Abstract
Novel drug targets are required in order to design new defenses against antibiotic-resistant pathogens. Comparative genomics provides new opportunities for finding optimal targets among previously unexplored cellular functions, based on an understanding of related biological processes in bacterial pathogens and their hosts. We describe an integrated approach to identification and prioritization of broad-spectrum drug targets. Our strategy is based on genetic footprinting in Escherichia coli followed by metabolic context analysis of essential gene orthologs in various species. Genes required for viability of E. coli in rich medium were identified on a whole-genome scale using the genetic footprinting technique. Potential target pathways were deduced from these data and compared with a panel of representative bacterial pathogens by using metabolic reconstructions from genomic data. Conserved and indispensable functions revealed by this analysis potentially represent broad-spectrum antibacterial targets. Further target prioritization involves comparison of the corresponding pathways and individual functions between pathogens and the human host. The most promising targets are validated by direct knockouts in model pathogens. The efficacy of this approach is illustrated using examples from metabolism of adenylate cofactors NAD(P), coenzyme A, and flavin adenine dinucleotide. Several drug targets within these pathways, including three distantly related adenylyltransferases (orthologs of the E. coli genes nadD, coaD, and ribF), are discussed in detail.
The growing number of antibiotic-resistant microbial pathogens (44, 46; http://www.cdc.gov/narms/default.htm) presents a serious challenge to modern medicine. The majority of existing antibiotics utilize a limited number of core chemical structures and target only a few cellular functions, such as cell wall biosynthesis, DNA replication, transcription, and translation (67). Identification of unexplored cellular functions as potential targets is a prerequisite for development of novel antibiotic chemotypes. Choosing an optimal target function is a crucial step in the long and expensive process of drug development and requires the best possible understanding of related biological processes in bacterial pathogens and their hosts.
Extensive programs utilizing genomic information to search for novel antimicrobial targets have been launched recently in industry and academia (14, 15, 69, 72, 91). Complete genome sequences of multiple bacterial species, including many major pathogens, have become available in the last few years, with many more such projects under way (16). The abundance of genomic data has enabled the development of novel postgenomic experimental and computational techniques aimed at drug target discovery. Experimental approaches to genome-wide identification of genes essential for cell viability in several microbial species have been reviewed recently (23, 43, 54, 59, 67, 77, 79). These techniques are based on either systematic gene inactivation by directed mutagenesis on a whole-genome scale or high-throughput random transposon mutagenesis. The major advantage of the latter technique is the parallel analysis of thousands of genes under multiple growth conditions.
A transposon-based approach termed “genetic footprinting” was originally described for Saccharomyces cerevisiae (84, 85). Genetic footprinting is a three-step process: (i) random transposon mutagenesis of a large number of cells, (ii) competitive outgrowth of the mutagenized population under various selective conditions, and (iii) analysis of individual mutants surviving in the population using direct sequencing or various hybridization and PCR-based techniques. Various modifications of genetic footprinting have been recently applied to several microorganisms, including Mycoplasma genitalium and Mycoplasma pneumoniae (49), Pseudomonas aeruginosa (99), Helicobacter pylori (52), and Escherichia coli (7, 45). Another version of this method, termed genomic analysis and mapping by in vitro transposition, has been developed for Haemophilus influenzae (1, 75) and Streptococcus pneumoniae (1). Direct application of this technique is usually limited to microbial species with natural competence, since transposon mutagenesis is performed in vitro on isolated DNA fragments, and mutations are introduced into the genome by transformation with linear DNA fragments followed by gene conversion. By targeting a specific genomic region, this approach increases insertion density, improving resolution of genetic footprinting. An elegant extension of this method beyond naturally competent species was described for P. aeruginosa (99).
Genetic footprinting in E. coli by mini-Tn10 mutagenesis in vivo has been recently reported (7, 45). In our systematic analysis of E. coli gene essentiality, we have chosen an approach based on the use of the “transposome,” precut transposon DNA in a stable complex with modified Tn5 transposase (40, 47). This approach has several advantages over mini-Tn10 mutagenesis: (i) only single irreversible insertions are produced, since the only source of transposase activity is the protein within a transposome complex; (ii) there is no need to assemble an elaborate transposon delivery vector system with tight regulation of replication and transposase expression; and (iii) a limit of one insertion per cell can be achieved based on the ratio of transposome complexes to competent cells at the time of transformation.
Computational techniques for the identification of potential drug targets based on genomic data have been reviewed recently (35, 37, 67, 79). In a few studies, in silico analysis was successfully combined with experimental techniques (4, 23, 36). For example, Arigoni and coauthors used comparative genome analysis to identify previously uncharacterized genes as potential broad-spectrum targets by emphasizing genes which are (i) broadly conserved in various bacteria, including pathogens; (ii) not conserved in humans; and (iii) likely to encode soluble proteins. The essentiality of selected genes was further assessed by directed knockouts in E. coli and Bacillus subtilis (4). Most in silico target identification techniques are focused on formal comparative sequence analysis, without attempting to assess conservation of the overall biological context in various pathogens and the human host. We believe that comparative analysis of pathways and biological subsystems may significantly improve our ability to select potential targets.
Here we describe an approach to the identification and prioritization of broad-spectrum antimicrobial targets with known functional roles. We use a list of essential genes inferred by genetic footprinting in a model microorganism (E. coli) as a starting point for analysis of the corresponding functional roles in the context of metabolic pathways in microbial pathogens. This kind of comparative analysis based on metabolic reconstruction from genomic data indicates suitable target pathways and target functions within these pathways, as well as the potential range of pathogens for each target. Comparison of microbial pathways and individual target proteins with their human counterparts allows further target prioritization. Finally, prospective targets are validated by directed knockouts in model pathogens (Staphylococcus aureus or H. influenzae). The corresponding proteins are cloned from the representative pathogens and human cDNA libraries, expressed and analyzed in detail. The most advanced target proteins from representative bacterial sources, as well as their human counterparts, are crystallized, and their structures are determined to assist in further drug development.
We illustrate this approach using examples from the biosynthesis of three adenylate cofactors, NAD, coenzyme A (CoA), and flavin adenine dinucleotide (FAD), presenting several promising targets for the development of novel broad-spectrum antibiotics.
MATERIALS AND METHODS
Strains.
E. coli strains MG1655 (F− lambda− ilvG rfb50 rph1) (53) and DH10B [F′ mcrA Δ(mrr-hsdRMS-msrBC) φ80lacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara-leu)7697 araD139 galU galK nupG rpsL] (42) and S. aureus strain RN4220 (55) were used in this work.
Transposome formation and transposon mutagenesis.
Plasmid pMOD<MCS> containing artificial transposon EZ::TN<KAN-2> (Epicentre Technologies, Madison, Wis.) was isolated from the same strain (MG1655 or DH10B) that was subsequently mutagenized to avoid restriction-modification problems. Transposon DNA was released by PvuII digestion, as recommended by the manufacturer, and gel purified using QIAquick gel extraction columns (Qiagen, Valencia, Calif.). Transposomes were preformed by incubating transposon DNA (7 ng/μl) with hyperactive Tn5 EZ::TN transposase (0.1 U/μl; Epicentre Technologies and generous gift from W. Reznikoff) in a solution containing 40 mM Tris-acetate (pH 7.5), 100 mM potassium glutamate, 0.1 mM EDTA, 1 mM dithiothreitol, and tRNA (0.1 mg/ml). Samples were incubated for 30 min at 37°C and dialyzed against 10 mM Tris-acetate, pH 7.5, plus 1 mM EDTA on 0.05-μm filters (Millipore, Bedford, Mass.) for 1 h. Dialyzed samples were mixed with electrocompetent E. coli in a 1:2 ratio (vol/vol) and transformed by electroporation. Cultures were immediately diluted with a Luria-Bertani-based rich medium (see below) without kanamycin and incubated at 37°C for 40 min with gentle agitation. The efficiency of electroporation for E. coli strains MG1655 and DH10B was 5 × 104 and 2 × 106 kanamycin-resistant colonies per 1 μg of transposon DNA, respectively.
Outgrowth of mutagenized population.
Half of the mutagenized population was immediately frozen and stored as the time zero sample. The rest of the culture was used to inoculate a BIOFLO 2000 fermentor (New Brunswick Scientific, Edison, N.J.) containing 950 ml of the following medium: tryptone, 10 g/liter; yeast extract, 5 g/liter; 50 mM NaCl, 9.5 mM NH4Cl, 0.528 mM MgCl2, 0.276 mM K2SO4, 0.01 mM FeSO4, 5 × 10−4 mM CaCl2, and 1.32 mM K2HPO4. This medium was supplemented with the following micronutrients: 3 × 10−6 mM (NH4)6(MoO7)24, 4 × 10−4 mM H3BO3, 3 × 10−5 mM CoCl2, 10−5 mM CuSO4, 8 × 10−5 mM MnCl2, and 10−5 mM ZnSO4 (68). The following vitamins were added (concentrations are in milligrams per liter): biotin, 0.12; riboflavin, 0.8; pantothenic acid, 10.8; niacinamide, 12.0; pyridoxine, 2.8; thiamine, 4.0; lipoic acid, 2.0; folic acid, 0.08; and p-aminobenzoic acid, 1.37. Kanamycin was added to 10 μg/ml. Throughout the fermentation temperature was held at 37°C, dissolved oxygen was held at 30 to 50% of saturation, and the pH was held at 6.95 (via titration with 5% H3PO4). Media and growth conditions were designed to minimize the number of genes required for cell survival. Cells were grown in batch culture for 23 population doublings (12 h) to a cell density of 1.4 × 109. Genomic DNA was isolated and used to generate genetic footprints.
Detection of transposon insertions by nested PCR.
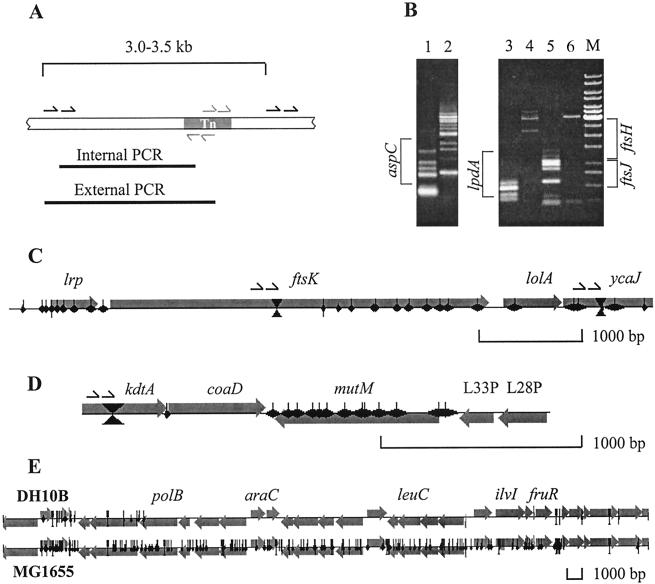
Two pairs of primers were used consecutively, with the second pair of primers nested within the first (see Fig. 2A). Each primer pair contained one transposon-specific primer and one chromosome-specific primer. Chromosome-specific landmark primers were designed as an ordered set of unidirectional primer pairs covering the entire E. coli genome by using custom software. Pairs were separated on average by 3,500 bp, while primers within each pair were separated by the shortest possible distance in the range −3 to 900 bp. Average primer length was 27 bp (sequences available upon request). Transposon-specific primers were chosen to avoid any significant similarity with the E. coli chromosome, using PrimerSelect software (DNASTAR, Inc., Madison, Wis.). Two pairs of nested, outwardly directed transposon-specific primers (one at each end) were used to detect transposons inserted in both orientations (see Fig. 2A). The forward primer pair includes an external primer, 5′-GTTCCGTGGCAAAGCAAAAGTTCAA-3′, and an internal primer, 5′-GGTCCACCTACAACAAAGCTCTCATCA-3′. The reverse primer pair includes an external primer, 5′-CCGACATTATCGCGAGCCCATTTAT-3′, and an internal primer, 5′-GCAAGACGTTTCCCGTTGAATATGGC-3′.
