Abstract
A novel spirochete strain, SPN1, was isolated from the hindgut contents of the termite Neotermes castaneus. The highest similarities (about 90%) of the strain SPN1 16S rRNA gene sequence are with spirochetes belonging to the genus Spirochaeta, and thus, the isolate could not be assigned to the so-called termite clusters of the treponemes or to a known species of the genus Spirochaeta. Therefore, it represents a novel species, which was named Spirochaeta coccoides. In contrast to all other known validly described spirochete species, strain SPN1 shows a coccoid morphology and is immotile. The isolated strain is obligately anaerobic and ferments different mono-, di-, and oligosaccharides by forming formate, acetate, and ethanol as the main fermentation end products. Furthermore, strain SPN1 is able to grow anaerobically with yeast extract as the sole carbon and energy source. The fastest growth was obtained at 30°C, the temperature at which the termites were also grown. The cells possess different enzymatic activities that are involved in the degradation of lignocellulose in the termite hindgut, such as β-d-glucosidase, α-l-arabinosidase, and β-d-xylosidase. Therefore, they may play an important role in the digestion of breakdown products from cellulose and hemicellulose in the termite gut.
Spirochetes are distinguished from all other bacteria by their unique morphology and mechanism of motility due to the helical shape of the cells and the location of the flagella (axial filaments). They form a coherent phylogenetic group at the phylum level. These axial filaments, ultrastructurally similar to bacterial flagella, are attached to the cell poles and wrapped around the protoplasmic cylinder, which consists of the cytoplasmic and nuclear regions. The flagella and the protoplasmic cylinder are surrounded by a multilayered membrane called the outer sheath or outer cell envelope (9). Comparisons of 16S rRNA sequences demonstrate that the spirochetes represent a monophyletic phylum within the bacteria (35). Spirochetes are widespread in several environments, either as free-living cells, mainly in marine and limnic sediments, or host associated as commensals or parasites of animals and humans.
One of the spirochetal habitats is the digestive tract of termites and wood-eating cockroaches. Termites have developed a unique hindgut flora consisting of bacteria, archaea, flagellates, and yeasts. This symbiotic microbial community supports the decomposition of complex organic compounds and thus enables the termites to feed on wood or soil (6, 13, 21, 44). Spirochetes are one of the most abundant bacteria present in the gut fluid of termites, (36). Spirochetes of several sizes (3 to 100 μm in length; 0.2 to 1.0 μm in width) are consistently present in the hindguts of all termites (5). Whereas most spirochetes usually exist free-living in the gut fluid, they have also been found as ectosymbionts attached to the surfaces of protists, such as Mixotricha paradoxa, that inhabit the gut of the lower wood-eating termite Mastotermes darwiniensis (10, 46). These observations suggest that this bacterial group plays an important role within the complex symbiotic system in the termite hindgut.
A few years ago, the first pure cultures of hindgut spirochetes were obtained from the lower termite Zootermopsis angusticollis (Treponema primitia strains ZAS1 and ZAS2 and Treponema azotonutricium strain ZAS-9) (16, 17). These isolates display physiological pathways which were previously unknown within the spirochetal group, including acetogenesis from H2 plus CO2 (23) and nitrogen fixation (24). Both processes are beneficial for termites, because acetate is their major carbon and energy source (30) and N2 fixation by symbiotic hindgut bacteria can supply up to 60% of the nitrogen in the termite biomass (24). These findings imply an important role of symbiotic spirochetes in the nutrition of the termites.
The evolutionary distance between spirochetes from termite guts and other members of this group has been well demonstrated by several studies based on culture-independent investigations of 16S rRNA gene sequences from different termite guts (2, 3, 31, 36). All spirochetal clones obtained from termite gut contents are only distantly related to known spirochetes from other habitats. However, phylogenetic analyses have shown that most symbiotic spirochetes obtained so far form their own cluster within the Treponema group of the spirochetes. In our present study, we isolated a novel spirochete from the hindgut contents of the lower dry-wood termite Neotermes castaneus (Kalotermitidae). The obtained organism shows an unexpected coccoid morphology, which differs from those of all known spirochete species. Strain SPN1 ferments different carbohydrates and produces formate, acetate, and ethanol as the main fermentation end products. Furthermore, the new isolate possesses glycolytic activities which are involved in the degradation of oligosaccharides derived from lignocelluloses.
MATERIALS AND METHODS
Termites.
The termite Neotermes castaneus was obtained from the Federal Institute for Material Research and Testing (Berlin, Germany). The animals were fed with poplar wood and cultured in metallic vessels containing humid vermiculite at 30°C.
Preparation and isolation.
