Abstract
An enzymatic in vitro alginate polymerization assay was developed by using 14C-labeled GDP-mannuronic acid as a substrate and subcellular fractions of alginate overproducing Pseudomonas aeruginosa FRD1 as a polymerase source. The highest specific alginate polymerase activity was detected in the envelope fraction, suggesting that cytoplasmic and outer membrane proteins constitute the functional alginate polymerase complex. Accordingly, no alginate polymerase activity was detected using cytoplasmic membrane or outer membrane proteins, respectively. To determine the requirement of Alg8, which has been proposed as catalytic subunit of alginate polymerase, nonpolar isogenic alg8 knockout mutants of alginate-overproducing P. aeruginosa FRD1 and P. aeruginosa PDO300 were constructed, respectively. These mutants were deficient in alginate biosynthesis, and alginate production was restored by introducing only the alg8 gene. Surprisingly, this resulted in significant alginate overproduction of the complemented P. aeruginosa Δalg8 mutants compared to nonmutated strains, suggesting that Alg8 is the bottleneck in alginate biosynthesis. 1H-NMR analysis of alginate isolated from these complemented mutants showed that the degree of acetylation increased from 4.7 to 9.3% and the guluronic acid content was reduced from 38 to 19%. Protein topology prediction indicated that Alg8 is a membrane protein. Fusion protein analysis provided evidence that Alg8 is located in the cytoplasmic membrane with a periplasmic C terminus. Subcellular fractionation suggested that the highest specific PhoA activity of Alg8-PhoA is present in the cytoplasmic membrane. A structural model of Alg8 based on the structure of SpsA from Bacillus subtilis was developed.
Bacterial alginates are linear exopolysaccharides consisting of β-1,4-linked β-d-mannuronic acid and its C5 epimer α-l-guluronic acid (10, 15). Only the two bacterial genera Pseudomonas (29) and Azotobacter (43) are known to produce alginates. Although the polymer is primarily synthesized as polymannuronate (62), the monomer distribution is variable throughout the polymer due to epimerization of mannuronic acid residues (58, 62). Bacterial alginates can be acetylated at the O2 and/or O3 positions of mannuronic acid residues (62). The best-characterized alginate-producing organism is Pseudomonas aeruginosa, an opportunistic human pathogen. Alginate is one important virulence factor, and the conversion of the nonmucoid to the alginate-overproducing mucoid form after infection of cystic fibrosis patients is associated with a decline of pulmonary function and survival rate (41). Alginate acts as an extracellular matrix material that allows the formation of differentiated biofilms, which restrict diffusion of clinical antibiotics and protect embedded cells against human antibacterial defense mechanisms (30, 39, 42).
Most of the genes involved in alginate biosynthesis are clustered in P. aeruginosa at 34 min of the bacterial chromosome (5) and are separated from other regulatory genes such as algU and the muc genes (68 min) (8, 33). The biosynthesis cluster is an operon and comprises 12 genes (algD, alg8, alg44, algK, algE, algG, algX, algL, algI, algJ, algF, and algA) under tight control of the alginate promoter upstream of algD (59, 60). algC is the only gene involved in alginate synthesis that is not located in the cluster, but it is also involved in lipopolysaccharide synthesis and expressed from its own promoter (17, 64). The alginate biosynthesis pathway can be divided into four different stages: (i) synthesis of precursor substrate, (ii) polymerization and cytoplasmic membrane transfer, (iii) periplasmic modification, and (iv) export through the outer membrane. The precursor synthesis is well characterized and starts from the central metabolite fructose-6-phosphate, which is converted to GDP-mannuronic acid in four enzymatic steps by the proteins AlgA, AlgC, and AlgD (45). The modifications of the polymannuronate chain are carried out by a number of periplasmic proteins. The three proteins AlgI, AlgJ, and AlgF form an enzyme complex that catalyzes the O acetylation of mannuronic acid residues (12-14). AlgG is a C5-mannuronan-epimerase (11), and AlgL is an alginate lyase (38, 55). The export or secretion of the polymer chain through the outer membrane is mediated by AlgE, an alginate-specific outer membrane channel (47, 48).
The polymerization step is still not understood. The proteins Alg8, putatively encoding a glycosyltransferase, and Alg44 are supposed to be transmembrane proteins and therefore possible subunits of the alginate polymerase (31, 35). The proteins AlgK and AlgX are periplasmic proteins, and deletion mutants showed secretion of free uronic acids presumably due to alginate lyase activity (24, 49). Together with AlgG, these proteins are supposed to be part of a scaffold surrounding the nascent alginate chain (16, 23, 49).
