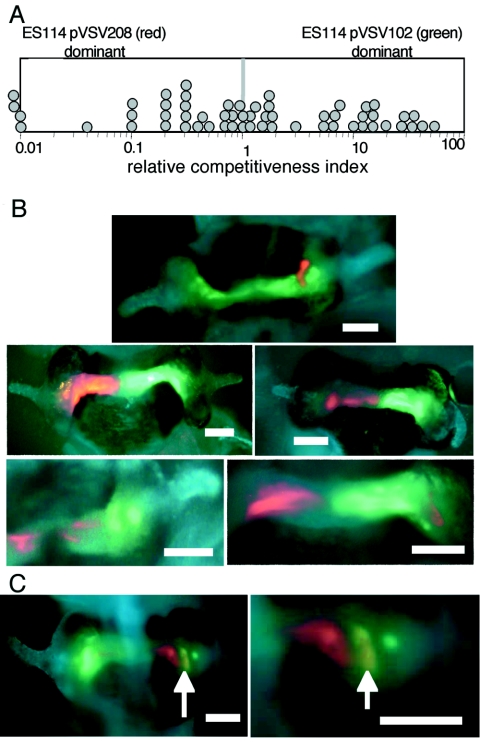
FIG. 5.
Competition during mixed infection by V. fischeri ES114 derivatives carrying pVSV102 (GFP) or pVSV208 (RFP). Juvenile squid were exposed to a ∼1:1 mixture of the strains at a total concentration of 2,000 to 7,000 CFU ml−1 for 14 h. (A) The relative competitiveness index of ES114(pVSV102), defined as the ratio of ES114(pVSV102) to ES114(pVSV208) in the squid divided by the ratio of these strains in the inoculum, was determined for bacteria in each animal 48 h after inoculation and plotted as a circle. Symbols to the left of the frame indicate that the RCI was <0.01. The average RCI of 1.2 (n = 57) was not significantly different from 1.0 (P > 0.25). (B and C) Epifluorescence images of E. scolopes light organs coinfected with ES114(pVSV102) (green) and ES114(pVSV208) (red), with animal tissue stained with CellTracker Blue (blue). Bars are 100 μm long. The light organs of five typical animals are shown in panel B, with red- and green-tagged cells appearing segregated. The light organ of one animal is shown at two magnifications in panel C, revealing a large pocket of mixed infection by red- and green-tagged cells appearing yellow (indicated by arrows).
We microscopically observed GFP- and RFP-expressing V. fischeri cells in squid light organs during such competition experiments and found that the two strains were predominantly segregated even though most animals were infected by both strains (Fig. 5B). In four similar experiments, both GFP- and RFP-expressing cells were readily visualized in 43 of a total of 51 animals observed; however, only 11 animals had a region that was clearly infected by a mixture of both strains (Fig. 5C). Even in animals where a pocket of mixed infection was visible, such as the animal shown in Fig. 5C, most symbionts in the light organ appeared to be segregated.
In these dual-label coinoculation experiments, the majority of the animals were viewed by epifluorescence microscopy, and our ability to discern individual cells was limited to the most peripheral symbiotic tissues. It was therefore difficult to judge the strictness of segregation. However, laser-scanning confocal microscopy performed on seven animals indicated that single crypts could be colonized clonally, by one strain or the other, even in a coinfected animal. Although we could not precisely quantify the extent of mixing, we estimate that fewer than 10% of symbionts with one label were closely associated with cells carrying the other label. This unexpected result illustrates that GFP and RFP markers on vectors pVSV102 and pVSV208 can be used in situ to elucidate fundamental spatial dynamics of symbiotic infection.
We also developed vectors that contain GFP and RFP markers to help elucidate gene expression in situ (Table 2). pVSV33 and pVSV209 each contain a transcriptionally isolated and promoterless cat-gfp reporter operon with an upstream multiple cloning site. Promoters cloned into the multiple cloning site will drive cat and gfp expression, with the promoter strength determining the degree of chloramphenicol resistance and green fluorescence. pVSV209 also carries a constitutive RFP marker, so that cells harboring this plasmid can be visualized in situ even when GFP is not expressed. In this way, symbionts could be localized by red fluorescence and specific gene expression could be determined by measuring green fluorescence.