FIG. 2.
Detection and mapping of transposon insertions. (A) Primer strategy for nested PCR. Transposon-specific primers are shown in gray; chromosome-specific landmark primers are shown in black. (B) The gel image shows analysis of the three chromosomal loci: aspC (nonessential) and lpdA and ftsJ (essential) at time zero (lanes 1, 3, and 5) and after outgrowth (lanes 2, 4, and 6). ORF locations are marked relative to each pair of lanes. Several inserts are visible in lpdA and ftsJ at time zero, while none can be detected after outgrowth. The nonessential aspC contains insertions at both time points. (C to E) Examples of genetic footprints. Note that the scale is different in each panel. The length and direction of each gene are indicated by the large horizontal gray arrows. Black diamonds represent transposon inserts. The width of each diamond corresponds to the mapping error introduced by gel electrophoresis. The positions of the landmark PCR primers are shown by bows crossing the genes, as well as by arrows above the genes. (C) Genetic footprinting of the ftsK locus in MG1655. Only the 3′ half of this essential gene contains inserts. (D) Genetic footprinting of the coaD locus in MG1655. A transposon insertion immediately upstream of this essential gene apparently does not interfere with its expression. (E) Mapping of the Δ(ara-leu)7697 deletion in DH10B. The genetic footprint of the corresponding region in MG1655 is shown for comparison.
The first of two consecutive PCR amplifications (the external PCR) was performed under the following conditions: 95°C for 1 min; 94°C for 12 s, 70°C for 6 min (2 cycles); 94°C for 12 s, 69°C for 6 min (2 cycles); 94°C for 12 s, 68°C for 6 min (36 cycles); 68°C for 6 min. Amplification reactions contained 0.3 μg of template DNA (equivalent of 6 × 107 E. coli genomes), a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each primer, PCR buffer (40 mM Tricine-KOH [pH 9.2], 15 mM potassium acetate, 3.5 mM magnesium acetate, bovine serum albumin [3.75 μg/ml]), and 0.4 μl of Advantage cDNA Polymerase Mix (Clontech Laboratories, Palo Alto, Calif.) in 20 μl. The second internal PCR was performed in the same reaction mix, except the DNA templates consisted of the products of the first PCR diluted 103-fold. Amplification conditions for internal PCR were as follows: 95°C for 1 min; 94°C for 12 s, 69°C for 6 min (2 cycles); 94°C for 12 s, 68°C for 6 min (9 cycles); 68°C for 6 min. The products of the internal PCRs (3-μl aliquots) were size separated on 0.65% agarose gels. All insert detection and analysis procedures were performed in a 96-well format.
Image capture and analysis were performed with 1D Image Analysis Software (Eastman Kodak Company, Rochester, N.Y.). Mapping of the detected Tn5 insertions was done using custom software, which calculates insert positions within a genome sequence using the addresses of the internal landmark primers and the size of the corresponding PCR products. Visualization of insert locations was done using custom software integrated into the ERGO database. The microbial genome database WIT, a predecessor of ERGO, has been described previously (71).
Optimization of experimental conditions for low-noise detection of transposon insertions.
Reaction conditions were optimized using a small set of control genes with known essentiality, in order to detect the maximum number of inserts, while keeping the level of PCR-introduced noise very low. First, the minimum amount of genomic DNA that contained the representative mix of all mutant chromosomes and consistently yielded reproducible patterns of bands in a PCR was determined. Second, the products of external and corresponding internal PCRs were analyzed side by side on an agarose gel. This comparison was used as a guide for optimizing the PCR parameters: cooling rate, number of cycles in external versus internal PCR, and the rate of PCR products' dilutions. PCR conditions were fine-tuned to yield the minimum number of false products, namely, internal PCR bands lacking the corresponding external PCR product. Third, genetic footprints of a 4-kb nadD locus were generated independently using identical template DNA samples and four different chromosome-specific nested primer pairs. All four footprints yielded coherent patterns, consistently visualizing 10 transposon inserts in the area (data not shown). Finally, 12 random internal PCR bands were gel purified and sequenced. All the bands originated from the expected chromosomal loci. Comparison of the calculated insert locations with the sequencing results indicated a mapping error introduced by gel electrophoresis of about 4.5% of the size of each band.
Metabolic reconstruction from genomic sequence data.
The versions of genomes used in this work are as follows: E. coli K-12 MG1655 (18) (GenBank accession no. U00096), P. aeruginosa PAO1 (88) (GenBank accession no. AE004091), Bacillus anthracis Ames (http://www.tigr.org/tdb/mdb/mdbinprogress.html), Mycobacterium tuberculosis H37Rv (24) (GenBank accession no. AL123456), H. pylori J99 (3) (GenBank accession no. AE001439), S. aureus COL (http://www.tigr.org/tdb/mdb/mdbinprogress.html/), S. pneumoniae (GenBank accession no. AE005672), M. genitalium (34) (GenBank accession no. L43967), H. influenzae Rd (32) (GenBank accession no. L42023), Chlamydia trachomatis (87) (GenBank accession no. AE001273), and Homo sapiens (http://www.ncbi.nlm.nih.gov/genome/guide/human/).
Comparisons of the specific biochemical capabilities of E. coli, relevant bacterial pathogens, and the human host were based on metabolic reconstructions performed in the ERGO database. Metabolic reconstructions in ERGO are tentative projections of all known biochemical pathways onto a specific organism with a completely sequenced and annotated genome (described in reference 82). These projections are based on the presence or absence of orthologs of specific genes known, on the basis of studies with other organisms, to be involved in corresponding pathways. This approach produces an estimate of the metabolic potential of a given organism (many asserted pathways may or may not be actually expressed under specific conditions) but without direct experimental data it falls short of defining actual metabolic fluxes.
Examples illustrating this type of analysis are given in Tables 2 to 4 for three metabolic subsystems (groups of pathways) related to biosynthesis of the adenylate cofactors NAD(P), CoA, and FAD. We define a pathway as a series of consecutive transformations without bifurcations (82). Merging and/or alternative pathways (numbered I and II, etc., in Table 2) are combined in higher hierarchical blocks, which ultimately form a subsystem. In contrast to pathways, subsystems do not have formally defined boundaries, and they are assembled based on conventions in the field (such as cofactor biosynthesis described in references 5, 12, and 13), as well as on biological expertise and the interests of a particular user.
TABLE 2.
Functional reconstruction of metabolic pathways of NAD/NADP biosynthesisa
| NAD(P) biosynthesis | EC no. | Protein example | Presence of ortholog or pathway in genome of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. colib | P. aeruginosa | B. anthracis | M. tuberculosis | H. pylori | S. pneumoniae | S. aureus | M. genitalium | H. influenzae | C. trachomatis | H. sapiens | |||
| De novo pathways to NaMN | Yes | Yes | Yes | Yes | Yes | No | No | No | No | No | Yes | ||
| (I) Tryptophan to quinolinate | − | − | − | − | − | − | − | − | − | − | + | ||
| (II) Aspartate to quinolinate | |||||||||||||
| Aspartate oxidase | 1.4.3.16 | gi|16130499 | nadB | + | + | + | + | − | − | − | − | − | − |
| Quinolinate synthase | gi|16128718 | nadA | + | + | + | + | − | − | − | − | − | − | |
| Quinolinate to NaMN | |||||||||||||
| Quinolinate phosphoribosyl transferase | 2.4.2.19 | gi|16128102 | nadC | + | + | + | + | − | − | − | − | − | + |
| Salvage: from niacin to NaMN/NMN | Yes | Yes | Yes | Yese | No | Yes | Yes | Yes | No | No | Yes | ||
| (I) Nicotinamide to NaMN | |||||||||||||
| Nicotinamide deamidase | 3.5.1.19 | gi|140602 | pncA | + | + | + | − | + | + | − | − | − | − |
| Nicotinate phosphoribosyl transferase | 2.4.2.11 | gi|16128898 | pncB | + | + | + | − | + | + | − | − | − | − |
| (II) Nicotinamide to NMN | |||||||||||||
| Nicotinamide phosphoribosyl transferase (nadVc) | 2.4.2.12 | gi|13629024 | − | − | − | − | − | − | − | + | − | − | + |
| Salvage: from exogenous NMN to NAD | Yes | Yes | No | No | No | No | No | No | Yes | No | No | ||
| NMN phosphohydrolase (extracellular) (nadNd) | 3.1.3.5 | gi|1573165 | ushA | − | + | − | − | − | − | − | + | − | + |
| Pyridine nucleoside transporter | gi|16273640 | pnuC | + | + | − | + | + | − | − | + | − | − | |
| Nicotinamide ribose kinasef | 2.7.1.22 | gi|16132207 | nadR | + | − | + | − | − | − | − | + | − | − |
| NMN adenylyltransferase | |||||||||||||
| (i) NMN specific (archaea, some bacteria) | 2.7.7.1 | gi‖16132207 | nadR | − | − | + | − | − | − | − | + | − | − |
| (ii) NaMN/NMN specific (eukarya) | 2.7.7.1/18 | gi|11245478 | − | − | − | − | − | − | − | − | − | − | + |
| Common pathway to NAD/NADP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | ||
| NaMN to NAD | |||||||||||||
| NaMN adenylyltransferase | |||||||||||||
| (i) NaMN specific (most bacteria) | 2.7.7.18 | gi|1723307 | nadD | + | + | + | + | + | + | + | − | − | − |
| (ii) NaMN/NMN specific (eukarya) | 2.7.7.1/18 | gi|11245478 | − | − | − | − | − | − | − | − | − | − | + |
| NAD synthetase | 6.3.5.1 | gi|16129694 | nadE | + | + | + | + | + | + | + | − | − | + |
| NAD to NADP | |||||||||||||
| NAD kinase | 2.7.1.23 | gi|8489010 | nadF | + | + | + | + | + | + | + | + | − | + |
Subsystems, pathways, and individual functional roles (specified by Enzyme Classification [EC] numbers) involved in adenylate cofactor biosynthesis in all relevant organisms are presented as nodes and branches (rows) of a hierarchical tree. Proteins are shown by representative examples (GenBank ID numbers). The presence (+) or absence (−) of an orthologous gene in a specific genome is marked in the column corresponding to this organism, except for E. coli, where actual gene names are given. Alternative pathways within a subsystem are indicated by uppercase roman numerals. Nonorthologous genes with the same functional role are shown in separate rows and indicated by lowercase roman numerals. Bifunctional (fused) proteins with more than one functional domain are shown in boldface type. “Yes” indicates that a particular subsystem can be asserted in a given organism by the presence of required genes (enzymatic functions) in its genome. “No” indicates that a particular subpathway cannot be asserted in an organism on the basis of genomic data alone. Functional roles inferred by metabolic context analysis but not associated with specific genes (“missing genes”) are indicated by question marks.
Identical patterns of pathways with very similar genes are found in E. coli, Salmonella enterica serovar Typhi, and Yersinia pestis in all of the subsystems considered.
Identified in Haemophilus ducreyi; present in V factor-independent Pasteurellaceae, but not in H. influenzae (62).
Identified in H. influenzae (76); closest homolog in E. coli is ushA.
Experimental data suggest that this pathway is not functional in M. tuberculosis (83).
The corresponding function in E. coli and H. influenzae is catalyzed by the C-terminal domain of the multifunctional protein NadR (O. Kurnasov et al., unpublished data).
TABLE 4.