Termites were surface sterilized in 70% ethanol and then washed in sterile water. All further steps were performed under aseptic conditions in an anaerobic chamber. The hindgut was removed from the abdomen, and the contents were transferred into tubes containing 5 ml medium. The tubes were sealed with butyl stoppers, flushed with H2-CO2 (80/20), and incubated at 28°C for up to 4 weeks. For enrichment and isolation, a modified medium, previously described by Leadbetter and Breznak (22), was used. The medium contained (per 1,000 ml) NaCl, 1.0 g; KCl, 0.5 g; MgCl2 · 6H2O, 0.4 g; CaCl2 · 2H2O, 0.1 g; NH4Cl, 0.3 g; KH2PO4, 0.2 g; Na2SO4, 0.15 g; glucose, 2.0 g; yeast extract, 2.0 g; peptone, 5.0 g; resazurin, 0.5 mg; NaHCO3, 5.0 g; and dithiothreitol at a final concentration of 5 mM and rifamycin SV at a final concentration of 50 μg/ml. Sodium hydrogen carbonate, dithiothreitol, and rifamycin were added to the autoclaved medium from sterile stock solution as described by Widdel and Pfennig (47). The isolated strain was obtained by repeated deep-agar dilution series. To ensure that the isolated strain was an often-encountered member of the hindgut community, we repeated the isolation procedure several months later. The obtained strain, SPN2, was morphologically and phylogenetically identical to the previously isolated strain, SPN1.
Physiological and enzymatic tests.
Utilization of different growth substrates was tested in screw-cap tubes with the medium described above, which was modified according to the carbon sources (without glucose, 4 g/liter yeast extract, 2 g/liter peptone) (S-YP medium). The gas phase was 80% H2 and 20% CO2 or 100% nitrogen. The utilization of carbohydrates and the production of fermentation end products were analyzed by using a high-performance liquid chromatograph (HPLC) (Shimadzu LC 10AD VP; Shimadzu Corp., Kyoto, Japan) with refractive index detection (RI Detector ERC-7515B; Erma CR, Inc., Kawaguchi City, Japan). Analysis of the substrates and fermentation products was performed by using an Aminex HPX 87 H column (300 by 7.8 mm; Bio-Rad Laboratories, California). The injection volume was 5 μl, the column temperature was 65°C, and the flow rate was 0.6 ml/min. Concentrations of ethanol determined with an HPLC were checked by enzymatic analysis. To determine the formation of end products without the background effect of yeast extract, we used dense cell suspensions, which were incubated without undefined carbon sources. Strain SPN1 was incubated with S-YP medium till the end of the exponential growth phase. Subsequently, cells were concentrated by centrifugation and washed twice in phosphate-buffered saline buffer (NaCl, 8.77 g/liter; KCl, 0.22 g/liter; Na2HPO4, 1.14 g/liter; KH2PO4, 0.2 g/liter; pH 7.4). The pellet was suspended to obtain a final titer of about 5 × 109 cells/ml and then incubated for 24 h in a mineral medium (S-YP medium without yeast extract and peptone), which was supplemented with maltose. The amounts of fermentation products were calculated from the average of three parallels. Growth rates, as well as the temperature and pH optima, were determined by measuring the increase of optical density by using a spectral photometer. The cell titer was determined at the end of exponential growth by direct counting after DAPI (4′,6′-diamidino-2-phenylindole) staining. The activities of enzymes which were involved in the degradation of oligosaccharides were tested qualitatively by using nitrophenol-labeled carbohydrates (see Table 2) (all from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany). Actively growing cells were incubated with the p-nitrophenol derivatives (final concentration, 0.3 mg/ml) over 24 h. Enzymatic activity affects the development of yellow color as a result of free nitrophenol in the medium. Negative controls were performed with cell-free medium and with inactivated cells (10 min at 80°C).
TABLE 2.
Enzymatic activities of strain SPN1
| Tested substrates | Corresponding enzyme | Reactiona |
|---|---|---|
| Nitrophenyl-β-l-arabinopyranoside | α-l-Arabinosidase | + |
| Nitrophenyl-β-d-fucopyranoside | β-d-Fucosidase | + |
| Nitrophenyl-α-d-galactopyranoside | α-d-Galactosidase | + |
| Nitrophenyl-β-d-galactopyranoside | β-d-Galactosidase | − |
| Nitrophenyl-α-d-glucopyranoside | α-d-Glucosidase | + |
| Nitrophenyl-β-d-glucopyranoside | β-d-Glucosidase | + |
| Nitrophenyl-β-d-glucuronide | β-d-Glucuronidase | − |
| Nitrophenyl-α-d-mannopyranoside | α-d-Mannosidase | − |
| Nitrophenyl-β-d-xylopyranoside | β-d-Xylosidase | + |
+, positive; −, negative.
Electron microscopy.
