Abstract
Transcription of two unfolded protein response genes, hacA and bipA, was examined in Aspergillus niger strains overproducing membrane proteins. Despite elevated bipA mRNA levels, no 5′-truncated hacA transcript was detected, raising the possibility of a hacA-independent induction of bipA mRNA under the stress of membrane protein overproduction in A. niger.
Although filamentous fungi such as Aspergillus niger have been used successfully for the commercial production of native enzymes like glucoamylase, they often suffer from low secreted yields for heterologous proteins (2, 8). This has sparked interest in the mechanism of secretion and the effects of stress from secreting heterologous proteins to allow identification of the production bottlenecks (8). A rise in the level of unfolded proteins and an increased requirement for chaperones can trigger the unfolded protein response (UPR), a stress response mediated at both transcriptional and posttranscriptional levels (9). The UPR was initially defined in Saccharomyces cerevisiae, and a similar but distinct pathway is now apparent in filamentous fungi. The major UPR transcription factor HacA (or Hac1) has been identified in A. niger, Trichoderma reesei, and Aspergillus nidulans, and all three hac genes have unconventional 20-nucleotide introns which are spliced under dithiothreitol (DTT) stress (5, 10). In addition to this splicing event, hacA/hac1 transcripts are shorter at the 5′ end following DTT stress. In both A. niger and A. nidulans, hacA mRNA truncation correlates with increased transcript levels for the gene encoding the major endoplasmic reticulum (ER) chaperone, bipA (5, 10). Here we present evidence that, under the stress of overproducing both membrane-bound components of a cytochrome P450 system, cytochrome P450 reductase (CprA) and benzoate p-hydroxylase (BphA), elevated levels of bipA mRNA occur independently of hacA truncation in A. niger.
A. niger strains N271 (csp21 fwn21 pdx21; derivative of A. niger ATCC 1015 containing single copies of bphA and cprA), T18 (derivative of N271 with ∼10 copies of bphA) (13) and T18-5 (derivative of T18 with ∼15 copies of cprA) (12) were grown in Aspergillus complete medium (ACM; 1% wt/vol glucose) (1) at 30°C, 200 rpm, for 21 h at an initial inoculum of 3 × 105 spores per ml. Mycelia were washed with sterile 0.9% (wt/vol) NaCl and aseptically transferred to AMMN 0.1% [wt/vol] glucose, 50 μg/liter pyridoxine) (1) ± 8.2 mM benzoic acid and incubated for another 3 h at 30°C, 200 rpm. The MIC of benzoic acid (pKa, 4.2) for these strains at pH 6.5 in both AMMN and ACM was greater than 70 mM, and both spore germination and growth appeared to be normal in the presence of 8.2 mM benzoic acid, suggesting that this concentration was not toxic to the cells. The pH of the medium did not alter significantly from 6.5.
Total RNA was extracted from freeze-ground and freeze-dried mycelia according to the RNeasy Plant Mini Protocol (QIAGEN) and was treated with 5 U RNase-free DNase (Promega) for 30 min at 37°C to remove residual DNA. Five to 15 μg glyoxalated total RNA was electrophoresed and hybridized at 65°C, and [32P]dATP-labeled bands were visualized, as previously reported (6). Signals from transcripts of interest were normalized to histone H2A RNA loading controls. Probes for Northern analysis were made by PCR amplification from A. niger AB4.1 genomic DNA. The 1,239-bp hacA and 450-bp histone H2A probes were amplified using primers hacFAW (5′-TTGCCATTGTTGACAGT-3′) with hacRAW (5′-CATAAGGAGAGACGTCAAC-3′) and AD30 (5′-GCAAGACTCGGAACATCATT-3′) with AD31 (5′-AAATCCTCGTCACTCTTGCG-3′), respectively. The bipA probe was as described previously (6). For reverse transcription-PCR (RT-PCR) studies, first-strand cDNA was made from 1 μg DNase-treated total RNA using 100 pmol oligo(dT)15 and 100 U M-MLV Reverse Transcriptase RNase H Minus, Point Mutant (Promega), according to the manufacturer's instructions. Five microliters of the above reaction mixture was used for second-strand cDNA synthesis and PCR amplification using the QIAGEN Hot Start Taq kit. Primers AD68 (5′-CTTCTCCTACCCTAACTCCT-3′) and AD72 (5′-TCAAAGAGAGAGAGGGCACT-3′) annealed to sites on either side of the 20-bp hacA intron. Amplification parameters were the following: 95°C for 15 min; 30 cycles consisting of 30 s at 95°C, 45 s at 56°C, and 45 s at 68°C; and 10 min at 68°C. Thus, Northern analysis of hacA mRNA detects both the full-length and 5′-truncated transcripts, whereas the RT-PCR detects the presence or absence of the 20-bp intron.