Functional reconstruction of metabolic pathways of FMN/FAD biosynthesisa
| FMN/FAD biosynthesis | EC no. | Protein example | Presence of ortholog or pathway in genome of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. colib | P. aeruginosa | B. anthracis | M. tuberculosis | H. pylori | S. pneumoniae | S. aureus | M. genitalium | H. influenzae | C. trachomatis | H. sapiens | |||
| De novo pathways to riboflavin | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No | ||
| Ribuloso-5-phosphate to l-3,4-dihydroxy-2-butanone-4-phosphate | |||||||||||||
| 3,4-Dioxy-2-butanone-4-phosphate synthetase | 4.1.2.- | gi|1789420 | ribBc | + | + | + | + | + | + | − | + | + | − |
| GTP to 5-amino-6-ribitylamino-2,4-(1H,3H)-pyrimidinedione | |||||||||||||
| GTP cyclohydrolase II | 3.5.4.25 | gi|16129238 | ribAc | + | + | + | + | + | + | − | + | + | − |
| Pyrimidine deaminase | 3.5.4.26 | gi|1786616 | ribDd | + | + | + | + | + | + | − | + | + | − |
| Pyrimidine reductase | 1.1.1.193 | gi|1786616 | ribDd | + | + | + | + | + | + | − | + | + | − |
| Pyrimidine phosphatase | 3.1.3 | ? | ? | ? | ? | ? | ? | ? | − | ? | ? | − | |
| l-3,4-Dihydroxy-2-butanone-4-phosphate, 5-amino-6-ribitylamino-2,4- (1H,3H)-pyrimidinedione to riboflavin | |||||||||||||
| 6,7-Dimethyl-8-ribityl-lumazine synthase | 2.5.1.9 | gi|16128400 | ribH | + | + | + | + | + | + | − | + | + | − |
| Riboflavin synthase | 2.5.1.9 | gi|16129620 | ribE | + | + | + | + | + | + | − | + | + | − |
| Riboflavin (vitamin B2) salvage | No | No | Yes | No | No | Yes | Yes | Yes | No | No | Yes | ||
| Riboflavin transporter (ypaA)e | gi|16079362 | − | − | + | − | − | + | + | ? | − | − | ? | |
| Common pathway to FMN/FAD | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | ||
| Riboflavin kinase | 2.7.1.26 | gi|16128019 | ribFf | + | + | + | + | + | + | + | + | + | + |
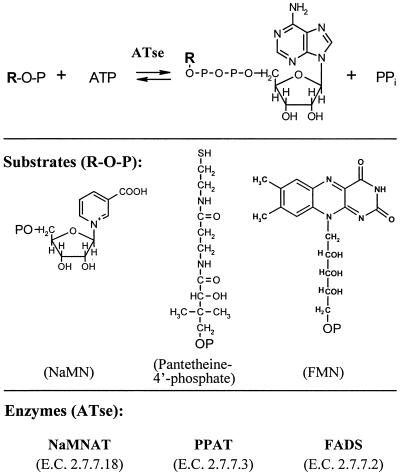
| FAD synthetase | 2.7.7.2 | ||||||||||||
| (i) Bacteria | gi|16128019 | ribFf | + | + | + | + | + | + | + | + | + | − | |
| (ii) Eukarya | gi|6320159g | − | − | − | − | − | − | − | − | − | − | + | |
See Table 2, footnote a, for details.
Identical patterns of pathways with very similar genes are found in E. coli, Salmonella enterica serovar Typhi, and Yersinia pestis in all of the subsystems considered.
RibA and RibB form a bifunctional (fused) protein in many bacteria and monofunctional separate proteins in E. coli and H. influenzae.
RibD is a bifunctional (fused) protein in most bacteria.
Riboflavin transporter originally identified in B. subtilis (56).
RibF is a bifunctional (fused) protein in most bacteria.
Eukaryotic FAD synthase originally identified in S. cerevisiae (100) has no detectable sequence similarity with corresponding bacterial enzymes.
Profile HMMs.
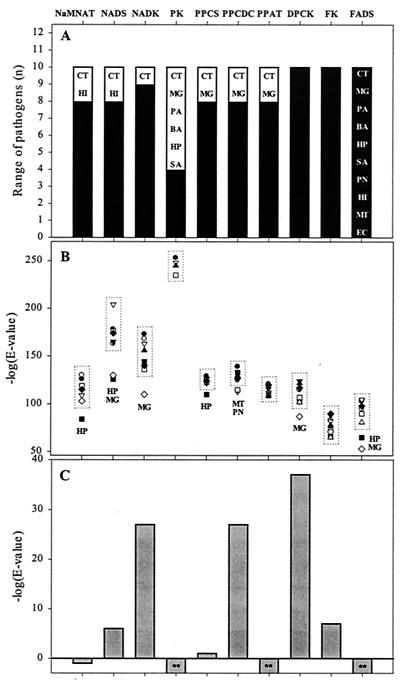
The comparisons between human and bacterial sequences were done using the HMMER 2.2 program (30; http://hmmer.wustl.edu/), with multiple-sequence alignments obtained from CLUSTAL-W (90). We first built a profile hidden Markov model (HMM) for each target enzyme, using the ortholog sequences from the pathogens of interest, as specified in Fig. 5A. HMMs for the following enzymes were built using the full-length protein sequences of nicotinic acid mononucleotide adenylyltransferase (NaMNAT), NAD synthetase (NADS), NAD kinase (NADK), pantothenate kinase (PK), phosphopantetheine adenylyltransferase (PPAT), and dephospho-CoA kinase (DPCK). Protein domains roughly corresponding to amino acid residues 1 to 212 and 213 to 413 of the E. coli CoaBC protein were used for the phosphopantothenoylcysteine decarboxylase (PPCDC) and phosphopantothenoylcysteine synthetase (PPCS) HMMs, respectively. Likewise, protein domains approximately corresponding to amino acid residues 1 to 176 and 177 to 214 of the E. coli RibF protein were used to build profile HMMs of FAD synthase (FADS) and flavokinase (FK), respectively. Obtained HMMs were then run against each bacterial and human sequence, using a database size of 106 and a high-cutoff E value of 104. The resulting E values give an indication of how closely the bacterial sequences are related and how remote the human sequence is from the conserved core of the bacterial group (the higher the E value is, the higher the divergence is).
FIG. 5.
Prioritization of potential broad-spectrum antibacterial drug targets in biosynthetic pathways of NAD(P), CoA, and FAD. Three criteria are shown for each target enzyme (marked by abbreviations along the x axis). (A) Range of pathogens. A number of pathogens (from the representative set) containing the given target enzyme (and included in building of the corresponding HMM) are represented by filled bars. Organisms not containing this target enzyme and therefore excluded from the spectrum are indicated inside open bars (by two-letter abbreviations). (B) Compact pathogen subsets and outliers. Negative logarithms of the E values indicate how closely each bacterial sequence is related to the probabilistic consensus of the corresponding profile HMM (larger values indicate higher similarity). Subsets of pathogens with target orthologs most closely related to the consensus are shown inside dotted boxes. Outliers are indicated by two-letter organism abbreviations. Organisms shown are P. aeruginosa (○) (PA), B. anthracis (▾) (BA), M. tuberculosis (▿) (MT), H. pylori (▪) (HP), S. pneumoniae (□) (PN), S. aureus (♦) (SA), M. genitalium (⋄) (MG), H. influenzae (▴) (HI), C. trachomatis (▵) (CT), and E. coli (•). The E. coli proteins are very similar to those of Yersinia pestis and Salmonella enterica serovar Typhi. (C) Human counterparts: distance from bacterial families. The negative logarithms of the E values of human counterpart sequences compared to the corresponding bacterial HMMs (smaller values indicate higher divergence of the human enzyme from the corresponding bacterial family). ∗∗, HMM E values for these human proteins are higher than the threshold (>10,000).
Verification of gene essentiality in E. coli.
For direct verification of gene essentiality in E. coli we used pKO3-based allelic exchange (58). This is briefly illustrated here using nadD as an example. Deletion of this gene (predicted to be essential) was attempted in parallel with nadR (a nonessential gene from the same metabolic subsystem) as a control. The E. coli nadD (12, 63) (formerly ybeN) and nadR genes with their flanking regions were PCR amplified using MG1655 genomic DNA as a template and the following primers: 5′-CAGCTGATTCGTAAGCTGCCAAGCATC-3′ and 5′-GGGGTCGACTCACTCACGGTGATAAGGATGGTTGGTGGTGATG-3′ for nadD and 5′-TGGCCTGCCGACTGACAATCTC-3′ and 5′-GCTGGAAAACGCCCTGCTGGAGT-3′ for nadR. Both PCR products were cloned into the gene replacement vector pKO3 (58). Next, the 650-bp HincII-ClaI fragment of nadR was replaced with the 1.7-kb Eco57I DNA fragment containing a tetracycline resistance cassette of pACYC184 (GenBank accession no. X06403). Similarly, the 434-bp ApaLIBglII fragment of nadD was excised and replaced with the kanamycin resistance cassette from pUC4K (GenBank accession no. X06404). E. coli strain MG1655 was transformed with these deletion plasmids. Following cointegration, resolution, and elimination of the plasmids, Kmr Sucr colonies were screened for sensitivity to chloramphenicol (58). Final verification of gene replacement was done by PCR analysis with primers flanking the region of recombination.
In the case of nadR, gene replacement was readily achieved: 98% of Kmr Sucr colonies were also Cms, and all of the Kmr Sucr Cms colonies tested by PCR contained the expected deletion in the chromosome. In the case of nadD, no Kmr Sucr Cms colonies were recovered (out of 192 Kmr Sucr colonies screened), suggesting that nadD is essential for E. coli growth in rich medium.
Verification of predicted gene essentiality in S. aureus.
Allelic exchange vector pBT2 was utilized for verification of gene essentiality in S. aureus (20). The nadD (gi|15927174) and pncA (gi|15927492) S. aureus orthologs were PCR amplified using primers 5′-ATTCCTTGTCGCCCGTTATGC-3′ and 5′-AACGCGCTTCATTGTATCCT-3′ for nadD and 5′-GTCCGTTAATCCCACAAGCATCA-3′ and 5′-CGCCGACTTTATCTTTTTCAGC-3′ for pncA. Genomic DNA isolated from S. aureus strain ATCC 29213 was used as a PCR template. Both PCR products were cloned into plasmid pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.). A 384-bp NheI-AvrII fragment of nadD was replaced with a tetracycline resistance marker (generous gift from M. Smeltzer, University of Arkansas for Medical Sciences, Little Rock), and the resulting 4.0-kb DNA fragment containing the inactivated nadD was cloned into pBT2 (20). This plasmid was introduced into S. aureus RN4220 by electroporation. Following cointegration, resolution, and elimination of the plasmid (20), tetracycline-resistant (Tetr) colonies were screened for sensitivity to chloramphenicol. No Tetr Cms colonies were found in the case of nadD, while pncA was successfully inactivated in the control experiment. In this case, 38% of the Tetr colonies were also Cms, and all of the Tetr Cms colonies tested by PCR contained the correct pncA deletion in the chromosome.
The S. aureus coaD ortholog (gi|15926709) with flanking regions was amplified by PCR using primers 5′-GATTGCCAGTTGTAGGGTTCATA-3′ and 5′-GCGTTGGCTTAATCACAGAATA-3′, cloned into pBT2 (to produce plasmid pMF32), and subjected to in vitro transposon mutagenesis. To this end an artificial transposon was constructed (M. Farrell et al., unpublished data) by cloning the Tet M marker derived from Tn916 (38) into pMOD-2<MCS> (Epicentre Technologies). Plasmid pMF32 was mixed in a 1:1 molar ratio with Tet M-transposon DNA, and EZ::TN transposase (Epicentre Technologies) was added to a 0.1-U/μl final concentration. The mixture was incubated for 2 h at 37°C in reaction buffer (50 mM Tris-acetate [pH 7.5], 150 mM potassium acetate, 10 mM magnesium acetate, and 40 mM spermidine) and used to transform E. coli strain DH10B. One hundred Tetr E. coli colonies were screened by whole-cell PCR, and five different transposon-containing pMF32 plasmids were selected, including two plasmids containing coaD inactivated by a single transposon insertion in the middle of the gene and three plasmids with a transposon in open reading frames (ORFs) immediately upstream or downstream of coaD. All five plasmids were used in parallel for pBT2-driven allelic exchange in S. aureus RN4220 (see above). No Tetr Cms (knockout) colonies were obtained for coaD disruptions, while the neighboring ORFs (encoding gi|15926708 and gi|15926710) were successfully inactivated.
Preliminary characterization of bacterial targets and their human counterparts.