Cells were harvested by centrifugation and then fixed in a solution of 4% paraformaldehyde and 2.5% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.2. After 60 min of incubation, the cells were washed three times in buffer and postfixed in 2% OsO4 for 1.5 h. The cells were dehydrated in a series of ethanol and embedded in Spurr's resin. Ultrathin sections were stained with uranyl acetate and lead citrate prior to their examination with a Philips CEM 120 Bio-Twin transmission electron microscope.
Determination of the G+C content of the DNA.
The G+C content of the DNA was determined as described by Mesbah et al. (27) after degradation of the DNA to nucleosides by P1 nuclease and alkaline phosphatase and subsequent separation of the nucleosides by high-performance liquid chromatography.
Preparation of DNA.
Bacteria from 5-ml cultures were harvested by centrifugation and washed twice in sterile phosphate-buffered saline buffer. Genomic DNA was extracted by the InstaGene protocol (InstaGene Matrix; Bio-Rad, Munich, Germany). The washed cells were suspended in 100 μl sterile water and, after addition of 500 μl of the InstaGene Matrix, incubated at 56°C for 20 min. Subsequently, the suspension was incubated at 98°C for 5 min and then shaken at 1,000 rpm for 1 min. Insoluble debris of the bacterial cells was separated from the DNA by centrifugation (2 min; 12,000 rpm). The supernatant was removed and used for PCR without further purification. The extraction of DNA from the hindgut bacteria of the termite Neotermes castaneus was performed as previously described by Berchtold and König (2).
Amplification of 16S rRNA gene and sequencing.
The bacterial small-subunit rRNA gene was amplified by PCR using the primers Eubak5 (3) and Eubak1392r (5′-CCA CGG GCG GTG TGT AC-3′). The reaction mixture (50 μl) was composed of 2 μl of the isolated DNA solution, 0.4 μM of each primer, 0.25 mM of each deoxynucleotide triphosphate, 0.2 mM MgCl2, and 0.2 μl of Taq polymerase (5 U/μl) in 1× PCR buffer (Peqlab, Erlangen, Germany). Reactions were carried out in a thermal cycler (Mastercycler gradient; Eppendorf, Hamburg, Germany) with a hot start. The enzyme was added after an initial denaturation step of 95°C for 5 min. The temperature profile consisted of 10 cycles of denaturation at 94°C for 1 min; annealing for 1 min, beginning at 58°C for the first cycle and then decreasing every cycle by 0.5°C until 53.5°C; followed by an extension at 72°C for 2 min. Then, 20 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min were performed, followed by a final extension step of 5 min at 72°C. The amplified DNA was purified by using the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany). Direct sequencing of the 16S rRNA gene PCR product was carried out by the didesoxy method (40) by the Genterprise Company (Mainz, Germany). Based on the obtained 16S rRNA gene sequence of strain SPN1, we constructed the strain-specific primer SPN1 167f (5′-CCA CGG GCG GTG TGT AC-3′), which was used for the molecular detection of strain SPN1 in the DNA extract from hindgut bacteria of Neotermes castaneus.
Phylogenetic analysis.
The novel small-subunit rRNA gene sequence was added to the ARB database and aligned against existing homologous sequences using the fast aligner of the ARB software package (26). The alignment was checked manually and corrected where necessary. Highly variable regions of the 16S rRNA gene were excluded from the phylogenetic analysis by using only those nucleotide positions that were identical in at least 50% of all sequences of the alignment. To determine the evolutionary relationships of the newly isolated organism, the maximum likelihood method, included in the PHYLIP 3.5 program package, was used (15). The reliability of the constructed tree was evaluated by bootstrap analysis (14).
RESULTS
Morphology.
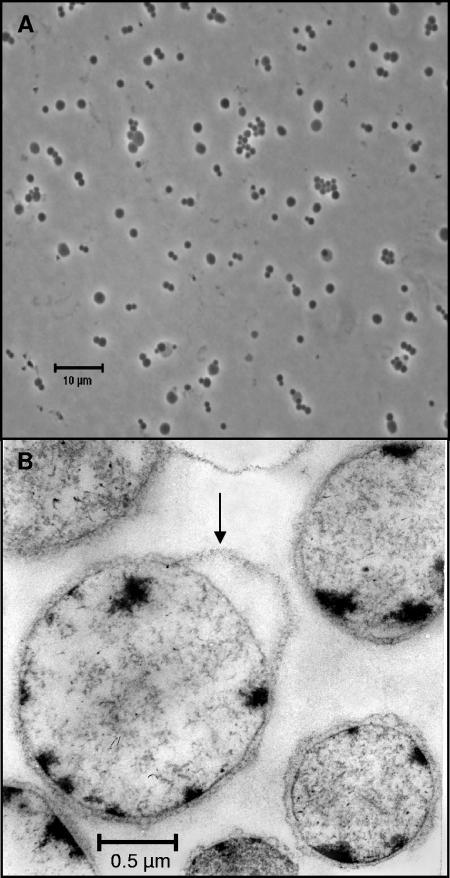
The new isolate was obtained by repeated deep-agar dilution series (11). Strain SPN1 formed yellow-white colonies with well-defined edges in S-YP medium, whereas most spirochetes form spherical colonies with diffuse edges (cotton ball-like), according to their ability to locomote through agar gels. The isolated strain SPN1 showed a coccoid cell form and was nonmotile (Fig. 1A). Axial filaments were not observed. The cells had an average diameter of about 1.5 μm and were surrounded by an outer envelope (Fig. 1B). In the early growth phase, aggregates are formed. Ultrathin sections show mainly coccoid cells with loose cytoplasma and small condensed areas (probably DNA) (Fig. 1B).