In the present study we establish for the first time an enzymatic in vitro alginate polymerase assay enabling the subcellular localization of the respective enzyme activity. Previous studies applied transposon mutagenesis and complementation studies to investigate the role of Alg8 in alginate biosynthesis (31, 63). Here we generated a nonpolar deletion mutant of alg8 to reveal the essential role of Alg8 in alginate polymerization. Evidence was provided that Alg8 is the bottleneck in alginate biosynthesis. Moreover, a structural model of Alg8 was developed, and evidence subcellular localization of Alg8 was obtained by using translational fusions with reporter enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used in the present study are listed in Table 1. Escherichia coli strains were grown in LB medium at 37°C. E. coli S17-1 was used for conjugative transfer of mob site containing plasmids (61). When required, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; and tetracycline, 12.5 μg/ml. P. aeruginosa FRD1 (40) and PDO300 (34) are used in the present study. P. aeruginosa strains were grown in LB or PI(A) medium (Pseudomonas isolation [agar] medium: 20 g of peptone, 10 g of K2SO4, 1.4 g MgCl2, 0.025 g of Triclosan, and 20 ml of glycerol per liter) at 37°C and, if required, antibiotics were added to appropriate concentrations. The antibiotic concentrations used for P. aeruginosa strains were as follows: gentamicin, 300 μg/ml; and carbenicillin, 300 μg/ml. All chemicals were purchased from Sigma-Aldrich (St. Louis, Mo.).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| FRD1 | Cystic fibrosis isolate; Alg+ | 40 |
| FRDΔalg8Gm | Δalg8::aacC1; Alg− | This study |
| FRDΔalg8 | Δalg8; Alg− | This study |
| PAO1 | Prototrophic wild-type strain; Alg− | 22 |
| PDO300 | ΔmucA22 variant of PAO1; Alg+ | 34 |
| PDO300Δalg8Gm | Δalg8::aacC1; Alg− | This study |
| PDO300Δalg8 | Δalg8; Alg− | This study |
| E. coli | ||
| TOP10 | E. coli cloning strain | Invitrogen |
| S17-1 | thi-1 proA hsdR17 (rK− mK+) recA1; tra gene of plasmid RP4 integrated in chromosome | 61 |
| Plasmids | ||
| pBBR1MCS-5 | Gmr; broad-host-range vector; Plac | 28 |
| pBBR1MCS-5:alg8 | HindIII-BamHI fragment comprising gene alg8 inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8His | HindIII-BamHI fragment encoding C terminally hexahistidine-tagged Alg8 inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8GFP | Translational Alg8-GFP fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8lacZ | Translational Alg8-LacZ fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8phoA | Translational Alg8-PhoA fusion, inserted into vector pBBR1MCS-5 | This study |
| pEX100T | Apr Cbr, gene replacement vector containing sacB gene for counterselection | 20 |
| pEX100TΔalg8Gm | Apr Cbr Gmr; vector pEX100T with SmaI-inserted alg8 deletion construct | This study |
| pPS856 | Apr Gmr; source of 1,100-bp BamHI fragment comprising aacC1 gene flanked by FRT signal sequences | 20 |
| pPFLP2 | Apr Cbr; broad-host-range vector encoding Flp recombinase | 20 |
| pPHO7 | Apr; phoA without signal sequence | 19 |
| pZsGreen | GFP translational fusion vector | BD Clontech |
| pJE608 | LacZ lacking the first 8 amino acids with promoter Ptac in pMMB67EH | 9 |
| Oligonucleotides | ||
| alg81N-Ec5 | CGCAGGATATCGGAAACTTACAAACGTGGCCTC | |
| alg81N-Ba | TAGAGGATCCGGTTCATCTTCTCCCACAGAG | |
| alg82C-Ba | TGATGGATCCGTTCACCATGCTGGTGCTGTTC | |
| alg82C-Ec5 | CCAGGATATCTCATACGATGGTCAGCAGCAC | |
| alg8up | AAGAACCTCTTTATCGCCCTCGGAC | |
| alg8down | TCACGGATCCCCAGGTAGGAGGTGATCAGGTAG | |
| alg8N(HiSDNd) | CCGGGAAGCTTGAGGAGCACAGCCATATGGAACTGATGATGGAAACTTACAAA | |
| alg8C(Ba) | CCCGGATCCTCATACGATGGTCAGCAGCACG | |
| alg8C(HisBa) | CTTGGATCCTCAATGGTGATGGTGATGGTGTACGATGGTCAGCAGCACGGCGAC | |
| alg8C(Δstop) | CCCGGATCCCATACGATGGTCAGCAGCACG |
Enzymatic synthesis and purification of GDP-mannuronic acid.
GDP-mannose dehydrogenase (GMD) was partially purified from P. aeruginosa 8822 (7) as previously described by Roychoudhury et al. (50). GMD activity was measured according to the method of Preiss (44) by monitoring reduction of NAD+ to NADH at a wavelength of 340 nm. A specific activity of 570 mU/mg of protein was obtained, and the substrate GDP-mannose was completely oxidized to GDP-mannuronic acid. One unit of GMD activity corresponds to the oxidation of 1 μmol of GDP-mannose to GDP-mannuronic acid per min. Proteins were either removed by ultrafiltration (cutoff = 10 kDa) or, when [14C]GDP-mannuronic acid was produced, by phenol-chloroform (1:1) extraction. GDP-mannuronic acid containing fractions were subjected to anion-exchange chromatography (MonoQ). GDP-mannuronic acid containing fractions were detected by uronic acid analysis and subjected to gel filtration chromatography using a Sephadex G15 column.
[14C]GDP-mannuronic acid and GDP-mannuronic acid were separated by thin-layer chromatography (TLC) with PEI-cellulose (Schleicher & Schuell) and the solvents 0.2 M LiCl, 1.0 M LiCl, and 1.6 M LiCl. The [14C]GDP-mannuronic acid was detected by autoradiography and use of a TLC scanner (Berthold LB 2760). As standards, purified GDP-mannuronic acid and GDP-mannose (Sigma-Aldrich, St. Louis, MO) were used and were detected by using 0.002% (wt/vol) fluorescein in methanol for staining.
Enzymatic in vitro alginate synthesis.
Crude extracts and cytoplasmic membrane (CM), outer membrane (OM), and envelope fractions were used as a source for alginate polymerase activity. The CM was obtained by sucrose gradient ultracentrifugation as described previously (48). All other fractions, including the OM were obtained as described below. The contamination of CM with OM was estimated from its 2-keto-3-desoxyoctonate (KDO) content and was given as a percentage of the total amount of KDO present in the CM and OM (48). The contamination was determined to be ca. 10% as has been previously published (48).
Protein fractions contained 50 mM Tris-HCl (pH 8.0), 0.5 mM phenylmethylsulfonyl fluoride, 0.1% (vol/vol) Triton X-100, and 2 mM dithiothreitol. The in vitro synthesis reaction contained 143 pmol [14C]GDP-mannuronic acid (286.1 mCi/mmol), 857 pmol of GDP-mannuronic acid, 50 mM Tris-HCl (pH 8.0), 10 μM MgCl2, 70 μg of alginate oligomers (n = 3 to 6), and 2.5 mg of protein sample (polymerase source) in a total volume of 260 μl. The alginate oligomers were obtained by acid hydrolysis as previously described (46). As a negative control, inactive enzyme preparation was used. Inactive enzyme was obtained by heat treatment applying 100°C for 10 min. The synthesis reaction was started by the addition of protein sample (polymerase source) and conducted at 37°C for 20 min. The reaction mixture (50 μl) was loaded onto anion-exchange filter (DE81; Whatman), and the filter was subsequently washed with 0.3 M NaCl (GDP-mannuronic acid was tested to be eluted from the filter by using 0.3 M NaCl), water, and then ethanol. A total of 5 ml of scintillation cocktail [0.5% (wt/vol) 2,5-diphenyloxazol; 0.02% (wt/vol) 2,2-p-phenylen-bis-(5-phenyloxazol)] was added, and the counts per minute (cpm) were measured. The specific alginate polymerase activity is given as follows: Δcpm [(a20-a0) − (i20-i0)]/mg of protein × min (where a is active enzyme, i is inactive enzyme, and 0 and 20 refer to t = 0 min and t = 20 min, respectively). In this reaction mixture, Δ100 cpm corresponded to 0.215 pg of alginate and a specific alginate polymerase activity of 1.15 μU/mg of protein. One unit corresponds to the conversion of 1 μmol of GDP-mannuronic acid into alginate per min.
