Abstract
The presence of Escherichia coli in water is used as an indicator of fecal contamination, but recent reports indicate that soil populations can also be detected in tropical, subtropical, and some temperate environments. In this study, we report that viable E. coli populations were repeatedly isolated from northern temperate soils in three Lake Superior watersheds from October 2003 to October 2004. Seasonal variation in the population density of soilborne E. coli was observed; the greatest cell densities, up to 3 × 103 CFU/g soil, were found in the summer to fall (June to October), and the lowest numbers, ≤1 CFU/g soil, occurred during the winter to spring months (February to May). Horizontal, fluorophore-enhanced repetitive extragenic palindromic PCR (HFERP) DNA fingerprint analyses indicated that identical soilborne E. coli genotypes, those with ≥92% similarity values, overwintered in frozen soil and were present over time. Soilborne E. coli strains had HFERP DNA fingerprints that were unique to specific soils and locations, suggesting that these E. coli strains became naturalized, autochthonous members of the soil microbial community. In laboratory studies, naturalized E. coli strains had the ability to grow and replicate to high cell densities, up to 4.2 × 105 CFU/g soil, in nonsterile soils when incubated at 30 or 37°C and survived longer than 1 month when soil temperatures were ≤25°C. To our knowledge, this is the first report of the growth of naturalized E. coli in nonsterile, nonamended soils. The presence of significant populations of naturalized populations of E. coli in temperate soils may confound the use of this bacterium as an indicator of fecal contamination.
Indicator bacteria, such as Escherichia coli, were originally thought to be present only in the intestinal tracts of warm-blooded animals, including humans (25, 29). The occurrence of E. coli in water and food has been used as an indicator of fecal contamination, signaling the possible presence of fecal pathogens such as Salmonella and Shigella species (37). This is due, in part, to the observed correlation between elevated E. coli counts in water and the rate of occurrence of gastrointestinal symptoms or disease (37). Due to this association, E. coli has been routinely used as an indicator of fecal contamination in drinking and recreational waters throughout the world.
The determination of the origin(s) of E. coli in water is of great interest to government regulatory agencies, beach managers, environmentalists, and operators of sewage treatment facilities. Since E. coli strains in water can originate from human and nonhuman sources, such as farm and wild animals, waterfowl, and pets, source determination may allow for proper risk assessment and abatement procedures. Several methods have been examined to track the potential source(s) of E. coli in waterways, including the use of antibiotic resistance patterns (18, 23, 27), hybridization with restriction fragments of 16S and 23S rRNA genes (ribotyping) (10), analysis of sequence variation in the uidA gene (28), and repetitive extragenic palindromic PCR (14, 20). Although experimental in nature, many of these techniques have shown that E. coli in water may originate from human and nonhuman hosts, suggesting that elevated levels of E. coli in water may not always indicate the presence of human-derived pathogens.
E. coli has been found in tropical and subtropical soils (8, 13, 16, 31) and has been shown to grow in tropical soils in laboratory studies if provided with amendments (7, 8). Recently, Byappanahalli and coworkers (9) reported that E. coli could also be isolated from coastal temperate forest soils in Indiana, suggesting that soilborne E. coli may be more ubiquitous than originally thought. However, while there is limited information concerning the survival of E. coli in riverine sediments and soils (2, 5, 6, 11, 30, 38), growth of these bacteria in the environment is not well understood or documented. Various stresses influence the survival of E. coli in soils, such as high and low temperatures (3, 34), limited moisture (4, 7, 9, 13, 31), variation in soil texture (13), low organic matter content (33), high salinity (32), and predation (5, 6, 7, 11, 31). Solo-Gabriele et al. (31) reported that E. coli counts were elevated after tidal events, suggesting that this bacterium can grow in riverbank soils and move back into water by erosion. Based on these results, soil and sand should perhaps be thought of as both a sink and a source of E. coli for waterways. Since the use of E. coli as an indicator of fecal contamination is based on the assumption that it inhabits only the intestinal tracts of warm-blooded animals (37), the existence of soilborne E. coli confounds the use of this bacterium as a reliable indicator of fecal contamination (16).
Since most studies that have examined E. coli in soils were done in tropical, subtropical, or moderate temperate environments, we were interested in determining if soilborne E. coli was also present, and grew, in northern temperate soils exposed to extreme environmental conditions, such as repetitive freeze-thaw cycles. Consequently, the objectives of this study were to (i) examine the survival and persistence of E. coli populations in three soils in several coastal Lake Superior watersheds and to determine if these E. coli strains have become naturalized to these soils, (ii) examine the genetic relatedness of soilborne E. coli strains from different locations, and (iii) determine if soilborne E. coli could actively multiply in the soils examined.
MATERIALS AND METHODS
Site description.