FIG. 1.
Phase-contrast micrograph of whole coccoid cells (A). Electron micrograph of ultrathin section (B). The thin section shows the outer envelope (arrow) surrounding coccoid forms with loose cytoplasma and small condensed areas.
Growth conditions, substrates, and enzymatic activities.
The obtained isolate SPN1 is a strictly anaerobic organism. Strain SPN1 exhibited no catalase activity and was not able to grow under either oxic or microaerophilic conditions. For enrichment and isolation, we used a substrate mixture containing glucose, yeast extract, and peptone. The isolate utilized no glucose but was able to grow with yeast extract as the sole carbon and energy source. The minimum concentration of yeast extract required was 0.2%, and growth was improved by addition of peptone. Consequently, we used a medium containing 0.4% yeast extract and 0.2% peptone for further cultivation (S-YP medium). In addition to yeast extract, strain SPN1 also fermented different carbohydrates. The utilization of amino acids or polysaccharides, like celluloses, xylan, and arabinogalactan, was not detected.
Obviously, yeast extract contains one or more growth factors which were not present in standard vitamin (seven-vitamin solution; DSMZ medium 503) or cofactor (23) solution, because no growth occurred if yeast extract was replaced by these mixtures. However, the addition of fermentable pentoses and oligosaccharides, but not hexoses, led to a significantly higher cell yield and a decrease in doubling time (Table 1). The fastest growth and highest cell yield were observed with the addition of maltose or maltotriose to S-YP medium. In contrast to the pentoses or oligosaccharides used, the utilization of the hexoses glucose and galactose was only weak or not detectable and the effect on cell yield and doubling time was not significant in comparison to S-YP medium without additional substrates. From both yeast extract and fermentable sugars, strain SPN1 produced formate, acetate, and ethanol as the main end products. To determine the ratios of fermentation products from sugars without the background effect from yeast extract, we used dense suspensions of washed cells. Maltose was fermented to ethanol (2.2 mol per mol maltose), formate (2.9 mol/mol), and acetate (1.4 mol/mol). Other fermentation end products were not detected by HPLC analysis. The recovery of carbon from maltose in the fermentation end products was 84%. SPN1 grew under atmospheres of both H2-CO2 and nitrogen without hydrogen. The formation of acetate from H2 plus CO2 by acetogenesis could not be found (23). The optimal growth temperature of the isolate was 30°C. No growth was observed above 40°C or below 15°C. The pH range of strain SPN1 was between 5.5 and 9.5. The optimum pH value for growth was 7.4.
TABLE 1.
Cell yield and doubling time of strain SPN1
| Substratea | Cell titer (108 cells/ml) | Doubling time (h) |
|---|---|---|
| Controlb | 3.9 | 29.1 |
| Arabinose | 5.1 | 25.7 |
| Xylose | 5.4 | 25.2 |
| Glucose | 4.2 | 28.8 |
| Galactose | 4.1 | 30.0 |
| Maltose | 6.4 | 22.6 |
| Cellobiose | 4.6 | 28.3 |
| Maltotriose | 6.6 | 21.4 |
| Maltotetraose | 5.7 | 24.9 |
The substrates were added to S-YP medium. Incubation was at 30°C. Final concentrations of substrates were 6 mM, except disaccharides (3 mM), maltotriose (2 mM), and maltotetraose (1.5 mM).
S-YP medium without additional substrate.
The di- and oligosaccharide-degrading capabilities of strain SPN1 were tested with different nitrophenol-labeled carbohydrates (Table 2). Strain SPN1 showed glycolytic activities, which are involved in the further breakdown of oligosaccharides derived from cellulose and hemicellulose in the termite gut. The tested enzymatic activities seemed to be cell bound, because no glycolytic activity was found in the supernatant of the growth medium.
G+C content of DNA.
The G+C content of the DNA of strain SPN1 was within the range 56.6 to 57.4 mol%.
Phylogenetic position.