Isolation, analysis, and manipulation of DNA.
General cloning procedures were performed as described previously (51). DNA primers, deoxynucleoside triphosphate, Taq, and Platinum Pfx polymerases were purchased from Invitrogen. DNA sequences of new plasmid constructs were confirmed by DNA sequencing according to the chain termination method using an ABI310 automatic sequencer.
Construction and confirmation of alg8 deletion mutants.
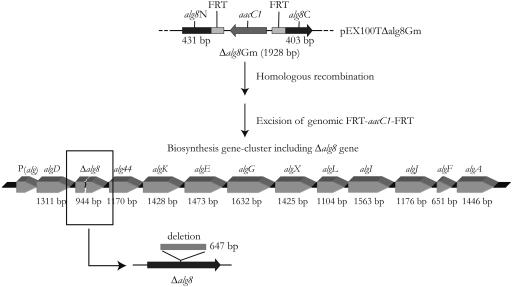
Two regions of the alg8 gene were amplified by using Taq polymerase and primers alg81N-Ec5, alg81N-Ba, alg82C-Ba, and alg82C-Ec5. Region alg8N (431 bp) comprised bases 40 to 470 and region alg8C (403 bp) comprised bases 1117 to 1519 relative to the designated alg8 coding region (31), respectively. Both PCR products were hydrolyzed by using BamHI and inserted into vector pGEM-TEasy (Promega). Vector pPS856 (20) was hydrolyzed with BamHI, releasing an about 1,100-bp fragment containing the aacC1 gene (encoding gentamicin acetyltransferase) flanked by two FRT (Flp recombinase target) sites. The 1,100-bp BamHI fragment (aacC1 gene) was inserted into the BamHI site of plasmid pGEM-TEasy:Δalg8NC, resulting in plasmid pGEM-TEasy:Δalg8Gm. The 1,949-bp Δalg8Gm comprising DNA fragment was amplified by Pfx polymerase using primers alg81N-Ec5 and alg82C-Ec5, and the corresponding PCR product was inserted into SmaI site of vector pEX100T (20), resulting in plasmid pEX100TΔalg8Gm.
E. coli S17-1 was used as donor for transfer of plasmid pEX100TΔalg8Gm into P. aeruginosa strains, and transconjugants were selected on mineral salt medium (56) containing 300 μg of gentamicin/ml and 5% (wt/vol) sucrose. Cells growing on this selective medium should have emerged from double-crossover events. Gene replacement was confirmed after subculture of cells on PIA medium containing 300 μg of gentamicin/ml and using PCR with primers alg8up and alg8down.
E. coli S17-1 was used to transfer the Flp recombinase encoding vector pFLP2 (20) into P. aeruginosa Δalg8Gm strains and after 24 h of cultivation on PIA medium containing 5% (wt/vol) sucrose, gentamicin- and carbenicillin-sensitive cells were analyzed by PCR for loss of the gentamicin-resistant cassette.
Complementation of isogenic alg8 deletion mutants.
For complementation of alg8 deletion mutants, the alg8 gene of P. aeruginosa PAO1 was amplified by PCR with the primers alg8N(HiSDNd) and alg8C(Ba). The PCR product was hydrolyzed with HindIII and BamHI and was inserted into HindIII and BamHI sites of broad-host-range vector pBBR1MCS-5 (28), resulting in plasmid pBBR1MCS-5:alg8. In addition, the 3′-end primer alg8C(HisBa) was used to generate an alg8 gene encoding a C-terminally hexahistidine-tagged Alg8, which was inserted into vector pBBR1MCS-5 as described above (Table 1) All inserts cloned into the multiple cloning site of vector pBBR1MCS-5 are under the control of the lac promoter.
Subcellular localization using PhoA, LacZ, and GFP fusions.
The 3′ end of the alg8 gene was amplified by PCR using Pfx polymerase and primers alg82C-Ba and alg8C(Δstop). The corresponding 422-bp PCR-fragment was inserted into SmaI sites of pBluescript KS(−) (Stratagene), resulting in plasmid pKS−:alg8(Δstop). After BglII and SacII hydrolysis, the resulting 203-bp fragment was used to replace the original 3′ end of the alg8 gene in plasmid pBBR1MCS-5:alg8, resulting in plasmid pBBR1MCS-5:alg8(Δstop). XbaI-BamHI fragments of vectors pPHO7 (19), pJE608 (9), and pZsGreen (BD Biosciences Clontech) were inserted into XbaI-BamHI-restricted pBBR1MCS-5:alg8(Δstop) to create translational PhoA, LacZ, and green fluorescent protein (GFP) fusions, respectively.
Subcellular fractionation.
An overnight culture of the respective P. aeruginosa strain in LB medium was diluted 1:50 in the same medium and grown for 4 h or until a optical cell density at 600 nm of 0.5 to 0.6 was reached. The cultures were harvested, and cell sediments were suspended and washed twice in 10 mM HEPES buffer (pH 7.4). The cells were resuspended in 1 ml of HEPES buffer (pH 7.4) and sonicated at 30% intensity for eight cycles of 15 s sonication, followed by 20 s of cooling down. Cellular debris and unlysed cells were sedimented by centrifugation (1 h at 5,000 × g). Then, 800 μl of the supernatant was centrifuged at 100,000 × g for 2 h. The supernatant (soluble fraction) was transferred into a clean tube, and the sediments were resuspended in 800 μl of 10 mM HEPES buffer (pH 7.4) and centrifuged under the same conditions. The supernatant was again transferred into a clean tube (wash fraction), the sediment (envelope fraction) was redissolved in 800 μl of 10 mM HEPES buffer (pH 7.4) containing 0.7% (wt/vol) N-lauroylsarcosine, and selective solubilization of the cytoplasmic membrane was achieved by incubation on a horizontal shaker for 1.5 h at 37°C. The mixtures were centrifuged for 2 h at 100,000 × g, and the supernatant was transferred into a clean tube (solubilized cytoplasmic membrane). This solubilization step was repeated, and the resulting membrane sediment (outer membrane) was resuspended in 800 μl of 10 mM HEPES buffer (pH 7.4).