Representative bacterial target enzymes and their human counterparts were expressed in E. coli BL21(DE3) as N-terminal fusions with a six-His tag using the pProEX-HT3a system (Invitrogen). Corresponding DNA fragments were amplified by PCR using bacterial genomic DNA (American Type Culture Collection, Manassas, Va.) or human brain cDNA (Clontech) and primers generating an NcoI or BspHI site at the 5′ end and a SalI or PstI site at the 3′ end. The DNA fragments were purified, digested, and cloned between NcoI and SalI (or PstI) restriction sites in the vector. The resulting constructs were verified by DNA sequencing. Recombinant proteins were purified to homogeneity using a two-step procedure consisting of chromatography on Ni-nitrilotriacetic acid agarose (Qiagen) and gel filtration (Superdex G-200) as described previously (28). Preliminary enzymatic characterizations were performed for representative bacterial and human enzymes involved in the biosynthesis of NAD (NMN and NaMNAT), CoA (PPAT), and FAD (FADS) using high-performance liquid chromatography analysis of the reaction products and/or coupled enzymatic assays, such as described in reference 27. Enzymatic reactions were performed in the presence of 0.1 mM ATP and a 0.1 mM concentration of the respective substrates: NMN (or NaMN), 4′-phosphopantetheine, or flavin mononucleotide (FMN). High-performance liquid chromatography analysis was performed using ion-pair chromatography under isocratic conditions: 100 mM sodium phosphate buffer (pH 3.5) with 8 mM tetrabutylammoniumbromide and methanol (8% for NaMNAT, 15% for PPAT, and 30% for FADS), on a column (50 by 4.6 mm [inner diameter]), packed with 5 μM C18 (Supelco).
RESULTS
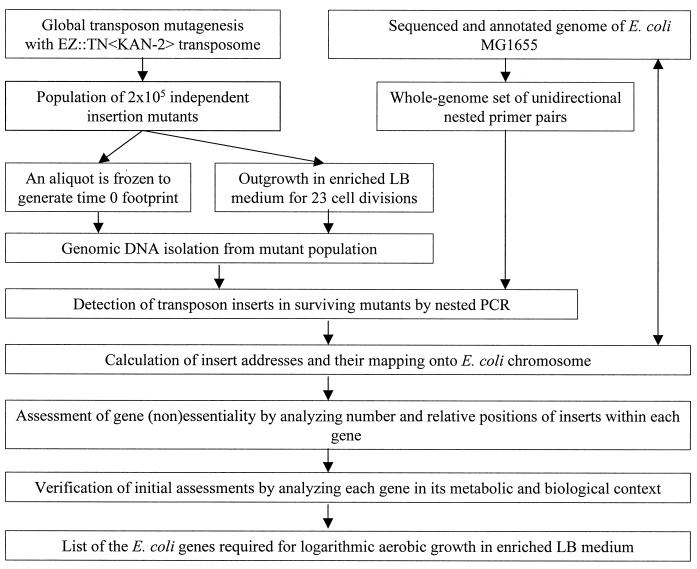
Genetic footprinting (Fig. 1) was systematically applied to determine the genes required for logarithmic aerobic growth of E. coli MG1655 in enriched Luria-Bertani medium. Genetic footprinting of a limited chromosomal region was also performed with E. coli strain DH10B under identical experimental conditions.
FIG. 1.
General scheme for E. coli genetic footprinting procedure.
Preliminary gene essentiality conclusions were made based on a semiautomatic analysis of the number and relative positions of inserts retained within each gene after selective outgrowth for 23 population doublings. Failure to recover inserts, or the presence of only a limited number of inserts at the very end of a coding sequence, suggested that cells carrying transposons in that gene were not viable under these growth conditions. However, this could also occur if a gene constituted a cold spot for transposition or if an insert had a polar effect on an essential downstream gene. We validated the technology with a combination of the following controls: (i) genetic footprinting of the mutagenized population prior to outgrowth (time zero sample) for a number of genes previously established to be essential (Fig. 2B), (ii) verification of consistency of preliminary essentiality assignments within the metabolic and biological context of each gene (see below), (iii) comparison of the observed essentiality with data reported in the literature, and (iv) genetic footprinting in the presence of a complementing DNA fragment (for an example, see reference 27).
Global transposon mutagenesis in E. coli.
Libraries of 2 × 105 independent insertion mutants were generated in E. coli strains MG1655 and DH10B using Tn5-based EZ::TN<KAN-2>Tnp transposome (Epicentre Technologies) as a transposon delivery system. It utilizes a mutant hyperactive form of the Tn5 transposase, which has been reported to have low insertion site specificity (41, 64). In our study EZ::TN transposase was found to be sufficiently random to yield 1 to 14 independent insertions within a majority of E. coli genes (for example, see Fig. 2). The average insertion density was experimentally determined to be one insert per 250 bp (after outgrowth). However, the total number of 105 independent insertion mutants analyzed after 23 cell divisions theoretically corresponds to one insert per 46 bp of genomic sequence (the E. coli genome size is 4,639 kb). We believe that this difference may be due to slight preferences in the target sequence recognition by the modified Tn5 transposase and can actually be used as a numerical measure of such a bias. In these data there is only a sixfold difference between the observed insertion density of one per 250 bp versus the frequency expected from completely random insertions (one per 46 bp). This insertion density allowed us to make essentiality assessments for 87% of E. coli ORFs, most of them larger than 80 amino acid residues in length.
To assess the reliability of our approach, we compared results from the first 50 min of the E. coli chromosome with gene essentiality data compiled from the literature by the Genetic Resource Committee of Japan (http://www.shigen.nig.ac.jp/ecoli/pec/Analyses.jsp). Of the 111 nonessential genes listed from this region of the chromosome, 98 genes (88%) were experimentally determined to be nonessential (retained inserts) by our procedure.
Of the 81 known essential genes located within the first 50 min of the E. coli chromosome, no transposon insertions were detected in 70 genes (86%) in our experiment. One gene out of the 81 analyzed, encoding essential cell division protein FtsK (11), contained 10 insertions. However, all of the inserts were clustered within the C-terminal half of the coding sequence, corresponding to amino acid residues 780 to 1329 (Fig. 2C). Only the N-terminal domain of FtsK is required for its role in cell division and viability (97). This suggests that genetic footprinting can be used to successfully detect (and even map) in vivo essential domains within ORFs, in cases where the essential region is immediately adjacent to a translational start. In the 10 remaining essential genes a single transposon insertion was detected after outgrowth in at least one of the strains (MG1655 or DH10B). This may be due to the fact that these genes can tolerate transposon inserts within certain restricted loci without a detrimental effect on the corresponding gene product. Similar phenomena have been reported in other genetic footprinting experiments in both E. coli (45) and H. influenzae (1).
Comparison of genetic footprints between two E. coli strains.
To test the reproducibility of our approach, we generated a limited genetic footprint in E. coli strain DH10B (covering the first 650 kb of the chromosome) and compared it with the corresponding region in MG1655. Unambiguous essentiality data in both strains were obtained for 530 genes (excluding those deleted in DH10B). Identical assessments of essentiality were produced for 487 genes (92%) of this group. The results of genetic footprinting experiments in MG1655 and DH10B for genes controlling biosynthesis of NAD, CoA, and FAD are presented in Table 1. For the majority of these genes the two sets of data are in good agreement. Of 13 genes that lacked transposon insertions after outgrowth in MG1655, 12 did not contain any inserts in strain DH10B as well. The only exception was nadE, which contained a single insert in the DH10B experiment. Six out of seven nonessential genes in these pathways that have been analyzed in both strains contained inserts in both MG1655 and DH10B. Only panD lacked any inserts in DH10B, while in the MG1655 experiment it contained two. This may reflect the fact that in the DH10B experiment only transposons inserted in one of two possible orientations were monitored, while both orientations were mapped in the MG1655 experiment.
TABLE 1.
Genetic footprinting of E. coli genes related to metabolism of adenylate cofactors NAD(P), CoA, and FAD
| Biosynthetic subsystem | Gene | ORF size (bp) | No. of inserts in ORF in strain:
|
Conclusionb | |
|---|---|---|---|---|---|
| MG1655 | DH10B | ||||
| NAD | nadB | 1,620 | 9 | ND | N |
| nadA | 1,041 | 12 | ND | N | |
| nadC | 891 | 1 | 4 | N | |
| ushA | 1,650 | 6 | ND | N | |
| pnuC | 717 | 4 | ND | N | |
| nadR | 1,251 | 6 | 4 | N | |
| pncA | 657 | 2 | ND | N | |
| pncB | 1,200 | 8 | ND | N | |
| nadD (ybeN) | 639 | 1 | 4 | Ec | |
| nadE | 825 | 0 | 1 | E | |
| nadF (yfiB) | 876 | 0 | 0 | E | |
| CoA | panD | 378 | 2 | 0 | N |
| panB | 792 | 1 | 3 | N | |
| panE | 909 | 5 | 2 | N | |
| panC | 849 | 1 | 5 | N | |
| coaA | 948 | 0 | 0 | E | |
| coaBC (dfp) | 1,290 | 0 | 0 | E | |
| coaD (kdtB) | 477 | 0 | 0 | E | |
| coaE (yacE) | 618 | 0 | 0 | E | |
| acpS | 378 | 0 | 0 | E | |
| FMN/FAD | ribA | 588 | 0 | 0 | E |
| ribD | 1,101 | 0 | 0 | E | |
| ribB | 651 | 0 | 0 | E | |
| ribH | 468 | 0 | 0 | E | |
| ribE | 639 | 0 | 0 | E | |
| ribF (yaaC) | 939 | 0 | 0 | E | |
The number of transposon insertions per gene in strains MG1655 and DH10B detected after logarithmic aerobic outgrowth for 23 population doublings in rich medium is shown.
Assessments of gene essentiality, based on results of genetic footprinting in strains MG1655 and DH10B. E, essential; N, nonessential; ND, not determined.
nadD appeared to be nonessential in the two genetic footprinting experiments but was then shown to be essential by a directed-knockout technique.
Comparison of the MG1655 and DH10B genetic footprints permitted detailed mapping of the two deletions in the DH10B chromosome: Δ(ara-leu)7697 (22) and ΔlacX74 (10, 89). Deletion Δ(ara-leu)7697 covers a 25.6-kb region, which corresponds to the area from 63.4 to 89.0 kb in the MG1655 genome and includes all the genes between polB and fruR (Fig. 2E). Deletion ΔlacX74 corresponds to the region from 340.3 to 369.5 kb (total of 29.2 kb) in the MG1655 sequence and includes all the genes between b0324 and b0347 and possibly mhpB (not shown).
Do transposon insertions produce polar effects?
Surprisingly, in many cases transposon insertions were detected upstream of known essential genes, where they are expected to destroy promoter sequences or otherwise have a polar effect on expression of downstream genes. This is the case with coaD (as shown in Fig. 2D), as well as with argS, frr, rplT, secA, and many other genes. We believe that the EZ::TN<KAN-2> transposon sequence inserted in either orientation is capable of initiating a level of transcription sufficient for cell survival in many cases, even though no specific promoter sequence was added to its structure. Analysis of the transposon sequence by the Neural Network Promoter Prediction program (95; http://www.fruitfly.org/seq_tools/promoter.html) reveals multiple putative E. coli-type promoters oriented outwards in both directions. This probably explains why polar effects of transposon insertions on downstream genes were rarely observed in our study. However, translational polarity on distal domains within genes is likely to be a significant limiting factor of this technique. In a multifunctional protein with an essential C-terminal domain, inserts in nonessential N-terminal regions of this protein will not be tolerated since they interfere with translation of downstream sequences. This limits the subgenic resolution of genetic footprinting to cases of multifunctional proteins with essential domains proximal to a translational start (as shown in Fig. 2C).
Analysis of gene essentiality data in the context of E. coli metabolic pathways.