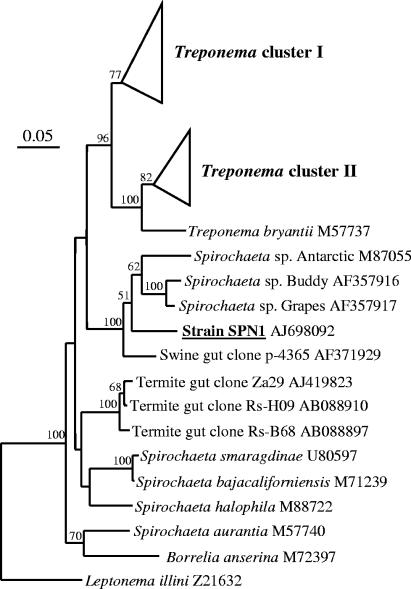
The morphology of strain SPN1 differs from those of all known validly described spirochetes, but based on its 16S rRNA gene sequence, the isolate could be assigned to the order Spirochetales. Based on phylogenetic analysis, strain SPN1 forms a monophyletic cluster with different free-living spirochetes and a clone obtained from swine intestine (Fig. 2). The highest similarities in the 16S rRNA gene sequence of strain SPN1 were to the spherical isolates Spirochaeta sp. strain Buddy (90.5%; AF357916) and Spirochaeta sp. strain Grapes (90.1%; AF357917) (37-39). Interestingly, both isolates also lacked the typical spiral morphology and were termed free-living pleomorphic spirochetes with a spherical cell shape (37-39). These strains are not host-associated organisms but were isolated from freshwater sediments and were assigned to the genus Spirochaeta. Consequently, strain SPN1 did not cluster in one of the so-called “termite Treponema clusters I and II” of the treponemes as illustrated in Fig. 2. However, the isolate seems to be permanently in the hindgut, because we could detect the 16S rRNA gene of strain SPN1 with semispecific PCR (primers SPN1 167f and Eub 1392r) in the hindgut contents of Neotermes castaneus.
FIG. 2.
Phylogenetic tree showing the relationship of strain SPN1 to other spirochetes. The tree was constructed by using the maximum likelihood method. Bootstrap values above 50 (from 100 resamplings) are shown for each node. A sequence of Escherichia coli (accession number AE000129) was used as the outgroup (not shown). For tree construction, the 16S rRNA gene sequences of additional spirochetes were included. Cluster I, clone NkS56 AB084967, clone RFS8 AF068343, Spirochaeta caldaria M71240, Treponema azotonutricium AF320287, and Treponema primitia strain ZAS-1 AF093251; cluster II, clone HsPySp20 AB032004, clone RFS9 AF068432, clone Rs-A53 AB088895, and clone Rs-G65 AB088858. The bar indicates 5% estimated sequence divergence.
DISCUSSION
The cell shape of strain SPN1 was unexpected, because spirochetes are usually characterized by their typical helical cell structure. Furthermore, all known spirochetes are motile via their axial filaments, which are attached to the cell poles. However, the formation of cocci by morphologically regular spirochetes in the stationary growth phase is known (9). Similar observations have been made in the case of Arthrobacter globiformis, which forms coccoid cells after the end of the exponential growth phase (20). In contrast to strain SPN1, which occurs only in round form, Arthrobacter globiformis is a dimorphic bacterium that exhibits reversible rod-sphere switching, and spirochetes usually form cocci only when the environmental conditions become unfavorable. Probably, this coccoid stage is the origin of the unusual morphology of strain SPN1. Recently, novel spirochetal isolates have been obtained from freshwater sediments which also reproduce only in round forms (37-39). This may indicate that “nonspiral spirochetes” are more widespread in nature and not restricted to specialized ecosystems like the termite hindgut.
Considering the environmental conditions in the hindgut, it is not surprising that the obtained strain SPN1 is an anaerobic organism, like other intestinal spirochetes. Because of the high oxygen consumption rates within the gut periphery, the central portion of the gut is predominantly anoxic (7). This anoxic central region of the termite hindgut is likely attributable to spirochetes, in contrast to the microoxic gut wall, which is colonized by methanogens and sulfate reducers (1, 22).