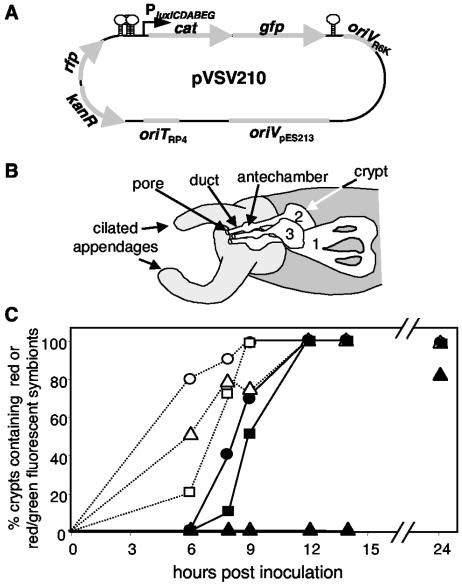
To test this approach, we cloned the promoter for the luxICDABEG bioluminescence operon upstream of the cat-gfp reporter in pVSV209, forming pVSV210 (Fig. 6A). In strain ES114 luminescence is expressed at very low levels in culture but is induced by quorum-sensing-dependent transcriptional regulation once symbionts reach a high cell density in the host light organ (4, 20, 34). Therefore, we predicted that during infection with ES114 carrying pVSV210, initial colonists should appear red fluorescent only, but they should appear red and green fluorescent shortly thereafter as they fill the light organ. However, we could not predict how lux expression might change as symbionts are exposed to different microenvironments as they pass through pores, ducts, and antechambers into three distinct crypts on each of the two light organ lobes (Fig. 6B).
FIG. 6.
In situ visualization of lux promoter expression by using pVSV210. (A) Map of pVSV210, which contains a constitutive RFP and has GFP expression driven by the luxICDABE promoter. Stem-loops indicate bidirectional transcriptional terminators flanking the reporter cassette. (B) Schematic representation of one lobe of a two-lobed light organ. Unshaded regions represent internal symbiotic tissues, with three distinct pores leading through ducts to widened antechambers and then into crypts where most symbionts ultimately reside. The three crypt types (29) are designated with numbers. (C) Rate of appearance of red-fluorescent symbiont cells (open symbols) or red- and green-fluorescent cells (filled symbols) in crypt 1 (circles), crypt 2 (squares), and crypt 3 (triangles). The light organs of between three and eight animals were visualized at each time point.
Using laser-scanning confocal microscopy, we visualized the fluorescence of ES114 cells carrying pVSV210 in different microenvironments during initiation of infection. As predicted, red-fluorescent symbionts lacking green fluorescence were detected first (Fig. 6C), and were visualized in the pores, ducts, antechambers, and crypts within 6 h after inoculation. From 6 h to 12 h postinoculation, crypts that contained cells expressing gfp, indicative of lux promoter activation, were observed (Fig. 6C). This corresponds well with the timing of luminescence induction (24, 34, 42). Surprisingly, this included only cells in crypt types 1 and 2. We did not see induction of the Plux-cat-gfp reporter in colonists of crypt 3 until later (Fig. 6C), even though the timing of crypt 3 colonization (Fig. 6C) and the density of symbionts appeared similar to those for the colonization of crypts 1 and 2. We also unexpectedly observed induction occurring asynchronously within a crypt, with cells expressing GFP in one region and not in another part of the same crypt. We did not observe Plux-cat-gfp-expressing cells in the ducts or antechambers until after the first morning, when E. scolopes expels most symbionts in the light organ back into the environment (6), and these GFP-expressing cells may have been pushed into the antechambers and ducts from deeper in the crypts. This observation could be explained by quorum-sensing regulation of the lux promoter, as symbionts did not achieve high cell densities in the ducts or antechambers. These data illustrate the utility of the reporter vectors we have constructed for analyzing V. fischeri gene expression patterns in situ.
DISCUSSION
In this study we found that pES213-derived plasmids are more stable than p15A-derived plasmids in V. fischeri (Fig. 1). This difference in stability may relate to the plasmids' evolutionary origins, in that pES213 is native to V. fischeri (12) whereas p15A was isolated from E. coli (10). Although plasmids derived from p15A have been used in V. fischeri in conjunction with antibiotic selection, this could pose complications in symbiotic studies (see the introduction). Such problems can be obviated by using pES213-derived vectors, which are stable during colonization of the squid (Fig. 1B). We therefore generated a series of pES213-derived shuttle vectors (Table 2) that will be valuable tools for studying V. fischeri and its association with E. scolopes. These include vectors containing various antibiotic resistance markers that can be used in complementation or other gene introduction experiments (Table 2), an increased-copy-number variant (Table 2 and Fig. 2), and vectors carrying tags for identifying strains (Table 2). Marking strains with lacZ constitutes a facile new approach for determining V. fischeri strain ratios in a mixed population (Fig. 3).