One of the major advantages of genetic footprinting over directed knockouts for identification of essential genes is the fact that footprinting is a fast and highly parallel method. As with any high-throughput technique, it has limitations, for example, due to cross-feeding during outgrowth in a complex mutant population. An efficient way to minimize erroneous conclusions with respect to the essentiality of a given gene is to consider the results in the context of the corresponding metabolic pathway or subsystem. Metabolic context analysis allows reconciliation of the data from genetic footprinting experiments, from the literature (if available), as well as predicted from metabolic reconstruction. This technique is most efficiently applied to E. coli, since it is one of the best-studied model microbial systems. When contradictions between these types of data are encountered for a certain gene, it is often possible to formulate the most probable assertion by inspecting the behavior of other E. coli genes within the same pathway.
The biosynthesis of adenylate cofactors provides a good example of the utility of this approach, since (i) these cofactors are essential metabolites in all types of organisms, (ii) the corresponding pathways in E. coli are rich in essential genes, and (iii) these subsystems have been thoroughly studied biochemically and genetically. On the other hand, some of the key genes were identified only recently, and their essentiality has not been directly confirmed. The results of genetic footprinting for the majority of the known E. coli genes involved with the biosynthesis and salvage pathways producing NAD(P), CoA, and FAD are summarized in Tables 2 to 4.
Genetic footprinting of E. coli genes involved in NAD(P) biosynthesis.
The behavior of the majority of NAD(P) biosynthetic genes in genetic footprinting experiments (Table 1) is consistent with previously published data and with the metabolic reconstruction of this system presented in Fig. 3A. As expected, all the genes of the de novo and salvage pathways appear nonessential, since in both experiments outgrowth occurred in rich media containing niacinamide. Nicotinamide riboside (NmR) is probably the most advanced NAD(P) precursor that can be transported by E. coli. Therefore, all the genes of the common pathway should be essential, since their inactivation cannot be compensated for by NAD(P) salvage. This is consistent with our observations, with the notable exception of nadD. Multiple insertions in this gene were observed, suggesting that nadD is dispensable for E. coli growth in rich media. This contradicted genetic data for Salmonella enterica serovar Typhimurium (48), as well as our own experiments in E. coli MG1655, which failed to produce a directed deletion of nadD in rich medium, even in the presence of 50 μM NAD or NMN. In our hands, nadD could only be successfully deleted in the presence of a plasmid containing a functional human nadD ortholog, pyridine nucleotide adenylyltransferase 1 (PNAT-1) (gi|12620200; O. Kurnasov, unpublished data).
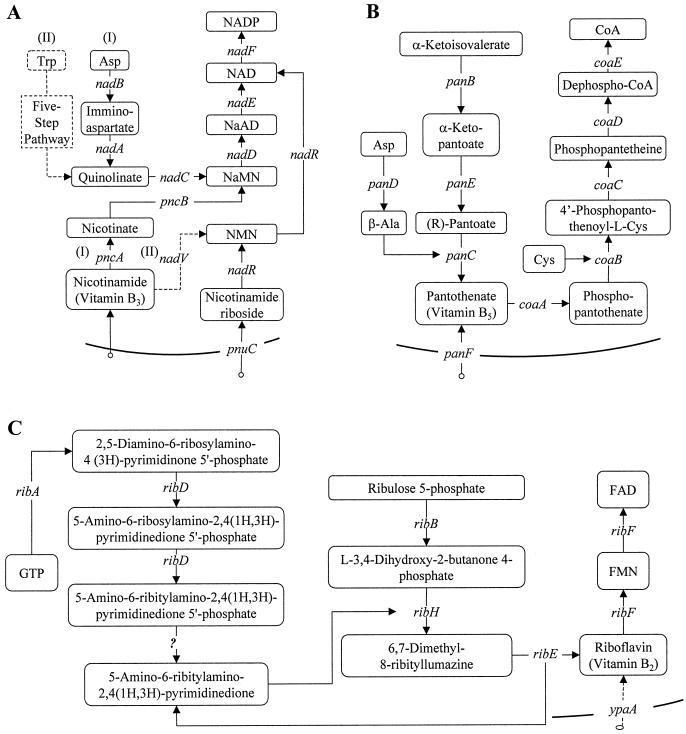
FIG. 3.
Simplified diagrams, illustrating the biochemical transformations directly involved in the biosynthesis of NAD(P) (A), CoA (B), and FMN/FAD (C). Most of the pathways and genes shown are those present in E. coli, with the few exceptions marked by dashed lines. Recycling pathways and other transformations related to genes that remain unknown (such as NMN deamidase) are not included.
One possible explanation of this contradiction is the presence of functional NadR in E. coli (Table 2). The nadR gene is a transcriptional regulator of the de novo biosynthesis and niacin salvage genes in E. coli (92). Recently NadR was shown to possess low levels of NMN adenylyltransferase activity (74) as well as NmR kinase activity (O. Kurnasov et al., unpublished data). This suggests that NadR may play a role in salvaging exogenous NMN. The NMN adenylyltransferase activity of NadR may also be involved in recycling intracellular NMN directly to NAD, bypassing the NAD synthase encoded by nadE (Fig. 3A). However, flux through the PnuC-NadR salvage pathway is unlikely to be sufficient to compensate for inactivation of nadD or nadE, which are responsible for the bulk of NAD production in E. coli (73). Also, this explanation contradicts the apparent essentiality of nadE observed in our genetic footprinting experiments, since the compensatory role of the PnuC-NadR bypass would imply nonessentiality of nadE as well as nadD (Fig. 3A). At present we cannot propose a single unambiguous interpretation of the contradiction between the classical genetic and genetic footprinting data with respect to nadD. However, this is the only point of disagreement in NAD(P) biosynthesis between the genetic footprinting data and other types of evidence.
Genetic footprinting of E. coli genes involved in pantothenate/CoA biosynthesis.
In contrast to the complexity of NAD(P) metabolism, the CoA biosynthetic pathway is topologically simple (Fig. 3B). All of the genetic footprinting data are consistent with other experimental data and with theoretical considerations (Table 1). Each enzymatic step of the common pathway from pantothenate to CoA is nonredundant and indispensable, since none of the phosphorylated intermediates can be transported into the cell. This eliminates potential cross-feeding and the possibility of exogenous CoA salvage or transport of any phosphorylated precursors from the growth medium. All the genes of the pantothenate biosynthetic pathway are nonessential and can be bypassed by exogenous pantothenate via a known transporter encoded by the panF gene (51, 93).
The coaA gene of E. coli, encoding pantothenate kinase, the first enzyme of the common five-step pathway from pantothenate to CoA, was previously characterized as essential (86, 94). Genes for the last four enzymatic steps of CoA biosynthesis (coaBC, coaD, and coaE) have only recently been discovered (39, 65). Two of these genes, coaE (formerly yacE) and coaD (kdtB), were previously shown to be essential (36, 45). As shown in Table 1, all four genes were found to be essential in our footprinting study, as was acpS, which produces the enzyme responsible for covalent attachment of CoA to acyl carrier protein, which is required for fatty acid biosynthesis (33).
Genetic footprinting of E. coli genes involved in FMN/FAD biosynthesis.
All of the FMN/FAD biosynthetic genes were found to be essential by genetic footprinting experiments in both E. coli strains (Table 1). The fact that the genes for de novo riboflavin biosynthesis were essential in the presence of riboflavin (0.8 mg/liter) in the medium is consistent with the absence of a riboflavin transporter in E. coli K-12 (6). Riboflavin auxotrophs can be obtained in E. coli using specific selection steps to facilitate riboflavin transport by uncharacterized mutations or on significantly higher concentrations of exogenous riboflavin (6, 8, 9).
Two enzymes of the common flavin pathway, consecutively producing the two cofactors, FMN and FAD, form a bifunctional protein encoded by the essential ribF gene. Both enzymatic activities are expected to be indispensable, although genetic footprinting data alone suggest only the essentiality of the C-terminal domain (encoding FK). This is a general limitation of the technique with respect to multifunctional proteins.
In summary, 13 genes encoding 16 enzymes within three vitamin/cofactor biosynthetic pathways were shown to be required for aerobic growth of E. coli in enriched media (Table 1). Twelve of these genes were identified as essential by genetic footprinting in agreement with published data and theoretical analysis. The only contradictory case (nadD) was reconciled by a directed gene deletion strategy. In addition, 12 more genes related to metabolism of NAD(P), CoA, and FAD were analyzed by genetic footprinting and found to be nonessential (Table 1). Again, these results are in good agreement with the available experimental data and metabolic reconstructions of the corresponding pathways.
Some essential genes representing potential broad-spectrum antibacterial targets were further analyzed with respect to the genomic data available for selected bacterial pathogens and for the human host (see below). This analysis allows extrapolation of the experimental gene essentiality data from E. coli onto several bacterial pathogens, including S. aureus.
Selected target validation.
The predicted essentiality of the S. aureus nadD ortholog (gi|15927174) was confirmed by an indirect approach. Deletion of nadD was attempted in parallel with the S. aureus pncA ortholog (gi|15927492), presumed to be dispensable in the presence of exogenous nicotinate, using allelic exchange vector pBT2 (20). No ΔnadD variants were obtained, even after extensive screening (2,248 colonies), while in the control experiment pncA was successfully inactivated in 38% of the colonies screened. This indicates that nadD is indeed essential for S. aureus growth in complex tryptic soy broth (Difco Laboratories, Detroit, Mich.).
A different strategy was used to verify the predicted essentiality of the S. aureus coaD ortholog (gi|15926709). In vitro transposon mutagenesis was used to disrupt coaD as well as two adjacent ORFs. The conclusion of the coaD essentiality was drawn based on the fact that coaD could not be disrupted while the neighboring ORFs (gi|15926708 and gi|15926710) were successfully inactivated by the Tet M transposon, with frequencies of 83 and 5%, respectively.
Selected bacterial targets and their human counterparts were overproduced in E. coli, purified, and characterized. The respective adenylyltransferase activities of the following recombinant enzymes were directly confirmed in vitro: NaMNATs from E. coli, S. aureus, and H. pylori (gi|1786858, gi|15924584, and gi|2314504, respectively); two isoforms of human PNAT, i.e., PNAT-1 (gi|12620200) and PNAT-2 (gi|11245478); PPATs from S. aureus, M. tuberculosis, H. influenzae, and H. pylori (gi|15924115, gi|560525, gi|1573650, and gi|15612433, respectively); the PPAT domain of the human PPAT/DPCK protein (27); FADS domain of the B. subtilis bifunctional FK/FADS protein (gi|16078730); and human FADS (F. Mseeh, unpublished results).
DISCUSSION
Our approach to identifying and ranking antibacterial drug targets based on a combination of genetic footprinting in a model system (E. coli) and comparative genome analysis is schematically illustrated in Fig. 4. Both major components of this approach, experimental and computational, are postgenomic techniques. Genetic footprinting in E. coli allows identification of essential genes as a function of growth conditions in a high-throughput format. This relatively new technique has multiple potential applications to functional genomics. For example, by monitoring changes in the pattern of gene essentiality while varying growth conditions and/or genetic background, it may be possible to assess various aspects of cell metabolism. This can allow functions of previously uncharacterized genes to be established and allows the functional roles of known genes to be refined. An example of such a study has been published recently (7). Here we consider application of genetic footprinting to identifying novel antibacterial drug targets (Fig. 4).
FIG. 4.
Selection of antibacterial drug targets by a combination of genetic footprinting in E. coli and comparative analysis of reconstructed metabolic subsystems, pathways, and individual genes in pathogens and humans.
Experimental gene essentiality data determined by genetic footprinting are analyzed in terms of the relevant E. coli metabolic subsystems and pathways using the ERGO database. Such a contextual analysis refines the essentiality assessment of each gene by addressing the following questions. Which metabolic pathway is this essential gene associated with? Does this pathway yield a metabolite essential for cell viability? Is there another route to produce the same metabolite? Can this metabolite be salvaged from the growth media in its final form? What is the most advanced precursor (the last point of salvage) that a cell can acquire from its environment? This kind of analysis generates hypotheses about the essentiality and nonessentiality of other known genes in the same pathways. Comparison of these models with actual genetic footprinting data and the data available from the literature (e.g., http://www.shigen.nig.ac.jp/ecoli/pec/Analyses.jsp) allows refinement of initial experimental assessments of gene essentiality or calls for further experimental examination (as in the case of nadD).