Strain SPN1 is able to grow on a variety of carbohydrates, including pentoses, disaccharides, and shorter oligosaccharides, which are present in the termite hindgut because of the hydrolytic breakdown of high-polymeric cellulose and hemicellulose. Strain SPN1 uses the glucose dimers maltose and cellobiose as carbon sources, but obviously not free glucose, which probably indicates a lack of the corresponding membrane transporter. Similarly, different rumen bacteria, like Ruminococcus flavefaciens, use cellobiose but not glucose (8). However, turnover rates of injected glucose in agarose-embedded hindguts (<0.5 nmol termite−1 h−1), determined by Tholen and Brune (43), may be a hint that free glucose is not a very important intermediate in the termite hindgut. In the past, numerous bacteria that could be involved in the different steps of lignocellulose degradation were isolated from termites based on their physiological properties (21, 41, 42, 45). It is well established that spirochetes are the most abundant group within the bacterial communities in all investigated termites. Thus, the spirochetes located in the termite hindgut should play an important role in the synergistic degradation of the main polymeric wood compounds cellulose and hemicellulose. Formally, the degradation of lignocellulose can be divided into three steps, a hydrolytic step, a fermentative step, and a methanogenic/acetogenic step (21). Our results indicate that spirochetes are involved in both the hydrolytic step of oligosaccharides and the fermentative step. This assumption is supported by the enzymatic spectrum of strain SPN1. The complete degradation of hemicelluloses like xylan requires a synergistically acting set of enzymes, including endo-β-1.4-xylanase, β-xylosidase, α-glucuronidase, and α-l-arabinosidase. The full set of enzymes has not been found in a single bacterial isolate of the termite gut, and thus, a community of intestinal bacteria has to be responsible for effective hemicellulose degradation. Strain SPN1 can be characterized as a member of this glycolytic community. The activities of α-glucosidase, α-galactosidase, and β-fucosidase indicate that strain SPN1 is also involved in the degradation of different plant carbon storage compounds, like amylose, raffinose, and stachyose. Strain SPN1 produces mainly ethanol, formate, and acetate during the fermentation of carbohydrates. The carbon recovery was 84%, and the electron recovery was 103%. During fermentation, Treponema saccharophilum (33, 34) incorporates about 15% of the carbon source into cell carbon (34). If this is also true for strain SPN1, the carbon recovery would be about 99%. Most spirochetes ferment carbohydrates via the Embden-Meyerhof pathway to pyruvate, which is metabolized to acetate, ethanol, CO2, and H2 by means of a clostridial-type clastic reaction (9). This pathway could also be proposed for Treponema azotonutricium, isolated from the termite Zootermopsis angusticollis (24), which produces acetate, ethanol, H2, and CO2 in a molar ratio of about 3:1:5:4 from maltose (17). The formation of formate in the case of strain SPN1 indicates some modifications in this pathway, probably including a pyruvate-formate lyase. The formation of formate in addition to ethanol and acetate is known from several spirochetes isolated from bovine rumen fluid, such as Treponema saccharophilum (33, 34). However, the ratio of fermentation products produced by strain SPN1 differs from that proposed for T. saccharophilum, which produces formate, acetate, and ethanol in a molar ratio of nearly 2:1:1 (34).
The fastest growth of strain SPN1 was obtained at the same temperature at which the termites were grown. The pH range for growth of strain SPN1 corresponded to the physicochemical conditions in the gut. The pH value of the hindgut of lower termites is in a range between 6.0 and 7.5, whereas in higher termites, parts of the midgut have an alkaline pH value up to 10.4 (4).
Because of the difficulties in the isolation of spirochetes from the termite hindgut, several investigations concentrated on culture-independent approaches. These investigations clearly verified that the termite hindguts are an enormous reservoir of novel spirochete species. Recently, more than 40 different spirochetal phylotypes were recognized in one termite species (28) in comparison to less than 30 well-characterized and named spirochetes of the genera Spirochaeta and Treponema (32). Corresponding to the phylogenetic positions of spirochetal clones obtained from termite gut contents, it was previously speculated that all termite spirochetes represent a separate phylogenetic branch of the treponemes (2, 36). Further investigations showed a second cluster of Treponema-related spirochetes in the termite hindgut (19, 25, 28, 31). This cluster, designated “termite Treponema cluster II,” is closely related to rumen spirochetes, such as Treponema bryantii and Treponema pectinovorum. However, demonstrating the phylogenetic variety within these clusters, large and free-living spirochete species like Spirochaeta stenostrepta and Spirochaeta caldaria were included in cluster I. According to Ochman and Wilson (29), 1% of sequence divergence in the 16S rRNA gene corresponds to roughly 50 million years of evolution. Concerning the establishment of the spirochete-termite symbiosis, the large genetic distances (>0.10) of spirochetes within individual termite species, as well as between different termite species, can lead to the speculation that the last common ancestor of termite spirochetes must have existed earlier than 500 million years ago and that the diversification of the gut spirochetes started long before the origin of termites (about 300 million years ago) (12). This may indicate that different spirochete species colonized the ancestral termites and evolved within the hindgut to their recent diversity. Recently, spirochetal clones from gut contents were obtained which could not be assigned to the treponema cluster of termite spirochetes (18, 28). The clone Za29 from Zootermopsis angusticolis was sequenced by our group. The same is true for the isolate SPN1, which is relatively closely related to the genus Spirochaeta. This indicates that the phylogenetic diversity of the gut spirochetes is greater than previously expected. It seems that, in addition to the treponema clusters I and II, the spirochetes from the termite hindgut also form two clusters related to the genus Spirochaeta.