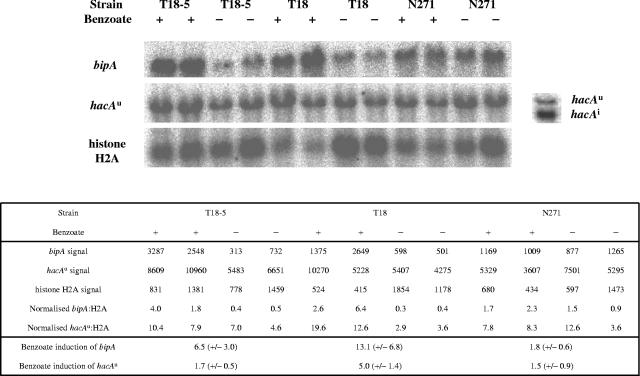
Northern blot analysis of the transcript levels for the UPR-specific transcription factor HacA and the ER chaperone BipA in the single (bphA and cprA) gene copy parental strain A. niger N271 and its multicopy membrane protein-overproducing daughter strains T18 and T18-5 is shown in Fig. 1. Previous work with these strains has shown that both the cprA and bphA genes show highly elevated mRNA levels in the presence of benzoate which correspond to moderate increases in activity at the protein level (11, 12, 13). Analysis of the bipA transcript levels shows a clear increase in benzoate-induced cultures of both T18 and T18-5 relative to either the uninduced controls or the parental strain, N271 (Fig. 1). That this increase is not mirrored in the parental strain, N271, indicates that the increased bipA transcript levels seen in the daughter strains, T18 and T18-5, result from protein overproduction rather than benzoate stress. Despite the elevated bipA transcript levels, only the longer, untruncated hacA transcript was observed in these cultures (Fig. 1). A shorter time course of benzoate treatment, restricted to 30, 60, or 120 min, was carried out to test if the 5′ truncation event was occurring earlier and being missed in the 3-h experiment, but shortened hacA mRNA was still not detected (data not shown).
FIG. 1.
Effect of overproducing CprA and BphA on bipA and hacA transcript levels in A. niger strains T18-5, T18, and N271. Northern blots show duplicate benzoate-treated and control cultures using histone H2A as an RNA loading control. The single panel to the right of the hacA data shows both truncated and untruncated hacA mRNAs routinely obtained from a DTT-stressed A. niger culture. Signals were quantified in a Fuji BAS 1500 phosphorimager, and benzoate induction was calculated relative to the untreated controls using mean normalized values ± standard deviations. hacAu, uninduced (nontruncated hacA mRNA; hacAi, induced (i.e., spliced and truncated) hacA mRNA.
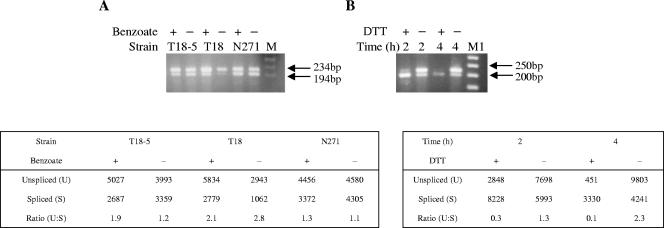
A more sensitive analysis by RT-PCR showed the presence of spliced and unspliced hacA mRNA at comparable levels in both benzoate-induced and uninduced cultures (Fig. 2). In contrast, under DTT stress conditions, most of the hacA transcript appeared to be spliced (Fig. 2). Although these data are not quantitative, the ratios of spliced to unspliced hacA were reproducible and suggest that splicing of the 20-nucleotide intron alone does not account for the elevated bipA mRNA levels. The lack of both an increase in intron splicing and a 5′-truncated transcript, despite elevated bipA transcript levels, suggests that the cellular response to the overproduction of membrane-bound proteins in filamentous fungi differs from that of a classical UPR.
FIG. 2.
RT-PCR to investigate hacA intron splicing. Effect of (A) benzoate treatment on A. niger strains T18-5, T18, and N271 and (B) DTT treatment on A. niger AB4.1. M and M1 represent the λ/HindIII-φX174/HaeII markers (Promega) and 50-bp ladder (Promega), respectively, with relevant marker bands indicated by arrows. DNA fragment intensity was determined densitometrically using TotalLab 1D software (Nonlinear Dynamics). The figure is representative of duplicate biological replicates.