Once consistent data are compiled for E. coli, the metabolic reconstruction of selected pathways is extended to a panel of representative pathogens. A hierarchical overview illustrated in Tables 2 to 4 provides a convenient way to perform a comparative cross-genome analysis of relevant pathways and metabolic subsystems. Questions addressed at this step of the analysis include the following. Are these pathways present in all the pathogens under consideration? Which pathways containing essential genes are conserved and nonredundant within the group? Which functions within the selected pathways are encoded by closely related genes? The last point may reflect the likelihood of finding a small molecule that will efficiently bind to a target protein in most pathogens of the spectrum suppressing its functional activity. We use two parameters as preliminary measures of this likelihood, the P values of pairwise sequence comparisons and the E values produced by comparing each bacterial sequence to the profile (HMM) built for a specific panel of pathogens (as illustrated in Fig. 5A and B). With more structural and functional data, more elaborate and productive criteria may be applied, such as conservation of the elements of the active site, relative substrate preferences, or affinities for a range of natural and synthetic ligands.
Next, the corresponding human pathways are reconstructed from genomic data to assess potential side effects that may arise due to inhibition of the human counterparts of antibacterial targets. One criterion often used in selecting antibacterial targets is the absence of the corresponding function and even the corresponding pathway in the human host (for instance, bacterial cell wall biosynthesis). This approach, although historically productive, often unnecessarily rejects promising targets. Many targets in universal biosynthetic pathways can now be reconsidered because of the new opportunities provided by comparative genomics and parallel chemical synthesis. Certain features revealed at the level of host-pathogen comparative genome analysis may be useful for evaluation of such targets. For instance, a pathway may be unique in pathogens but potentially redundant in humans (as with NAD biosynthesis), or the human functional counterpart may be structurally unrelated to bacterial proteins. In cases where bacterial orthologs of a target enzyme and the human version share some sequence similarity, we prioritize targets based on (i) the lowest of the P values generated from pairwise comparisons of target protein homologs from each bacterial pathogen with their human functional counterpart and (ii) the E value derived from comparison of a human protein sequence with the profile HMM built for the corresponding pathogen proteins (as in Fig. 5C).
To illustrate this approach, essential and conserved genes with known functions in the biosynthesis of the key adenylate cofactors NAD(P), CoA, and FAD were investigated. These biosynthetic pathways fit most of the criteria listed above: (i) each cofactor is utilized by multiple enzymes in all pathogens; (ii) most bacteria are unable to import these cofactors or their phosphorylated precursors without prior degradation; (iii) the last steps in each of the biosynthetic pathways are universal and nonredundant and contain a number of promising targets.
NAD(P) biosynthesis is characterized by a high level of complexity and diversity in various organisms (60, 66), including two versions of de novo biosynthesis and at least three routes for salvage of exogenous precursors (Table 2 and Fig. 3A). The absence of any recognizable genes for NAD(P) biosynthesis in the genome of C. trachomatis (Table 2) suggests that this obligate intracellular parasite has unique transport machinery for salvage of NAD and NADP. Therefore, C. trachomatis must be excluded from the panel of pathogens used to assess targets in NAD biosynthesis.
Genes controlling de novo NAD biosynthesis from aspartate and the salvage of niacin in E. coli are nonessential in rich media. This conclusion can be projected onto P. aeruginosa, B. anthracis, and possibly M. tuberculosis based on metabolic reconstructions (Table 2). The last of these cases is controversial. In spite of the fact that both genes required for niacinamide salvage (pncA and pncB) are present in the genome, M. tuberculosis has been reported to be strictly dependent on de novo NAD biosynthesis (83). Since pncA is expressed in M. tuberculosis (81, 102), one interpretation is that the pncB ortholog is functionally inactive in M. tuberculosis, at least under laboratory conditions.
Theoretically, bacterial de novo NAD biosynthesis from aspartate is an attractive target pathway, since it is absent in humans, where de novo biosynthesis occurs by a five-step oxidative degradation of tryptophan (66). However, within our set of bacterial pathogens, only H. pylori is expected to have useful targets in this pathway, since it lacks niacin salvage genes (Table 2). The two-step conversion of aspartate to quinolinate would make an excellent target pathway for anti-infective agents specific to H. pylori (Table 2 and Fig. 3A). A systematic analysis of essential genes conserved exclusively in H. pylori (23) did not recognize the potential of this pathway as a target for drug development, because it did not take into account the metabolic context of this organism.
The PnuC-NadR pathway of nicotinamide riboside salvage in H. influenzae provides another example of a narrow spectrum target. The complete pathway is present in only a limited number of bacteria, but it is the sole route for NAD biosynthesis in H. influenzae. H. influenzae requires so-called V factors for growth (25, 76), the simplest of which is nicotinamide riboside. All other acceptable V factors, NADP, NAD, and NMN (if present in the growth medium), are gradually degraded to nicotinamide riboside by membrane-bound or periplasmic phosphohydrolases, such as the nadN gene product (76). In addition, the two-step niacinamide salvage pathway in gram-positive pathogens such as S. pneumoniae and S. aureus may constitute a possible narrow-spectrum target. Both organisms lack all the genes for de novo NAD biosynthesis and for the PnuC-NadR pathway of nicotinamide riboside salvage (Table 2).
The three-step pathway from NaMN to NAD and NADP, requiring NaMNAT, NADS, and NADK, is conserved in the majority of bacterial pathogens (Table 2), representing potentially broad-spectrum targets. This pathway is nonredundant, and the corresponding genes are essential in E. coli (Table 2). NaMNAT and NADS are not present in C. trachomatis and H. influenzae, which excludes these pathogens from the target profile. Inhibition of NADK would cover the broadest range of pathogens, excluding only C. trachomatis (see Fig. 5A). However, NADK has been recently characterized in H. sapiens (57), revealing a very high sequence similarity with many bacterial enzymes (the P value of 2 · 10−25 for NADK from P. aeruginosa is significantly better than P values of pairwise comparison between some bacterial orthologs). The C-terminal synthase domain of the human NADS ortholog (gi|10433831) reveals a much lower but significant sequence similarity with some of its bacterial counterparts (the best P value is 6 · 10−13 for NADS from M. genitalium). Finally, the human counterpart of bacterial NaMNAT has only marginal sequence similarity with bacterial enzymes of the nadD family (the best P value is ∼0.2 for NaMNAT from H. pylori). E values derived from comparing human PNAT, NADS, and NADK to profile HMMs of the corresponding bacterial protein families support these conclusions (Fig. 5C). Prioritization of the three potentially broad-spectrum targets in NAD biosynthesis by a combination of these and other criteria summarized in Fig. 5 yields a target preference order as follows: NaMNAT is better than NADS and much better than NADK.
NaMNAT activity was initially characterized in E. coli (26), and the chromosomal locus was mapped in Salmonella (48). However, the identity of the structural gene remained unknown until recently. We have identified this gene based on its clustering on the chromosome with nadE and pncB gene orthologs in some microbial genomes (12). This gene has been independently identified in E. coli by Mehl and coworkers (63). We have cloned and expressed nadD from E. coli and have cloned and expressed its orthologs from gram-positive (S. aureus) and gram-negative (H. pylori) pathogens (12), and we have purified the NadD proteins. Analysis of the substrate specificity of the purified recombinant proteins revealed strong preferences among the bacterial enzymes for NaMN over NMN (up to >17,000-fold in the case of S. aureus NaMNAT). This bias is consistent with the major flux in NAD biosynthesis of most bacteria going through nicotinic acid mononucleotide (NaMN) to NaAD.
At least three isoforms of NadD can be identified in the human genome (gi|12620200, gi|11245478, and gi|14029540). We have cloned, purified, and characterized the first two forms and designated them PNAT-1 and PNAT-2, since both of them perform the adenylyltransferase reaction equally efficiently with both pyridine nucleotides NMN and NaMN (O. Kurnasov et al., unpublished data). Human PNAT-2 has been independently identified and characterized by two other groups (31, 80). The observed dual specificity of human PNATs is consistent with previous experimental data (61) and with the reconstruction of the putative human NAD biosynthetic pathway from genomic data (Table 2).
The difference in substrate preferences between human PNATs and bacterial NaMNATs is additional incentive for pursuing NadD as a selective, yet relatively broad-spectrum antibacterial target. The three-dimensional structures of NaMNATs from E. coli (101) and B. subtilis (70) have been solved in association with NaMN. Comparison of these structures with the structure of human PNAT-2 (103) reveals significant differences in the organization of their active sites, which may facilitate identification of a selective antibacterial inhibitor.
Similar comparative analysis of the CoA and FMN/FAD biosynthetic pathways in humans and bacteria produced additional broad-spectrum antibacterial targets (Fig. 5). The de novo pathway for CoA biosynthesis is present in many bacterial pathogens and absent in humans (Table 3), but the corresponding enzymes do not seem to be attractive targets due to the universal presence of pantothenate symporter orthologs. Some pathogens that lack de novo pantothenate biosynthetic genes (Table 3) depend solely on salvage of pantothenate (S. pneumoniae and H. influenzae) or dephospho-CoA (M. genitalium and C. trachomatis). Bacterial pantothenate symporters (panF family) share high sequence similarity with a human protein implicated in vitamin transport (96). No panF orthologs are present in the genomes of M. genitalium or C. trachomatis. Specific transporters allowing salvage of host dephospho-CoA by these intracellular pathogens (which might represent narrow spectrum targets) are still unknown.
TABLE 3.
Functional reconstruction of metabolic pathways of CoA biosynthesisa
| CoA biosynthesis | EC no. | Protein example | Presence of ortholog or pathway in genome of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. colib | P. aeruginosa | B. anthracis | M. tuberculosis | H. pylori | S. pneumoniae | S. aureus | M. genitalium | H. influenzae | C. trachomatis | H. sapiens | |||
| De novo pathway to pantothenate | Yes | Yes | Yes | Yes | Yes | No | Yes | No | No | No | No | ||
| Aspartate to β-alanine | |||||||||||||
| Aspartate 1-decarboxylase | 4.1.1.11 | gi|1786323 | panD | + | + | + | + | − | + | − | − | − | − |
| Ketovalerate to pantoate | |||||||||||||
| 3-Methyl-2-oxobutanoate hydroxymethyl transferase | 2.1.2.11 | gi|1786326 | panB | + | + | + | + | − | + | − | − | − | − |
| 2-Dehydropantoate 2-reductase | |||||||||||||
| (i) 2-Dehydropantoate 2-reductase | 1.1.1.169 | gi|1100871 | panE | + | + | + | − | − | + | − | − | − | − |
| (ii) Ketol-acid reductoisomerase | 1.1.1.86 | gi|146477 | ilvC | + | + | + | + | + | + | − | + | − | − |
| β-Alanine, pantoate to pantothenate | |||||||||||||
| Pantoate-β-alanine ligase | 6.3.2.1 | gi|1786325 | panC | + | + | + | + | − | + | − | − | − | − |
| Pantothenate (vitamin B5) salvage | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | ||
| Sodium/pantothenate symporter | gi|455654 | panF | + | + | + | + | + | + | − | + | − | + | |
| Common pathway to CoA | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | ||
| Pantothenate kinase | ?f | ?f | |||||||||||
| (i) Most bacteria | 2.7.1.33 | gi|1790409 | coaA | − | − | + | − | + | − | − | + | − | − |
| (ii) Eukarya | 2.7.1.33 | gi|6320740c | − | − | + | − | − | − | + | − | − | − | + |
| Phosphopantothenoylcysteine synthetase | 6.3.2.5 | gi|1790070 | coaBe | + | + | + | + | + | + | − | + | − | + |
| Phosphopantothenoylcysteine decarboxylase | 4.1.1.36 | gi|1790070 | coaCe | + | + | + | + | + | + | − | + | − | + |
| Pantetheine-phosphate adenylyltransferase | |||||||||||||
| (i) Bacteria | 2.7.7.3 | gi|1790065 | coaD | + | + | + | + | + | + | − | + | − | − |
| (ii) Eukarya | 2.7.7.3 | gi|632171d | − | − | − | − | − | − | − | − | − | − | + |
| Dephospho-CoA kinase | 2.7.1.24 | gi|1786292 | coaE | + | + | + | + | + | + | + | + | + | + |
See Table 2, footnote a, for details.