Description of Spirochaeta coccoides sp. nov.
Spirochaeta coccoides sp. nov. (coc.coi′des. Gr. n. coccos, a berry; Gr. n. eidos, shape; N.L. adj. coccoides, berry-shaped).
Cells are cocci, 0.5 to 2 μm in diameter. Cell aggregates are formed. Axial filaments were not observed. Nonmotile. Anaerobic. Catalase negative. Yeast extract is required for growth and could serve as a sole energy and carbon source. Optimal temperature for growth is 30°C. The pH range for growth was between 5.5 and 9.5 with an optimum at 7.4. Pentoses (arabinose, xylose) and oligosaccharides (maltose, cellobiose, maltotriose, and maltotetraose) stimulated growth in a medium containing 0.4% yeast extract and 0.2% peptone. Maltose is fermented to ethanol, formate, and acetate as the main fermentation products. Glucose, galactose, lactate, pyruvate, amino acids, and polysaccharides are not utilized. Activities of β-d-glucosidase, α-d-glucosidase, α-d-galactosidase, α-l-arabinosidase, β-d-fucosidase, and β-d-xylosidase are exhibited. G+C content is 56.6 to 57.4 mol%. Based on the nucleotide sequence of the 16S rRNA gene, this spirochete belongs to the genus Spirochaeta (accession number AJ698092; EMBL).
Source: strain SPN1 was isolated from the hindgut contents of the lower dry-wood termite Neotermes castaneus (Burmeister) (Isoptera: Kalotermitidae).
Type strain is strain SPN1 deposited with the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany (DSM 17374) and with the American Type Culture Collection (ATTC), Manassas, Va. (ATTC BAA-1237).
Acknowledgments
We thank H. Hertel from the Bundesanstalt für Materialprüfung und Materialforschung, Berlin, Germany, for supplying us with termite cultures.
Footnotes
This paper is dedicated to Otto Kandler on the occasion of his 85th birthday.
REFERENCES
- 1.Berchtold, M., A. Chatzinotas, W. Schönhuber, A. Brune, R. Amann, D. Hahn, and H. König. 1999. Differential enumeration and in situ localisation of microorganisms in the hindgut of the lower termite Mastotermes darwiniensis. Arch. Microbiol. 172:407-416. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold, M., and H. König. 1996. Phylogenetic analysis and in situ identification of uncultivated spirochetes from the hindgut of the termite Mastotermes darwiniensis. Syst. Appl. Microbiol. 19:66-73. [Google Scholar]
- 3.Berchtold, M., W. Ludwig, and H. König. 1994. 16S rDNA sequence and phylogenetic position of an uncultivated spirochete from the hindgut of the termite Mastotermes darwiniensis Froggatt. FEMS Microbiol. Lett. 123:269-273. [DOI] [PubMed] [Google Scholar]
- 4.Bignell, D. E., and J. M. Anderson. 1980. Determination of pH and oxygen status in the guts of lower and higher termites. J. Insect Physiol. 26:183-188. [Google Scholar]
- 5.Breznak, J. A. 1984. Hindgut spirochetes of termites and Cryptocercus puntulatus, p. 67-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 6.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 7.Brune, A., D. Emerson, M. Kühl, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant, M. P. 1984. Genus Ruminococcus Sijpesteijn 1948, 152AL, p. 1093-1097. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 9.Canale-Parola, E. 1984. Order I. Spirochaetales Buchanan 1917, 163AL, p. 38-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 10.Cleveland, L. R., and A. V. Grimstone. 1964. The fine structure of the flagellate Mixotricha paradoxa and its associated microorganisms. Proc. R. Soc. Lond. B 159:668-686. [Google Scholar]
- 11.Dröge, S., J. Fröhlich, and H. König. 2004. Isolation of a novel spirochete from the termite gut, p. 74. Biospektrum Spec. Abstr. Vol. Annu. Meet. Vereinigung Allg. Angew. Mikrobiol. Elsevier, Heidelberg, Germany.
- 12.Emerson, A. E. 1965. A review of the Mastotermitidae (Isoptera), including a new fossil genus from Brazil. Am. Mus. Novit. 2236:1-46. [Google Scholar]
- 13.Eutick, M. L., P. Veivers, R. W. O'Brian, and M. Slaytor. 1978. Dependence of the higher termite Nasutitermes exitiosus and the lower termite Coptotermes lacteus on their hindgut flora. J. Insect Physiol. 24:363-368. [Google Scholar]
- 14.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1993. Phylip (Phylogeny Inference Package) version 3.5. University of Washington, Seattle.