Alkaline phosphatase-β-galactosidase activity assays.
Alkaline phosphatase and β-galactosidase enzymatic assays were performed according to the methods of Miller (36) and Manoil (32), respectively. Cells of P. aeruginosa were grown overnight in LB with the appropriate antibiotic, and the cultures were diluted 1:50 in LB medium. The cells were allowed to grow until the cultures reached a optical cell density at 600 nm of 0.4 to 0.6. The enzyme assays were performed in 1 ml of these cultures, and 1 ml was used for freeze-drying to determine the cellular dry weight. PhoA activity was determined by the rate of p-nitrophenylphosphate hydrolysis, taking the amount of enzyme hydrolyzing 1 μmol of substrate per 1 min at 37°C as a unit of enzymatic activity. The results are given as average values of at least four independent experiments.
Alginate production assays.
A total of 2 ml of bacterial overnight cultures was harvested at 4°C and washed twice with saline. Then, 200 μl of cell suspension was plated onto PIA medium and incubated 72 h at 37°C. Cells of two agar plates were scraped off by using a sterile spatula and washed twice with 40 ml of saline. When viscosity of the solution was too high for separation of cells (complemented mutants), saline was added to a total volume of 300 ml to allow sedimentation of cells during centrifugation. Cellular sediments were freeze-dried, and the final weight was determined. Alginate supernatants were precipitated with 1 vol of ice-cold isopropanol, and alginate was harvested and freeze-dried. For further purification, the precipitated alginate was redissolved in 0.05 M Tris-HCl-10 mM MgCl2 (pH 7.4) to a final concentration of 0.5% (wt/vol), followed by incubation with 15 μg of DNase I/ml and 15 μg of RNase A/ml at 37°C for 6 h. Pronase E was added to a final concentration of 20 μg/ml, and this solution was incubated for further 18 h at 37°C. Alginate solutions were dialyzed against 5 liters of ultrapure H2O for 48 h. Alginate was precipitated with 1 volume of ice-cold isopropanol and freeze-dried for quantification and uronic acid analysis.
Uronic acid assays.
Alginate concentrations were assayed by a modification of the Blumenkrantz and Asboe-Hansen protocol (3), using purified P. aeruginosa PDO300 alginate (100% [wt/wt] uronic acid content) as a standard. Briefly, alginate samples were dissolved in 200 μl of ultrapure H2O at concentrations between 0.25 and 0.05 mg/ml. The sample was mixed with 1.2 ml of tetraborate solution (0.0125 M disodium tetraborate in concentrated sulfuric acid) and incubated on ice for 10 min. The mixtures were incubated at 100°C for 5 min and then cooled down on ice for further 5 min. Then, 20 μl of m-hydroxybiphenyl reagent (0.15% m-hydroxybiphenyl in 0.125 M NaOH) was added, and the reactions were mixed for 1 min. For each sample or dilution a negative control was assayed by using 0.0125 M NaOH instead of the hydroxybiphenyl reagent. Uronic acid concentrations were determined spectrophotometrically at a wavelength of 520 nm.
1H-NMR analysis of alginates.
The alginate samples were deacetylated and partially degraded by mild, acid hydrolysis in order to reduce the viscosity of the solutions. Alginate samples were analyzed by high-field 1H-nuclear magnetic resonance (NMR) spectroscopy at 90°C by using a Bruker AM-300 (300-MHz) spectrometer. 3-(Trimethylsilyl)propanesulfonate was used as an internal standard in the samples. Prior to the NMR spectroscopy, the samples were desalted on Bio-Gel P-4 (Bio-Rad), freeze-dried, and dissolved in D2O. The removal of salt resulted in a better signal-to-noise ratio. The composition, given as molar fraction of the monomers G (FG) and M (FM) and the dyads (FGG, FGM, and FMM) were determined from the spectra as described by Grasdalen et al. (18). In this procedure, the area under each peak, which is proportional to the amount of residues giving rise to the signal, is used to calculate the above parameters.
RESULTS
Enzymatic in vitro synthesis of alginate. An enzymatic in vitro polymerization assay was established by using 14C-labeled GDP-mannuronic acid as substrate, alginate oligomers as primer, and subcellular fractions of alginate-overproducing P. aeruginosa FRD1 as polymerase source. [14C]GDP-mannuronic acid is commercially not available and was enzymatically synthesized by using partially purified GDP-mannose dehydrogenase and GDP-mannose as substrate. Complete oxidation of [14C]GDP-mannose to [14C]GDP-mannuronic acid was monitored by TLC in combination with autoradiography and densitometry. Complete oxidation of GDP-mannose to GDP-mannuronic acid was monitored by TLC and uronic acid specific staining (data not shown). Anion-exchange filters were applied to selectively bind polymerized alginate. The binding of GDP-mannuronic acid to the filter was analyzed, revealing that 0.3 M NaCl is the lowest NaCl concentration completely eluting GDP-mannuronic acid from the filter (data not shown). Alginate and alginate oligomers (n = 3 to 6) did not elute after a wash with 0.3 M NaCl, enabling selective binding of polymerized GDP-mannuronic acid, which was detected by scintillation analysis. To subcellularly localize the alginate polymerase, various subcellular fractions were analyzed with respect to alginate polymerase activity (Table 2). The highest specific alginate polymerase activity was detected in the envelope fraction, suggesting that cytoplasmic and outer membrane proteins together constitute the functional alginate polymerase complex. Accordingly, no alginate polymerase activity was detected by using cytoplasmic membrane or outer membrane proteins, respectively. The omission of the detergent Triton X-100 in the envelope fraction caused a decrease in specific alginate polymerase activity (Table 2).
TABLE 2.