Identical patterns of pathways with very similar genes are found in E. coli, Salmonella enterica serovar Typhi, and Yersinia pestis in all of the subsystems considered.
Eukaryotic pantothenate kinase (21), structurally unrelated to the corresponding E. coli enzyme (CoaA family).
Eukaryotic pantetheine-phosphate adenylyltransferase is only distantly related to the corresponding bacterial enzymes (beyond recognition by Psi-Blast) (27); it forms a fusion protein with dephospho-CoA kinase in higher eukaryotes.
In most bacteria (except S. pneumoniae in this set), CoaB is the C-terminal domain, and CoaC is the N-terminal domain of a bifunctional protein.
Pantothenate kinase is still a “missing gene” in a number of bacterial pathogens.
All five enzymatic steps in the common pathway of CoA biosynthesis from pantothenate are encoded by essential genes in E. coli (Table 3). Four of these enzymes are conserved in a broad range of bacterial pathogens. The first enzyme of the pathway, PK, encoded in E. coli by coaA (86) belongs to a structural family different from that of the recently identified eukaryotic PKs (21). This makes coaA-related PKs an attractive target for a relatively narrow subset of our list of pathogens, including H. influenzae, M. tuberculosis, and S. pneumoniae. S. aureus (as well as other staphylococci and enterococci) and B. anthracis lack coaA orthologs, and this enzymatic function is most likely performed by remote orthologs of the eukaryotic PK (such as tr|Q99SC8 in S. aureus). Neither of the two known forms of PK can be identified by sequence similarity analysis in P. aeruginosa, H. pylori, and a few other pathogens not included in our set, which suggests the existence of at least one uncharacterized form of this enzyme.
All the remaining enzymes in this pathway represent potential antibacterial drug targets of relatively broad spectrum (Table 3). In most bacteria, two enzymatic activities, PPCS and PPCDC, are located within the C-terminal and N-terminal domains of a bifunctional (fused) protein (gene coaBC, previously dfp in E. coli). The exception in our list of pathogens is S. pneumoniae (as well as other streptococci and enterococci) containing two separate ORFs, coaB and coaC, in one operon (in reversed order relative to the fusion proteins). Interestingly, B. anthracis (as well as Bacillus cereus) contains both a bifunctional PPCDC/PPCS and a monofunctional PPCS ortholog but no monofunctional PPCDC. Monofunctional PPCSs from S. pneumoniae and B. anthracis reveal very high sequence similarity to each other, but they are quite divergent from the PPCS domain of the bifunctional PPCDC/PPCS proteins. In contrast to other bacterial orthologs, they produce a low but reliable similarity score compared with the human monofunctional PPCS (3 · 10−23 between H. sapiens and B. anthracis). The last enzyme of the common pathway, DPCK, is ubiquitous in all pathogens in our set.
The last four enzymes in human CoA biosynthesis were recently identified and characterized (27). Two of them, PPCDC and DPCK, are relatively similar to their bacterial counterparts (Fig. 5C). The human monofunctional PPCS is very distant from PPCS domains of bacterial bifunctional PPCDC/PPCS proteins, but it is more closely related to the bacterial monofunctional PPCDCs. The PPAT domain of the human bifunctional PPAT/DPCK protein reveals no significant sequence similarity with any bacterial counterpart, and it is likely to be quite dissimilar in the overall structure. PPAT probably represents the most attractive target in the common CoA biosynthetic pathway for a number of reasons: (i) it constitutes a target in a broad range of pathogens (Fig. 5A), (ii) all the bacterial orthologs of this enzyme are closely related to the consensus (no outliers; Fig. 5B), and (iii) the human PPAT is very dissimilar from the bacterial HMM profile (Fig. 5C). PPAT from E. coli (encoded by coaD, previously kdtB) was previously described, and its three-dimensional structure was solved (39, 50).
The majority of bacteria (with the exception of M. genitalium) considered here have a conserved de novo riboflavin biosynthetic pathway, which is not present in humans (Table 4 and Fig. 3C). Moreover, all of the genes in this de novo pathway are essential in E. coli (Table 4). These observations have brought attention to many of these gene products as potential antibacterial drug targets (17, 29, 98). However, direct experimental analysis of riboflavin transport and salvage in these pathogens is necessary in order to define how useful these targets may be. B. subtilis is known to effectively salvage exogenous riboflavin, bypassing the requirement for the de novo pathway (19). Orthologs of one of the riboflavin transporters, gene ypaA, recently identified in B. subtilis (56) are present in many gram-positive pathogens, including B. anthracis, S. aureus, and S. pneumoniae, although their functionality has not been directly confirmed. Additional structurally dissimilar riboflavin transporters are likely to exist in other pathogens. For example, riboflavin transport must occur in M. genitalium, since a de novo riboflavin biosynthesis pathway cannot be asserted in this organism (Table 4). However, no ypaA orthologs can be found in any of the available mycoplasma or ureaplasma genomes.
A common pathway for FMN/FAD biosynthesis is represented in all bacteria by a bifunctional enzyme that catalyzes two consecutive reactions. The first reaction is formation of FMN, catalyzed by the FK domain (C terminus). The second reaction is conversion of FMN to FAD by the FADS domain (N terminus). Both enzymes represent antibacterial targets with a very broad range (Fig. 5A). The HMM E values in the group of pathogens as a whole are high (Fig. 5B), indicating that these targets are rather divergent. However, for FADS this parameter can be improved by removing some outliers, such as H. pylori (often the source of the most divergent proteins within our sample group of pathogens) and M. genitalium (Fig. 5B).
In contrast to those in all known bacteria, eukaryotic FK and FADS identified in S. cerevisiae (78, 100) are monofunctional proteins. We have overexpressed, purified, and characterized both human enzymes of the common FMN/FAD biosynthetic pathway (F. Mseeh, unpublished data). Sequence comparison of human FK and FADS with the profile HMMs clearly indicates that FADS is potentially a more selective target (Fig. 5C). In agreement with this, no reliable P values are produced in pairwise comparisons of human FADS with any of its bacterial counterparts, while the human FK has sequence similarity to some bacterial FK domains higher than the similarity between some of the bacterial FK domains (for example, the M. tuberculosis FK domain gives a P value of 10−9 with human FK and a P value of 4 · 10−6 with the H. pylori FK domain).
In conclusion, genetic footprinting in E. coli in combination with comparative genome analysis of reconstructed metabolic subsystems and pathways has allowed us to identify a number of antibacterial targets involved in the biosynthesis of the adenylate cofactors NAD(P), CoA, and FAD. Among the most attractive targets in all three pathways are the adenylyltransferases: NaMNAT, PPAT, and FADS (Fig. 6). Two of these targets (NaMNAT and PPAT) have been validated by directed-knockout strategy in S. aureus. Some orthologs of the target enzymes from representative pathogens, as well as their human counterparts, were overproduced, purified, and characterized by direct in vitro assays. Comparative analysis of adenylate cofactor biosynthesis provides an illustration of a general approach that can be extended to other functional subsystems to reveal novel antibacterial targets.
FIG. 6.
General scheme of chemical transformations catalyzed by the three adenylyltransferases (ATse) in the biosynthesis of NAD(P), CoA, and FAD. These enzymes—NaMNAT (encoded in E. coli by nadD), PPAT (gene coaD), and FADS (ribF)—were selected as the most promising antibacterial drug targets within these pathways.
ADDENDUM
While the manuscript was being reviewed, a genome-wide analysis of essential genes in H. influenzae was published (2). Comparing essential genes in divergent organisms will be useful “for defining both the common essential pathways of life and potential targets for development of antimicrobial therapeutics” (2). Our genome-scale list of essential and nonessential genes in E. coli is currently in preparation.
Acknowledgments
We are grateful to William Reznikoff and Igor Goryshin (University of Wisconsin, Madison); Jerry Jendrisak and Les Hoffman (Epicentre Technologies); Tadhg Begley (Cornell University, Ithaca, N.Y.); Zoltan Oltvai (Northwestern University, Chicago, Ill.); and Henry Burd, Niels Larsen, Gregory Kogan, Yuri Kuniver, Yuri Grechkin, and Robert Haselkorn (Integrated Genomics) for valuable discussions, help, and support throughout this project. We thank George Church (Harvard Medical School, Boston, Mass.) for the gift of pKO3, Mark Smeltzer (University of Arkansas for Medical Sciences) for the gift of pCRII::tet and RN4220, Reinhold Brückner (Universität Tübingen, Tübingen, Germany) for pBT2, and Don Clewell (University of Michigan, Ann Arbor) for the plasmid pAM120 containing Tn916.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trost. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. [DOI] [PubMed] [Google Scholar]
- 5.Bacher, A., S. Eberhardt, W. Eisenreich, M. Fischer, S. Herz, B. Illarionov, K. Kis, and G. Richter. 2001. Biosynthesis of riboflavin. Vitam. Horm. 61:2-49. [DOI] [PubMed] [Google Scholar]
- 6.Bacher, A., S. Eberhardt, and G. Richter. 1996. Biosynthesis of riboflavin, p. 657-664. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 7.Badarinarayana, V., P. W. Estep, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 8.Bandrin, S. V., M. Y. Beburov, P. M. Rabinovich, and A. I. Stepanov. 1979. Riboflavin auxotrophs of Escherichia coli. Genetika 15:2063-2065. (In Russian.) [PubMed] [Google Scholar]
- 9.Bandrin, S. V., P. M. Rabinovich, and A. I. Stepanov. 1983. 3 linkage groups of the genes of riboflavin biosynthesis in Escherichia coli. Genetika 19:1419-1425. (In Russian.) [PubMed] [Google Scholar]
- 10.Beckwith, J. R., and E. R. Signer. 1966. Transposition of the Lac region of Escherichia coli. I. Inversion of the Lac operon and transduction of Lac by φ80. J. Mol. Biol. 19:254-265. [DOI] [PubMed] [Google Scholar]
- 11.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell-division gene, ftsk. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-119. [DOI] [PubMed] [Google Scholar]
- 13.Begley, T. P., C. Kinsland, and E. Strauss. 2001. The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61:158-171. [DOI] [PubMed] [Google Scholar]
- 14.Benton, B. L., J. Ving, F. Malouin, P. K. Martin, M. B. Schmid, and D. Sun. March 2000. Methods of screening for compounds active on Staphylococcus aureus target genes. U.S. patent 6,037,123.
- 15.Benton, B. L., J. Ving, F. Malouin, P. K. Martin, M. B. Schmid, and D. Sun. May 2001. Methods of screening for compounds active on Staphylococcus aureus target genes. U.S. patent 6,228,588.
- 16.Bernal, A., U. Ear, and N. Kyrpides. 2001. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 29:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black, M. T., L. K. Shilling, R. K. Stodola, R. L. Warren, A. L. Kosmatka, R. O. Nicholas, L. M. Palmer, M. A. Lonetto, J. C. Fedon, J. E. Hodgson, and D. J. C. Knowles. June 2001. RibB. U.S. patent 6,252,044.
- 18.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. ColladoVides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 19.Bresler, S. E., E. A. Glazunov, and D. A. Perumov. 1972. Study of the operon of riboflavin biosynthesis in Bacillus subtilis. IV. Regulation of the synthesis of riboflavin synthetase. Investigation of riboflavin transport through the cell membrane. Genetika (USSR) 8:109-117. [PubMed] [Google Scholar]
- 20.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 21.Calder, R. B., R. S. B. Williams, G. Ramaswamy, C. O. Rock, E. Campbell, S. E. Unkles, J. R. Kinghorn, and S. Jackowski. 1999. Cloning and characterization of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans. J. Biol. Chem. 274:2014-2020. [DOI] [PubMed] [Google Scholar]
- 22.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 23.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 25.Cynamon, M. H., T. B. Sorg, and A. Patapow. 1988. Utilization and metabolism of NAD by Haemophilus parainfluenzae. J. Gen. Microbiol 134:2789-2799. [DOI] [PubMed] [Google Scholar]
- 26.Dahmen, W., B. Webb, and J. Preiss. 1967. The deamido-diphosphopyridine nucleotide and diphosphopyridine nucleotide pyrophosphorylases of Escherichia coli and yeast. Arch. Biochem. Biophys. 120:440-450. [DOI] [PubMed] [Google Scholar]
- 27.Daugherty, M., B. Polanuyer, M. Farrell, A. Lykidis, V. de Crécy-Lagard, and A. Osterman. 2002. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 277:21431-21439. [DOI] [PubMed] [Google Scholar]
- 28.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debouck, C., J. C. Fedon, D. D. Jaworski, J. Mooney, L. M. Palmer, C. M. Traini, M. Wang, R. L. Warren, and Y. Y. Zhong. December 2001. RibH. U.S. patent 6,326,462.