- 16.Graber, J. R., and J. A. Breznak. 2004. Physiology and nutrition of Treponema primitia, an H2/CO2-acetogenic spirochete from termite hindguts. Appl. Environ. Microbiol. 70:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graber, J. R., J. R. Leadbetter, and J. A. Breznak. 2004. Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the first spirochetes isolated from termite guts. Appl. Environ. Microbiol. 70:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 19.Iida, T., M. Ohkuma, K. Ohtoko, and T. Kudo. 2000. Symbiotic spirochetes in the termite hindgut: phylogenetic identification of ectosymbiotic spirochetes of oxymonad protists. FEMS Microbiol. Ecol. 34:17-26. [DOI] [PubMed] [Google Scholar]
- 20.Keddie, R. M., M. D. Collins, and D. Jones. 1984. Genus Arthrobacter Conn and Dimmick 1947, 300AL, p. 1288-1301. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 21.König, H., J. Fröhlich, M. Berchtold, and M. Wenzel. 2002. Diversity and microhabitats of the hindgut flora of termites. Recent Res. Dev. Microbiol. 6:125-156. [Google Scholar]
- 22.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 24.Lilburn, T. G., K. S. Kim, N. E. Ostrom, K. R. Byzek, J. R. Leadbetter, and J. A. Breznak. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495-2498. [DOI] [PubMed] [Google Scholar]
- 25.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 28.Noda, S., M. Ohkuma, A. Yamada, Y. Hongoh, and T. Kudo. 2003. Phylogenetic position and in situ identification of ectosymbiotic spirochetes on protists in the termite gut. Appl. Environ. Microbiol. 69:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 30.Odelson, D. A., and J. A. Breznak. 1983. Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl. Environ. Microbiol. 45:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkuma, M., T. Iida, and T. Kudo. 1999. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181:123-129. [DOI] [PubMed] [Google Scholar]
- 32.Olsen, I., B. J. Paster, and F. E. Dewhirst. 2000. Taxonomy of spirochetes. Anaerobe 6:39-57. [Google Scholar]
- 33.Paster, B. J., and E. Canale-Parola. 1981. Physiological diversity of rumen spirochetes. Appl. Environ. Microbiol. 43:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paster, B. J., and E. Canale-Parola. 1985. Treponema saccharophilum sp. nov., a large pectinolytic spirochete from the bovine rumen. Appl. Environ. Microbiol. 50:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paster, B. J., and F. E. Dewhirst. 2001. Phylogenetic foundations of spirochetes, p. 5-9. In M. H. Saier, Jr., and J. Garcia-Lara (ed.), The spirochetes: molecular and cellular biology. Horizon Scientific Press, Wymondham, United Kingdom.
- 36.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritalahti, K., and F. E. Löffler. 2002. Ecology and characterization of novel, free-living, non-spiral spirochetes, abstr. I-14. Abstr. Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, D.C.
- 38.Ritalahti, K., and F. E. Löffler. 2003. Non-spiral spirochetes (NSS), sticky denizens in anoxic environments, abstr. I-133. Abstr. Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, D.C.
- 39.Ritalahti, K. M., and F. E. Löffler. 2004. Characterisation of novel free-living pleiomorphic spirochetes (FLiPS), abstr. 539. Abstr. 10th Int. Symp. Microb. Ecol. International Society for Microbial Ecology, Geneva, Switzerland.
- 40.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxena, S., J. Bahadur, and A. Varma. 1993. Cellulose and hemicellulose degrading bacteria from the termite gut and mound soils of India. Ind. J. Microbiol. 33:55-60. [Google Scholar]
- 42.Schäfer, A., R. Konrad, T. Kuhnigk, P. Kämpfer, H. Hertel, and H. König. 1996. Hemicellulose-degrading bacteria and yeasts from the termite gut. J. Appl. Bacteriol. 80:471-478. [DOI] [PubMed] [Google Scholar]
- 43.Tholen, A., and A. Brune. 2000. Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ. Microbiol. 2:436-444. [DOI] [PubMed] [Google Scholar]
- 44.Varma, A., B. K. Kolli, J. Paul, S. Saxena, and H. König. 1994. Lignocellulose degradation by microorganisms from termite hills and termite guts: a survey on the present state of art. FEMS Microbiol. Rev. 15:9-28. [Google Scholar]
- 45.Wenzel, M., M. Schönig, M. Berchtold, P. Kämpfer, and H. König. 2002. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J. Appl. Microbiol. 92:32-40. [DOI] [PubMed] [Google Scholar]
- 46.Wenzel, M., R. Radek, G. Brugerolle, and H. König. 2002. Identification of the ectosymbiotic bacteria of Mixotricha paradoxa involved in movement symbiosis. Eur. J. Protistol. 39:11-23. [Google Scholar]
- 47.Widdel, F., and N. Pfennig. 1981. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch. Microbiol. 129:395-400. [DOI] [PubMed] [Google Scholar]