A major goal of this work was to develop RFP as a fluorescent tag that could be used in tandem with GFP in dual-labeling experiments. In preliminary studies we were unable to detect DsRed2 in V. fischeri above the background autofluorescence of the host squid tissue. A similar problem was reported with Salmonella enterica serovar Typhimurium, where DsRed expression could not be detected over autofluorescent mouse tissue by using flow cytometry (36). However, upon testing several newer DsRed alleles (Table 3), we found brighter variants. DsRed.T3(DNT) was the most fluorescent in both log- and stationary-phase V. fischeri cells and was clearly visible by epifluorescence microscopy in each growth phase. This bright fluorescence is consistent with reports that the S4T, A2D, and S3N substitutions near the 5′ end of the gene in this variant enhance expression in prokaryotes (36; D. Strongin and B. Glick, personal communication). Each of the variants yielded greater fluorescence in stationary-phase cells (Fig. 4B) than in log-phase cells (Fig. 4A), suggesting that RFP maturation is still a limiting factor. Given this lengthy maturation, these RFP variants may not make suitable reporters for gene expression studies.
Despite the power of tagging bacteria with fluorescent labels and the limitations of DsRed2, we are not aware of another study that has compared these new DsRed variants in prokaryotes under isogenic expression conditions. Although our comparison was performed in V. fischeri, qualitatively similar results were seen with these constructs in E. coli (data not shown), suggesting that our results should be useful for researchers interested in utilizing RFP in other proteobacteria. Moreover, while DsRed.T3(DNT) was the optimal variant for our current purposes based on its superior fluorescence intensity, other variants may also be useful. For example, monomeric forms of DsRed might be preferable for generating translational fusions for protein tagging within cells. Based on our fluorescence assays (Fig. 4A and B), DsRed.M1 would be the preferred monomer for translational fusions and may be a useful tool for future applications involving protein localization in V. fischeri and other bacteria.
We found no selective disadvantage of pVSV102 or pVSV208 carriage in V. fischeri and were therefore able to mark strains with GFP or RFP without significantly attenuating the strains. In other proteobacteria, GFP (33) or RFP (22) expression can impose a fitness cost; however, either V. fischeri reacts differently to these proteins or GFP and RFP expression levels are lower under our experimental conditions. If the fitness costs of DsRed2 are due to the abundance of unfolded protein and/or the high levels of protein required to visualize fluorescence, then the improved maturation rate and high specific fluorescence of DsRed.T3(DNT) may account for why its expression in V. fischeri did not seem detrimental to cells.
Using GFP- and RFP-labeled wild-type cells, we visualized two strains simultaneously infecting E. scolopes individuals (Fig. 5B and C). Our results were consistent with many experiments wherein E. scolopes was presented with mixed inocula of thousands of CFU per milliliter and most animals became coinfected (1, 13, 19, 21, 24, 27, 28, 38, 42). However, in the current study we were able to visually determine that the two strains were largely segregated even when infecting the same animal, and we estimate that fewer than 10% of the cells with one marker were in close proximity to cells carrying the other marker. This discovery has important implications for interpreting previous experiments with mixed inocula. For example, we reported that conjugative plasmid transfer occurred at equal frequencies in culture and in the host light organ (12); however, our current data suggest that most donor and recipient cells were segregated in the light organ. Thus, the conjugative frequency may have actually been much higher in the light organ if only recipients in contact with potential donors were considered. Our finding that initial colonists tend to be segregated also parallels a similar phenomenon observed in the establishment of Xenorhabdus nematophila in its symbiotic host nematode Steinernema carpocapsae (22) and may reflect a larger trend in the establishment of highly specific animal-bacterium interactions. Future work will help determine whether the patchiness of initial V. fischeri infections represents a founder effect or whether mechanisms in the host or bacteria actively maintain regional clonality.