In vitro alginate synthesis using subcellular fractions of P. aeruginosa FRD1 as a polymerase sourcea
| Subcellular fraction | cpm/reaction mixture
|
Alginate polymerase sp act (μU/mg of protein) | In vitro-synthesized alginate (pg) | |
|---|---|---|---|---|
| No. | % | |||
| Crude extract | 161 | 3.8 | 0.7 | 0.36 |
| Cytoplasmic membrane | ND | ND | ND | ND |
| Outer membrane | ND | ND | ND | ND |
| Envelope | 3,037 | 100 | 13.2 | 6.5 |
| Envelopeb | 582 | 19 | 2.5 | 1.25 |
All experiments were performed in triplicates. Average values are given. The standard deviation was <30%. ND, not detectable.
Triton X-100 was omitted.
Construction of an isogenic knockout mutant of alg8.
To investigate the requirement of Alg8 in alginate biosynthesis, marker-free alg8 deletion mutants of alginate-overproducing strains P. aeruginosa PDO300 and P. aeruginosa FRD1 were generated, respectively (Fig. 1). Both mutant strains showed a nonmucoid phenotype on agar plates, suggesting that Alg8 is required for alginate synthesis. Recent publications suggested that the nonmucoid phenotype of alg deletion mutants (algK, algX, and algG) was associated with the secretion of uronic acid oligomers generated by degradation of the alginate polymer by the alginate lyase AlgL (23, 49). To investigate whether the loss of mucoidy of the alg8 mutants was associated with production of uronic acid oligomers, the respective supernatants were dialyzed and/or ultrafiltrated. No free uronic acids (<5,000 Da) were detected, suggesting that the alginate-deficient phenotype of the mutants was due to a loss of alginate formation.
FIG. 1.
Schematic view of alg8 knockout construct of plasmid pEX100TΔalg8Gm used for homologous recombination and the alginate biosynthesis operon after replacement of native alg8 gene with Δalg8.
Alg8 plays a key role in alginate production.
To verify that the deletion had no downstream effects within the biosynthesis operon, a plasmid containing only the ORF of alg8 (pBBR1MCS-5:alg8) was used to restore the mucoid phenotype. Interestingly, the mucoid phenotype was not only restored, but alginate production was also at least 20-fold increased compared to P. aeruginosa PDO300 (Table 3). Plasmid pBBR1MCS-5:alg8 mediated a 15-fold increased alginate production in P. aeruginosa PDO300, suggesting that Alg8 is the bottleneck in alginate production. A twofold increase of alginate production was observed when P. aeruginosa PDO300(pBBR1MCS-5) was grown in the presence of gentamicin compared to the absence of gentamicin. The alginate overproduction of alg8 mutants harboring plasmid pBBR1MCS-5:alg8 was associated with a decrease of cellular dry mass to 54% (wt/wt) ± 10% (wt/wt) compared to cellular dry mass production of strain P. aeruginosa PDO300. A similar effect was found in P. aeruginosa PDO300 harboring plasmid pBBR1MCS-5:alg8 (Table 3). Further complementation and production analyses with mutant P. aeruginosa PDO300Δalg8 harboring plasmids that encode C terminally tagged Alg8 proteins indicated that the polymer production yield was not affected by translational fusions of Alg8. The translational hexahistidine, PhoA, GFP, and LacZ fusions mediated an at least 20-fold-increased alginate production compared to P. aeruginosa PDO300 (Table 3).
TABLE 3.
Alginate and cellular dry mass production by different P. aeruginosa strains harboring various plasmids
| Strain | Mean ± SD
|
|
|---|---|---|
| Alginate production (g/g CDM) | Cellular dry mass (CDM [g]) | |
| PAO1 | NDa | 0.227 ± 0.021 |
| PAO1(pBBR1MCS-5) | ND | 0.223 ± 0.029 |
| PAO1(pBBR1MCS-5:alg8) | ND | 0.202 ± 0.007 |
| PDO300 | 0.091 ± 0.028 | 0.103 ± 0.006 |
| PDO300(pBBR1MCS-5) | 0.192 ± 0.004 | 0.115 ± 0.012 |
| PDO300(pBBR1MCS-5:alg8) | 1.314 ± 0.045 | 0.076 ± 0.011 |
| PDO300Δalg8 | ND | 0.101 ± 0.003 |
| PDO300Δalg8(pBBR1MCS-5) | ND | 0.091 ± 0.002 |
| PDO300Δalg8(pBBR1MCS-5:alg8) | 2.039 ± 0.613 | 0.057 ± 0.002 |
| PDO300Δalg8(pBBR1MCS-5:alg8His) | 2.770 ± 0.545 | 0.046 ± 0.008 |
| PDO300Δalg8(pBBR1MCS-5:alg8GFP) | 3.092 ± 0.241 | 0.053 ± 0.007 |
| PDO300Δalg8(pBBR1MCS-5:alg8phoA) | 2.852 ± 0.387 | 0.046 ± 0.007 |
| PDO300Δalg8(pBBR1MCS-5:alg8lacZ) | 2.263 ± 0.192 | 0.056 ± 0.011 |
ND, not detectable.
Transfer of plasmid pBBR1MCS-5:alg8 into alg8 mutant of P. aeruginosa FRD1 restored alginate production, but a free plasmid was not detectable. Interestingly, PCR analysis and DNA sequencing confirmed that the intact alg8 gene was inserted into the genome (data not shown).
Alg8 impacts on alginate composition.
Differences in alginate solubility and viscosity of alginates from P. aeruginosa PDO300 and the respective complemented alg8 mutants indicated that polymer composition might be different. 1H-NMR analysis of alginates isolated from P. aeruginosa PDO300 and P. aeruginosa PDO300Δalg8(pBBR1MCS-5:alg8) revealed that, due to additional alg8 gene copies, the degree of acetylation increased from 4.7 to 9.3%, whereas the guluronic acid content of the polymer decreased from 38 to 19%, and the frequency of the mannuronic acid doublet (FMM) increased from 24 to 62% (data not shown).
Construction and analysis of translational fusion proteins of Alg8.