The respective contributions of the 5′ truncation and unconventional intron splicing to the cellular stress response in filamentous fungi remain unresolved and may reflect both the nature and degree of stress involved. Several circumstantial lines of evidence suggest that the 5′ truncation provides a translational control mechanism regulating hacA/hac1 transcription (5, 10). Of three filamentous fungal hac genes examined to date, all show the presence of upstream open reading frames (uORFs) in the 5′-truncated sequence. cDNA constructs with the 5′ leader truncated also show a stronger induction of bipA, suggesting that the leader regulates hacA transcription (10). Finally, the filamentous fungal hacA intron also appears unable to form a secondary structure to attenuate translation in the way that the yeast system allows, necessitating an alternative strategy to fulfill this role. It is possible that significant 5′ truncation may occur only when a strong UPR is elicited, such as on DTT treatment, and very high levels of HacA are required. If the uORF in hacA indeed regulates the efficiency of translation, then spliced, as opposed to truncated, hacA mRNA may elicit only those basal levels of components needed to provide the machinery to allow transport of cell membrane or cell wall proteins during normal vegetative growth. Such a strategy may be better suited to dealing with conditions of extraordinary stress in a system designed for high secretory throughput where basal levels of chaperones and/or foldases may already be high. Although the data do not rule out the possibility that increased bipA mRNA levels are the result of decreased bipA transcript turnover (3), it also suggests the possibility of a pathway distinct from the classical UPR. The UPR signal is transduced from the ER to the nucleus via a transmembrane kinase, Ire1, which activates hacA/hac1 transcription. Recent studies in S. cerevisiae have shown an increasingly sophisticated interplay between IRE1-dependent and -independent pathways which modulates the cellular response to stress (4, 7). These include IRE1-independent regulation of total HAC1 transcript levels and a previously unidentified requirement for a second bZIP transcription factor, Gcn4p. Tellingly, these mechanisms reveal similarities to the PERK- and ATF-6-mediated mammalian ER-to-nuclear signaling pathways, suggesting that all three pathways show some conservation among eukaryotes. Although the detail of the mechanisms adopted by filamentous fungi in response to stress differ from those found in S. cerevisiae, it seems likely that they will share similar IRE1-independent features. Our preliminary data showing raised bipA transcript levels in the absence of truncated hacA in A. niger strains overproducing membrane proteins may thus be a case in point.
Acknowledgments
We gratefully acknowledge funding for this research from the European Commission (Eurofung: QLRK3 1999-00729) and the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Archer, D. B., D. J. Jeenes, D. A. MacKenzie, G. Brightwell, N. Lambert, G. Lowe, S. E. Radford, and C. M. Dobson. 1990. Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Bio/Technology 8:741-745. [DOI] [PubMed] [Google Scholar]
- 2.Conesa, A., P. J. Punt, N. van Luijk, and C. A. M. J. J. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 3.Hyde, M., L. Block-Alper, J. Felix, P. Webster, and D. I. Meyer. 2002. Induction of secretory pathway components in yeast is associated with increased stability of their mRNA. J. Cell Biol. 156:993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leber, J. H., S. Bernales, and P. Walter. 2004. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2:1197-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulder, H. J., M. Saloheimo, M. Penttilä, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Gen. Genet. 271:130-140. [DOI] [PubMed] [Google Scholar]
- 6.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. M. J. J. van den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patil, K. P., H. Li, and P. Walter. 2004. Gcn4p and novel upstream sequences regulate targets of the unfolded protein response. PLoS Biol. 2:1208-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Punt, P. J., N. van Biezen, A. Conesa, A. Albers, J. Mangnus, and C. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 9.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 10.Saloheimo, M., M. Valkonen, and M. Penttilä. 2003. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 11.van den Brink, J. M., C. A. M. J. J. van den Hondel, and R. F. M. van Gorcom. 1996. Optimization of the benzoate-inducible benzoate p-hydroxylase cytochrome P450 enzyme system in Aspergillus niger. Appl. Microbiol. Biotechnol. 46:360-364. [DOI] [PubMed] [Google Scholar]
- 12.van den Brink, J. M., P. J. Punt, R. F. M. van Gorcom, and C. A. M. J. J. van den Hondel. 2000. Regulation of expression of the Aspergillus niger benzoate para-hydroxylase cytochrome P450 system. Mol. Gen. Genet. 263:601-609. [DOI] [PubMed] [Google Scholar]
- 13.van Gorcom, R. F. M., J. G. Boschloo, A. Kuijvenhoven, J. Lange, A. J. van Vark, C. J. Bos, J. A. M. van Balken, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1990. Isolation and molecular characterisation of the benzoate-para-hydroxylase gene (bphA) of Aspergillus niger: a member of a new family of the cytochrome P450 superfamily. Mol. Gen. Genet. 223:192-197. [DOI] [PubMed] [Google Scholar]