Dual labeling with GFP and RFP also proved useful for analyzing gene expression in situ. Using pVSV210 (Table 2 and Fig. 6A), we visualized symbionts based on their constitutive red fluorescence while also monitoring lux promoter activity based on green fluorescence from a Plux-cat-gfp reporter. Colonization preceded Plux induction (Fig. 6C), consistent with previous observations (34), but there was an unexpected delay before Plux induction for cells colonizing crypt 3 (Fig. 6C). Crypt 3 is the smallest crypt type, and its development in embryos and hatchlings lags behind that of crypts 1 and 2 (29). Because crypt 3 is smaller, a greater proportion of colonists are proximal to the animal epithelium, and if autoinducer is removed from the infection via the epithelial cells, this could result in delayed autoinduction of luminescence in crypt 3. It is also possible that the host cells provide a signal that triggers the feed-forward autoinduction cycle in V. fischeri and that this signal develops later in crypt 3, resulting in delayed onset of luminescence. Whatever the mechanism, our results demonstrate that the host microenvironment can influence gene expression and that we can monitor this in situ using pVSV209.
The pES213-derived plasmids described here should have value beyond their use in V. fischeri. pVSV2 (originally designated pMSB6) is stable in Vibrio campbellii and was used as a marker in strain 90-69B3 to monitor clearance of this pathogen from tissues of the Pacific white shrimp (7). We also found that pES213Kn was at least as stable in Vibrio splendidis ATCC 33869 as it is in V. fischeri (data not shown). Interestingly, the pES213 replication origin (12) shares similarity with a plasmid from Vibrio shiloi (GenBank accession number AF009904) and with pSIO1, which was recently isolated from Vibrio cholerae (32). This raises the intriguing possibility that pES213 represents a widespread family of vibrio-borne plasmids. Given its remarkable stability in the three vibrios tested to date, it seems likely that pES213 derivatives will be useful for studying many of the fascinating and important species encompassed by the Vibrionaceae.
Supplementary Material
Acknowledgments
We thank Melissa Butler for technical assistance and Roger Tsien and Karen Visick for providing plasmids. We are especially grateful to Benjamin Glick for generously providing DsRed variants prior to publication and for helpful advice.
This work was supported by a career award from the National Science Foundation (MCB-0347317), by the National Institutes of Health (grant R01 AI 50661), and by the University of Georgia Research Foundation.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87.11753367 [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., and E. G. Ruby. 1994. Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Curr. Microbiol. 29:279-286. [Google Scholar]
- 6.Boettcher, K. J., E. G. Ruby, and M. J. McFall-Ngai. 1996. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. 179:65-73. [Google Scholar]
- 7.Burgents, J. E., L. E. Burnett, E. V. Stabb, and K. G. Burnett. 2005. Localization and bacteriostasis of Vibrio introduced into the Pacific white shrimp, Litopenaeus vannamei. Dev. Comp. Immunol. 29:681-691. [DOI] [PubMed] [Google Scholar]
- 8.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182:2811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLoney, C. R., T. M. Bartley, and K. L. Visick. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J. Bacteriol. 184:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 13.Fidopiastis, P. M., C. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 14.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 17.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 19.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 22.Martens, E. C., K. Heungens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matz, M. V., A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969-973. [DOI] [PubMed] [Google Scholar]
- 24.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery, M. K., and M. J. McFall-Ngai. 1993. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol. Bull. 184:296-308. [DOI] [PubMed] [Google Scholar]
- 30.Nyholm, S. V., and M. J. McFall-Ngai. 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl. Environ. Microbiol. 69:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purdy, A., F. Rohwer, R. Edwards, F. Azam, and D. H. Bartlett. 2005. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J. Bacteriol. 187:2992-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rang, C., J. E. Galen, J. B. Kaper, and L. Chao. 2003. Fitness cost of the green fluorescent protein in gastrointestinal bacteria. Can. J. Microbiol. 49:531-537. [DOI] [PubMed] [Google Scholar]
- 34.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 35.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of the Escherichia coli 16S ribosomal RNA: complementary to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen, M., C. Lippuner, T. Kaiser, A. Misslitz, T. Aebischer, and D. Bumann. 2003. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 552:110-114. [DOI] [PubMed] [Google Scholar]
- 37.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stabb, E. V., and E. G. Ruby. 2003. Contribution of pilA to competitive colonization of Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 69:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 40.Stabb, E. V., K. L. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. G. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction. In N. Saxena (ed.), Recent advances in marine science and technology 2000. PACON International, Honolulu, Hawaii.
- 41.Strongin, D., B. J. Bevis, and B. S. Glick. Unpublished data.
- 42.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. John Wiley and Sons, New York, N.Y.
- 44.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.