- 30.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 31.Emanuelli, M., F. Carnevali, F. Saccucci, F. Pierella, A. Amici, N. Raffaelli, and G. Magni. 2000. Human NMN adenylyltransferase: molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties. J. Biol. Chem. 276:406-412. [DOI] [PubMed] [Google Scholar]
- 32.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. Fitzhugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 33.Flugel, R. S., Y. Hwangbo, R. H. Lambalot, J. E. Cronan, and C. T. Walsh. 2000. Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J. Biol. Chem. 275:959-968. [DOI] [PubMed] [Google Scholar]
- 34.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 35.Freiberg, C. 2001. Novel computational methods in anti-microbial target identification. Drug Discov. Today 6:S72-S80. [Google Scholar]
- 36.Freiberg, C., B. Wieland, F. Spaltmann, K. Ehlert, H. Brotz, and H. Labischinski. 2001. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 3:483-489. [PubMed] [Google Scholar]
- 37.Galperin, M. Y., and E. V. Koonin. 1999. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 10:571-578. [DOI] [PubMed] [Google Scholar]
- 38.Gawron-Burke, C., and D. B. Clewell. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J. Bacteriol. 159:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geerlof, A., A. Lewendon, and W. V. Shaw. 1999. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J. Biol. Chem. 274:27105-27111. [DOI] [PubMed] [Google Scholar]
- 40.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 41.Goryshin, I. Y., J. A. Miller, Y. V. Kil, V. A. Lanzov, and W. S. Reznikoff. 1998. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95:10716-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamer, L., T. M. DeZwaan, M. V. Montenegro-Chamorro, S. A. Frank, and J. E. Hamer. 2001. Recent advances in large-scale transposon mutagenesis. Curr. Opin. Chem. Biol. 5:67-73. [DOI] [PubMed] [Google Scholar]
- 44.Hancock, R. E. W., and D. Knowles. 1998. Are we approaching the end of the antibiotic era? Curr. Opin. Microbiol. 1:493-494. [Google Scholar]
- 45.Hare, R. S., S. S. Walker, T. E. Dorman, J. R. Greene, L. M. Guzman, T. J. Kenney, M. C. Sulavik, K. Baradaran, C. Houseweart, H. Y. Yu, Z. Foldes, A. Motzer, M. Walbridge, G. H. Shimer, and K. J. Shaw. 2001. Genetic footprinting in bacteria. J. Bacteriol. 183:1694-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinemann, J. A. 1999. How antibiotics cause antibiotic resistance. Drug Discov. Today 4:72-79. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman, L. M., J. J. Jendrisak, R. J. Meis, I. Y. Goryshin, and W. S. Reznikoff. 2000. Transposome insertional mutagenesis and direct sequencing of microbial genomes. Genetica 108:19-24. [DOI] [PubMed] [Google Scholar]
- 48.Hughes, K. T., D. Ladika, J. R. Roth, and B. M. Olivera. 1983. An indispensable gene for NAD biosynthesis in Salmonella typhimurium. J. Bacteriol. 155:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 50.Izard, T., and A. Geerlof. 1999. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 18:2021-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackowski, S., and J. H. Alix. 1990. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J. Bacteriol. 172:3842-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenks, P. J., C. Chevalier, C. Ecobichon, and A. Labigne. 2001. Identification of nonessential Helicobacter pylori genes using random mutagenesis and loop amplification. Res. Microbiol. 152:725-734. [DOI] [PubMed] [Google Scholar]
- 53.Jensen, K. F. 1993. The Escherichia coli K-12 wild types W3110 and MG1655 have rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Judson, N., and J. J. Mekalanos. 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8:521-526. [DOI] [PubMed] [Google Scholar]
- 55.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O’Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 56.Kreneva, R. A., M. S. Gel'fand, A. A. Mironov, Y. A. Yomantas, Y. I. Kozlov, A. S. Mironov, and D. A. Perumov. 2000. Inactivation of the ypaA gene in Bacillus subtilis; analysis of the resulting phenotypic expression. Russ. J. Genet. 36:972-974. [Google Scholar]
- 57.Lerner, F., M. Niere, A. Ludwig, and M. Ziegler. 2001. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 288:69-74. [DOI] [PubMed] [Google Scholar]
- 58.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loferer, H., A. Jacobi, A. Posch, C. Gauss, S. Meier-Ewert, and B. Seizinger. 2000. Integrated bacterial genomics for the discovery of novel antimicrobials. Drug Discov. Today 5:107-114. [DOI] [PubMed] [Google Scholar]
- 60.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 61.Magni, G., M. Emanuelli, A. Amici, N. Raffaelli, and S. Ruggieri. 1997. Purification of human nicotinamide-mononucleotide adenylyltransferase. Methods Enzymol. 280:241-247. [DOI] [PubMed] [Google Scholar]
- 62.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehl, R. A., C. Kinsland, and T. P. Begley. 2000. Identification of the Escherichia coli nicotinic acid mononucleotide adenylyltransferase gene. J. Bacteriol. 182:4372-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meis, R. 2000. EZ::TN transposon insertions into target DNA in vitro are highly random. Epicentre Forum 7:5. [Online.] http://www.epicentre.com/forum.asp. [Google Scholar]
- 65.Mishra, P. K., P. K. Park, and D. G. Drueckhammer. 2001. Identification of yacE (coaE) as the structural gene for dephosphocoenzyme A kinase in Escherichia coli K-12. J. Bacteriol. 183:2774-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moat, A. G., and J. W. Foster. 1987. Biosynthesis and salvage pathways of pyridine nucleotides, p. 1-20. In D. Dolphin and O. Avramovich (ed.), Pyridine nucleotide coenzymes, part B. John Wiley & Sons, New York, N.Y.
- 67.Moir, D. T., K. J. Shaw, R. S. Hare, and G. F. Vovis. 1999. Genomics and antimicrobial drug discovery. Antimicrob. Agents Chemother. 43:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nilsen, T. W. June2001. Method for identifying essential or functional genes. U.S. patent 6,248,525.
- 70.Olland, A. M., K. W. Underwood, R. M. Czerwinski, M. C. Lo, A. Aulabaugh, J. Bard, M. L. Stahl, W. S. Somers, F. X. Sullivan, and R. Chopra. 2002. Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase. J. Biol. Chem. 277:3698-3707. [DOI] [PubMed] [Google Scholar]
- 71.Overbeek, R., N. Larsen, G. D. Pusch, M. D'Souza, E. Selkov, Jr., N. Kyrpides, M. Fonstein, N. Maltsev, and E. Selkov. 2000. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 28:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palmer, L. M., J. M. Pratt, and M. Rosenberg. October2000. Method for determining gene essentiality in a pathogen. U.S. patent 6,139,817.
- 73.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-730. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 74.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reich, K. A., L. Chovan, and P. Hessler. 1999. Genome scanning in Haemophilus influenzae for identification of essential genes. J. Bacteriol. 181:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reidl, J., S. Schlor, A. Kraiss, J. Schmidt-Brauns, G. Kemmer, and E. Soleva. 2000. NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 35:1573-1581. [DOI] [PubMed] [Google Scholar]
- 77.Rosamond, L., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973-1976. [DOI] [PubMed] [Google Scholar]
- 78.Santos, M. A., A. Jimenez, and J. L. Revuelta. 2000. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J. Biol. Chem. 275:28618-28624. [DOI] [PubMed] [Google Scholar]
- 79.Schmid, M. B. 1998. Novel approaches to the discovery of antimicrobial agents. Curr. Opin. Chem. Biol. 2:529-534. [DOI] [PubMed] [Google Scholar]
- 80.Schweiger, M., K. Hennig, F. Lerner, M. Niere, M. Hirsch-Kauffmann, T. Specht, C. Weise, S. L. Oei, and M. Ziegler. 2001. Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett. 492:95-100. [DOI] [PubMed] [Google Scholar]
- 81.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 82.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 83.Sharma, V., C. Grubmeyer, and J. C. Sacchettini. 1998. Crystal structure of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis: a potential TB drug target. Struct. Fold. Des. 6:1587-1599. [DOI] [PubMed] [Google Scholar]
- 84.Smith, V., D. Botstein, and P. O. Brown. 1995. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. USA 92:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith, V., K. N. Chou, D. Lashkari, D. Botstein, and P. O. Brown. 1996. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science 274:2069-2074. [DOI] [PubMed] [Google Scholar]
- 86.Song, W. J., and S. Jackowski. 1992. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J. Bacteriol. 174:6411-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. X. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 88.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 89.Sung, Y. C., D. Parsell, P. M. Anderson, and J. A. Fuchs. 1987. Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K-12. J. Bacteriol. 169:2639-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Timberlake, W., and V. Gavrias. November 1999. Identification of essential survival genes. U.S. patent 5,976,828.
- 92.Tritz, G. J. 1974. Characterization of nadR locus in Escherichia coli. Can. J. Microbiol. 20:205-209. [DOI] [PubMed] [Google Scholar]
- 93.Vallari, D. S., and C. O. Rock. 1985. Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J. Bacteriol. 164:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vallari, D. S., and C. O. Rock. 1987. Isolation and characterization of temperature-sensitive pantothenate kinase (coaA) mutants of Escherichia coli. J. Bacteriol. 169:5795-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waibel, A., T. Hanazawa, G. Hinton, K. Shikano, and K. J. Lang. 1989. Phoneme recognition using time-delay neural networks. IEEE Transact. Acoustics Speech Signal Process. 37:328-339. [Google Scholar]
- 96.Wang, H., W. Huang, Y.-J. Fei, H. Xia, T. L. Yang-Feng, F. H. Leibach, L. D. Devoe, V. Ganapathy, and P. D. Prasad. 1999. Human placental Na+-dependent multivitamin transporter. Cloning, functional expression, gene structure, and chromosomal location. J. Biol. Chem. 274:14875-14883. [DOI] [PubMed] [Google Scholar]
- 97.Wang, L. L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 98.Wang, M., J. M. Ward, R. L. Warren, R. O. Nicholas, L. M. Palmer, M. Leslie, J. M. Pratt, D. J. C. Knowles, M. A. Lonetto, J. Mooney, M. T. Black, M. K. Burnham, C. Debouck, J. C. Fedon, J. E. Hodgson, D. D. Jaworski, R. W. Reichard, R. M. Rosenberg, C. M. Traini, M. Christopher, and Y. Y. Zhong. August 2001. Polynucleotides encoding GTP cyclohydrolase II (RIBA). U.S. patent 6,280,971.
- 99.Wong, S. M., and J. J. Mekalanos. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:10191-10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu, M., B. Repetto, D. M. Glerum, and A. Tzagoloff. 1995. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang, H., T. J. Zhou, O. Kurnasov, S. Cheek, N. V. Grishin, and A. Osterman. 2002. Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD. Structure 10:69-79. [DOI] [PubMed] [Google Scholar]
- 102.Zhang, Y., A. Scorpio, H. Nikaido, and Z. Sun. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou, T., O. Kurnasov, D. R. Tomchick, D. D. Binns, N. V. Grishin, V. E. Marquez, A. L. Osterman, and H. Zhang. 2002. Structure of human nicotinamide/nicotinic acid mononucleotide adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin. J. Biol. Chem. 277:13148-13154. [DOI] [PubMed] [Google Scholar]