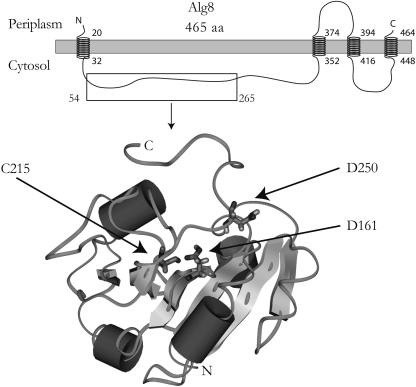
Alg8 is supposed to be a membrane protein that putatively encodes a glycosyltransferase linking the cytosolic precursor synthesis to polymer formation (45). Topological analyses of Alg8 using the SMART database (57) and the topology prediction tool TMHMM2 (37) suggested a signal sequence at the N terminus (1 to 32 amino acids) and four transmembrane helices (Fig. 2). To analyze the topology and subcellular localization of Alg8, C-terminal translational fusions of Alg8 to the reporter proteins LacZ, PhoA, and GFP were constructed. All Alg8 fusions were functional and restored alginate production in P. aeruginosa PDO300Δalg8 (Table 3). Reporter protein assays revealed a specific alkaline phosphatase activity (PhoA units) of 7.56 U/mg of cellular dry weight and a β-galactosidase activity of 0.48 U/mg of cellular dry weight. Alg8 fused to GFP did not enable localization of GFP foci using fluorescence microscopy. These data suggested a periplasmic localization of the C terminus. A recent publication reported an improved topology prediction algorithm using HMM (hidden Markov model) and experimentally verified localization of the C terminus (2). Thus, the HMM-based topology tool Phobius (http://phobius.cgb.ki.se) was used to further analyze the membrane topology using constrained prediction with a periplasmic C terminus (25). These results strongly support the model of a membrane protein containing a N-terminal signal sequence and 4 transmembrane helices (Fig. 2). Cell fractionation experiments were performed with P. aeruginosa PDO300Δalg8(pBBR1MCS-5:alg8phoA) and the envelope fraction, as well as the solubilized cytoplasmic membrane fraction, showed the highest specific alkaline phosphatase activity (PhoA units) of 13.9 ± 0.2 U/mg of protein and 18.4 ± 0.3 U/mg of protein, respectively. The cytoplasmic membrane proteins were selectively enriched by solubilization of the membrane with 0.7% (wt/vol) N-lauroylsarcosine, which also solubilized the Alg8-PhoA fusion protein. The cytosolic fraction showed a PhoA activity of 9.7 ± 0.1 U/mg, and the outer membrane fraction showed an activity of 9.9 ± 0.3 U/mg.
FIG. 2.
Predicted membrane topology of Alg8 based on different HMM-based algorithms (Phobius, SMART, and TMHMM2) of the processed Alg8. Numbers represent the location of the amino acids in the processed form starting with first N-terminal amino acid after the predicted signal peptide cleavage site with number 1. The threading model was developed based on the SAM-T02 alignment of Alg8 with SpsA (1qg8). Cylinders represent α-helical structures. Big arrows represent β-strands. The putative catalytic residues are given as stick side chains and indicated by arrows. N, N terminus of the structural Alg8 model; C, C terminus of the structural Alg8 model.
Development of a threading model of Alg8.
Hydrophobic cluster analysis showed that Alg8 shares significant homologies with β-glycosyltransferases of class II (54). A SMART database search also suggested that Alg8 contains the β-glycosyltransferase domain, although the result was less significant with respect to the required threshold according to the HMM model. Submission of the Alg8 sequence to algorithms that search structural databases SAM-T02 (26) and 3D-PSSM (27) showed 19.6% similarity of Alg8 to the spore coat polysaccharide biosynthesis protein SpsA (1qg8a) from Bacillus subtilis, which represents a nucleotide-diphospho-sugar transferase (4). A threading model of Alg8 was developed with the aid of the SpsA structure as previously described (21). Inspection of the protein model showed that homologues of amino acid residues presumably forming a catalytic triad in SpsA are also adjacent to the core structure in the Alg8 model. The SpsA structure exhibits several amino acid residues serving as potential base functions that might be involved in catalytic function: such as C160 and D191. The homologous amino acid residues C215 and D250 are supposed to be candidates for catalytic residues of Alg8. Another aspartic acid residue is located in a conserved DXD motif that can be found in many glycosyltransferases (53). This motif can be found twice in Alg8 at positions 150 to 152 and 159 to 161. In SpsA the DXD motif is suggested to be reduced to a single aspartic acid residue (52) that corresponds to D161 in the Alg8 model.
DISCUSSION
In this study, we describe the establishment of an enzymatic in vitro alginate synthesis assay using [14C]GDP-mannuronic acid as activated alginate precursor. An efficient method for enzymatic oxidation of GDP-mannose and purification of GDP-mannuronic acid was developed. For the first time alginate polymerase activity could be detected in vitro using subcellular fractions of P. aeruginosa. Evidence was provided that an envelope preparation containing cytoplasmic membrane, outer membrane, and associated proteins constitute the alginate polymerase complex, which was in addition stabilized by the nonionic detergent Triton X-100 (Table 2). This was further confirmed by separation of cytoplasmic membrane and outer membrane, which led to inactivation of alginate polymerase. These findings also support previous suggestions (16, 23, 49) of a protein scaffold in the periplasm related to alginate polymerization and/or modification. Additionally, these data suggest that the outer membrane export protein AlgE is attached to the protein scaffold composed of cytoplasmic membrane proteins (Alg8 and Alg44) and periplasmic proteins (AlgK, AlgL, AlgG, and AlgX). Thus, we propose that alginate polymerization and export through the outer membrane are coordinated via the formation of a protein complex involving cytoplasmic and outer membrane proteins, as well as periplasmic proteins.
In previous studies, only transposon mutagenesis has been used to characterize the putative alg8 gene within the alginate biosynthesis operon and DNA fragments comprising more than one open reading frame (ORF) were applied for complementation studies (31, 63). The transposon insertions showed polar effects on other biosynthesis genes (6, 63). Thus, to evaluate the requirement of the designated ORF of alg8 (31), we generated a marker-free, nonpolar alg8 deletion mutant by using homologous recombination (Fig. 1). The mutant P. aeruginosa PDO300Δalg8 showed a nonmucoid phenotype but, in contrast to the also-reported nonmucoid phenotypes of algK, algG, and algX deletion mutants, suggested to be caused by degradation of the nascent alginate chain by alginate lyase AlgL (23, 49), we were unable to detect uronic acids oligomers (<5,000 Da) or monomers in the respective culture supernatants. Jain et al. (23) suggested that the proteins AlgK and AlgG were part of a scaffold surrounding and therefore protecting the nascent alginate chain, and the findings of Robles-Price et al. (49) suggested that AlgX is also involved in the scaffold formation. The culture supernatants of the respective mutants contained unsaturated uronic acid oligomers, which indicated alginate lyase degradation. Since no uronic acid mono- or oligomers have been found in the supernatant of the alg8 deletion mutant, our findings suggest that deletion of alg8 abolishes alginate polymerization. Further studies will reveal whether alginate production deficiency is caused by a lack of polymerization or membrane translocation of the precursor GDP-mannuronic acid. Many glycosyltransferases of the class II are involved not only in membrane translocation but also in polymerization itself (52, 53). Thus, Alg8 might be the catalytic subunit of the alginate polymerase as previously suggested (45). This hypothesis was supported by the 20-fold overproduction of alginate by P. aeruginosa PDO300Δalg8(pBBR1MCS-5:alg8) compared to P. aeruginosa PDO300, indicating that Alg8 is the bottleneck in alginate production (Table 3). Unlike previous studies (31, 63), we used a defined PCR fragment comprising only the designated alg8 ORF of nonmucoid P. aeruginosa PAO1 and not DNA fragments that originated from subcloning of the alginate biosynthesis operon of P. aeruginosa FRD1. We were therefore able to limit the complementing DNA sequence to the defined ORF of Alg8.
Additional copy numbers of alg8 enhanced the alginate production of strain P. aeruginosa PDO300 by a factor of 15. P. aeruginosa PDO300 was used for further complementation studies because of plasmid stability problems associated with clinical alginate-overproducing isolate P. aeruginosa FRD1. Recent publications demonstrate that strain P. aeruginosa FRD1:pJLS3, in which the alginate biosynthesis operon is under control of the strong IPTG-inducible Ptac promoter, produced 0.55 g of alginate per g of cellular dry weight (1). Comparison of these alginate production data with the alginate production of strain P. aeruginosa PDO300Δalg8 pBBR1MCS-5:alg8, which produced ca. 2.5 g of alginate per g of cellular dry weight, suggests that Alg8 is the bottleneck in alginate production. This enabled us to functionally assign the ORF of alg8 as a complementary unit to restore alginate production in the alg8 deletion mutant.
Interestingly, not only was the alginate production of the complemented mutant P. aeruginosa PDO300Δalg8(pBBR1MCS-5:alg8) strongly enhanced but also the polymer composition was altered as indicated by 1H-NMR analysis. Overproduction of Alg8 seems to influence polymer composition and properties. The 1H-NMR data indicated a slightly increased degree of acetylation, whereas the amount of guluronic acid residues was found to be significantly reduced. Further experiments are required to explain how Alg8 impacts on polymer acetylation and composition.
Fusion protein analysis and the predicted topology model suggest that Alg8 is a transmembrane protein, with a N-terminal signal sequence and four transmembrane helices. The C terminus is presumably located in the periplasm, as indicated by alkaline phosphatase activity of the respective fusion protein. Since the C terminus appears to be very hydrophobic, we suggest that the untagged C-terminal end just crosses the cytoplasmic membrane and/or stays embedded in the cytoplasmic membrane. Our Alg8 protein model prediction suggests a large cytosolic loop at the N terminus (Fig. 2). This N-terminal domain shares homology with class II β-glycosyltransferases enabling development of a structural model of Alg8 based on the known structure of glycosyltransferase SpsA. Moreover, the cytosolic localization of the putative active site is consistent with the availability of GDP-mannuronic acid in the cytosol. Cellular fractionation experiments with the alkaline phosphatase fusion protein of Alg8 and analysis of specific alkaline phosphatase activity suggested localization in the cytoplasmic membrane. Although PhoA assays of the different cellular fractions showed PhoA activity, the highest specific activity was found to be associated with the cytoplasmic membrane fraction. The structural model of Alg8 indicated that the proposed residues Asp 161, Asp 250, and Cys 215 are located in or adjacent to the core structure (Fig. 2). These residues might be involved in substrate binding, because homologous amino acids are responsible for the nucleotide-sugar binding in SpsA (4). These and other amino acids that are proposed to be involved in catalytic function are currently being investigated by site-specific mutagenesis. The identification of catalytic residues might shed light into the alginate polymerization reaction and might enable the design of inhibitors that are able to block polymerization and therefore impair biofilm formation in cystic fibrosis patients. Furthermore, inhibitors of alginate polymerization could be identified using the in vitro alginate synthesis assay as screening tool.
Acknowledgments
We thank Gudmund Skjåk-Bræk for NMR spectroscopic analysis of alginates.
This study was supported by a research grant to B.H.A.R. from the Institute of Molecular Biosciences at Massey University and the Deutsche Forschungsgemeinschaft (Re 1097/6-1 to B.H.A.R.). U.R. received a doctoral scholarship from Massey University.
REFERENCES
- 1.Albrecht, M. T., and N. L. Schiller. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernsel, A., and G. Von Heijne. 2005. Improved membrane protein topology prediction by domain assignments. Protein Sci. 14:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenkrantz, N., and G. Asboe-Hansen. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484-489. [DOI] [PubMed] [Google Scholar]
- 4.Charnock, S. J., and G. J. Davies. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA, from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380-6385. [DOI] [PubMed] [Google Scholar]
- 5.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-593. [DOI] [PubMed] [Google Scholar]
- 6.Darzins, A., and A. M. Chakrabarty. 1984. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J. Bacteriol. 159:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deretic, V., J. F. Gill, and A. M. Chakrabarty. 1987. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 15:4567-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deretic, V., M. J. Schurr, J. C. Boucher, and D. W. Martin. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 176:2773-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ethier, J., and J. M. Boyd. 2000. Topological analysis and role of the transmembrane domain in polar targeting of PilS, a Pseudomonas aeruginosa sensor kinase. Mol. Microbiol. 38:891-903. [DOI] [PubMed] [Google Scholar]
- 10.Evans, L. R., and A. Linker. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin, M. J., C. E. Chitnis, P. Gacesa, A. Sonesson, D. C. White, and D. E. Ohman. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, M. J., and D. E. Ohman. 1993. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, M. J., and D. E. Ohman. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacesa, P., and N. J. Russell. 1990. The structure and property of alginate, p. 29-49. In P. Gacesa and N. J. Russell (eds.), Pseudomonas infection and alginates. Chapman & Hall, London, England.
- 16.Gimmestad, M., H. Sletta, H. Ertesvag, K. Bakkevig, S. Jain, S.-J. Suh, G. Skjåk-Bræk, T. E. Ellingsen, D. E. Ohman, and S. Valla. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J. Bacteriol. 185:3515-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg, J. B., K. Hatano, and G. B. Pier. 1993. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J. Bacteriol. 175:1605-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasdalen, H., B. Larsen, and O. Smidsrod. 1979. A P.M.R. study of the composition and sequence of uronate residues in alginates. Carbohydr. Res. 68:23-31. [Google Scholar]
- 19.Gutierrez, C., and J. C. Devedjian. 1989. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 17:3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, N., A. A. Amara, B. B. Beermann, Q. Qi, H. J. Hinz, and B. H. Rehm. 2002. Biochemical characterization of the Pseudomonas putida 3-hydroxyacyl ACP:CoA transacylase, which diverts intermediates of fatty acid de novo biosynthesis. J. Biol. Chem. 277:42926-42936. [DOI] [PubMed] [Google Scholar]
- 22.Holloway, B. W., H. Matsumoto, and P. V. Phibbs, Jr. 1986. The chromosome map of Pseudomonas aeruginosa PAO. Acta Microbiol. Pol. 35:161-164. [PubMed] [Google Scholar]
- 23.Jain, S., M. J. Franklin, H. Ertesvag, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 24.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 26.Karchin, R., M. Cline, Y. Mandel-Gutfreund, and K. Karplus. 2003. Hidden Markov models that use predicted local structure for fold recognition: alphabets of backbone geometry. Proteins Struct. Function Genet. 51:504-514. [DOI] [PubMed] [Google Scholar]
- 27.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 28.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 29.Linker, A., and R. S. Jones. 1966. A new polysaccharide resembling alginic acid isolated from pseudomonads. J. Biol. Chem. 241:3845-3851. [PubMed] [Google Scholar]
- 30.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maharaj, R., T. B. May, S. K. Wang, and A. M. Chakrabarty. 1993. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene 136:267-269. [DOI] [PubMed] [Google Scholar]
- 32.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell. Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 33.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathee, K., C. Sternberg, O. Ciofu, P. Jensen, J. Campbell, M. Givskov, D. E. Ohman, N. Hoiby, S. Molin, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiol. 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 35.Mejia-Ruiz, H., J. Guzman, S. Moreno, G. Soberon-Chavez, and G. Espin. 1997. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene 199:271-277. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Moller, S., M. D. Croning, and R. Apweiler. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646-653. [DOI] [PubMed] [Google Scholar]
- 38.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, S. S., N. Hoiby, F. Espersen, and C. Koch. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pier, G. B., F. Coleman, M. Grout, M. Franklin, and D. E. Ohman. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pindar, D. F., and C. Bucke. 1975. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem. J. 152:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preiss, J. 1966. GDP-mannose-dehydrogenase from Arthrobacter. Methods Enzymol. 8:285-287. [Google Scholar]
- 45.Rehm, B. H. A. 2002. Alginates from bacteria, p. 179-211. In E. J. Vandamme, S. DeBaets, and A. Steinbüchel (ed.), Biopolymers, vol. polysaccharides. Wiley VCH, New York, N.Y.
- 46.Rehm, B. H. A. 1998. Alginate lyase from Pseudomonas aeruginosa CF1/M1 prefers the hexameric oligomannuronate as substrate. FEMS Microbiol. Lett. 165:175-180. [DOI] [PubMed] [Google Scholar]
- 47.Rehm, B. H. A. 1996. The Azotobacter vinelandii gene algJ encodes an outer membrane protein presumably involved in export of alginate. Microbiology 142:873-880. [DOI] [PubMed] [Google Scholar]
- 48.Rehm, B. H. A., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robles-Price, A., T. Y. Wong, H. Sletta, S. Valla, and N. L. Schiller. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roychoudhury, S., T. B. May, J. F. Gill, S. K. Singh, D. S. Feingold, and A. M. Chakrabarty. 1989. Purification and characterization of guanosine diphospho-d-mannose dehydrogenase: a key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J. Biol. Chem. 264:9380-9385. [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Saxena, I. M., and R. M. Brown, Jr. 2000. Cellulose synthases and related enzymes. Curr. Opin. Plant Biol. 3:523-531. [DOI] [PubMed] [Google Scholar]
- 53.Saxena, I. M., R. M. Brown, Jr., and T. Dandekar. 2001. Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57:1135-1148. [DOI] [PubMed] [Google Scholar]
- 54.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209-222. (In German.) [PubMed] [Google Scholar]
- 57.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schurks, N., J. Wingender, H. C. Flemming, and C. Mayer. 2002. Monomer composition and sequence of alginates from Pseudomonas aeruginosa. Int. J. Biol. Macromol. 30:105-111. [DOI] [PubMed] [Google Scholar]
- 59.Schurr, M. J., D. W. Martin, M. H. Mudd, N. S. Hibler, J. C. Boucher, and V. Deretic. 1993. The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis. Cell. Mol. Biol. Res. 39:371-376. [PubMed] [Google Scholar]
- 60.Shankar, S., R. W. Ye, D. Schlictman, and A. M. Chakrabarty. 1995. Exopolysaccharide alginate synthesis in Pseudomonas aeruginosa: enzymology and regulation of gene expression. Adv. Enzymol. Relat. Areas Mol. Biol. 70:221-255. [DOI] [PubMed] [Google Scholar]
- 61.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 62.Skjåk-Bræk, G., H. Grasdalen, and B. Larsen. 1986. Monomer sequence and acteylation pattern in some bacterial alginates. Carbohydr. Res. 154:239-250. [DOI] [PubMed] [Google Scholar]
- 63.Wang, S. K., I. Sa'-Correia, A. Darzins, and A. M. Chakrabarty. 1987. Characterization of the Pseudomonas aeruginosa alginate (alg) gene region II. J. Gen. Microbiol. 133:2303-2314. [DOI] [PubMed] [Google Scholar]
- 64.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]