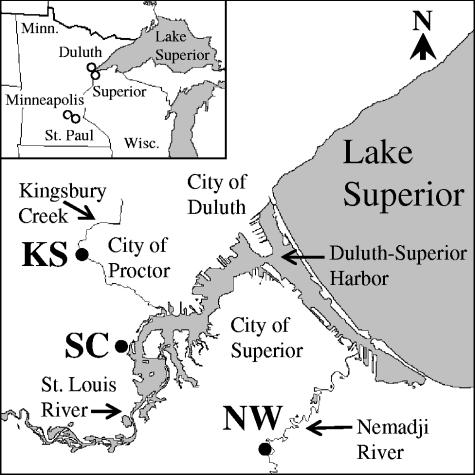
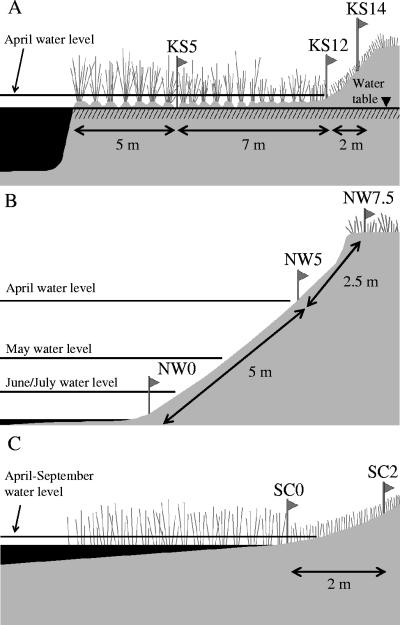
Field investigations were performed at three sampling sites located in Lake Superior watersheds near Duluth, Minnesota: Kingsbury Stark (KS), Nemadji Weinstein (NW), and St. Louis Clyde (SC). The KS site is adjacent to the intersection between Kingsbury Creek and Stark Avenue (46°44′59′′N, 92°14′18′′ W) in Proctor, MN (Fig. 1). The KS site is a partial wetland and rich in organic matter (Table 1). Samples were taken 10 times from three different landscape positions at the KS site (Fig. 2A), 5, 12, and 14 m from the shoreline (KS5, KS12, and KS14, respectively), from October 2003 to October 2004. The KS5 and KS12 site positions were saturated year-round, while the KS14 site position was located on a hill slope and was rarely saturated. The Kingsbury Creek water level increased in April due to the melting of snow, and the KS5 and KS12 positions were under river water during this month.
FIG. 1.
Lake Superior watersheds used in this study and relation to Duluth, Minnesota. Sampling locations are marked (•). Soil and water samples were taken from KS, NW, and SC sites.
TABLE 1.
Chemical and physical properties of soils used in this study
| Sample name | Textural class | Soil component (%)a
|
OMb (%) | Available Nc (mg/kg) | Total P (mg/kg) | pH | CECd [cmol/kg] | ||
|---|---|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | |||||||
| KS5 | Organic soil | 56.6 | 7.5 | 1,484.5 | 4.9 | 103.2 | |||
| KS12 | Organic soil | 20.8 | 9.6 | 798.7 | 5.1 | 57.5 | |||
| KS14 | Loamy sand | 85.9 | 8.3 | 5.8 | 4.1 | 20.2 | 558.8 | 5.8 | 10.4 |
| NW0 | Sandy loam | 73.8 | 14.1 | 12.1 | 1.3 | 8.5 | 318.2 | 7.7 | 10.7 |
| NW5 | Sand | 87.2 | 5.8 | 7.0 | 0.7 | 5.2 | 256.4 | 7.7 | 19.4 |
| NW7.5 | Sand | 90.0 | 4.4 | 5.6 | 0.5 | 10.7 | 215.8 | 8.0 | 8.1 |
| SC0 | Sandy loam | 79.7 | 7.7 | 12.6 | 2.9 | 3.7 | 605.0 | 7.5 | 25.4 |
| SC2 | Sandy loam | 74.0 | 10.5 | 15.5 | 5.2 | 5.2 | 614.8 | 8.3 | 25.4 |
Soil component determined by hydrometer method.
Organic matter content determined by loss of ignition method.
Summation of NH4-N and NO3-N measured on moist soil samples and calculated for oven-dry soil basis.
Cation exchange capacity (unit of charge per weight of soil) was calculated by summation of exchangeable cations (K, Ca, Mg, Na, and H).
FIG. 2.
Topography of sampling sites used. (A) KS; (B) NW; (C) SC.
The NW site is adjacent to the intersection between the Nemadji River and Weinstein Avenue (46°38′01′′N, 92°05′39′′W) in Superior, WI (Fig. 1). This site has a sandy soil that is low in organic matter (Table 1). Three different landscape positions were sampled at the NW site (nine times), at the waterline (NW0), 5 m upshore of the waterline on an eroded bank (NW5), and 7.5 m upshore of the waterline (NW7.5), from October 2003 to October 2004 (Fig. 2B). The water level of the Nemadji River increased from April to July, mainly due to melting snow. The NW0 and NW5 positions were under river water in April. A sediment clay layer was exposed at the NW0 position when the water level was low and may have contributed to bacterial retention due to a large, negatively charged surface area.
The SC site is adjacent to the intersection between the St. Louis River, whose estuary is part of Duluth-Superior Harbor, and Clyde Avenue (46°42′04"N, 92°12′26"W) in Duluth, MN (Fig. 1). Part of the City of Duluth's wastewater treatment facility was located near the SC site more than 20 years ago (personal communication, Joe Stepun, Western Lake Superior Sanitary District). Two different landscape positions (Fig. 2C) were used for sampling at this site: the waterline (SC0) and 2 m upshore of the waterline (SC2). Ten samples were taken from this site from October 2003 through October 2004. The water level of the St. Louis River increased from April to September, and the SC0 location during this period was under river water. Due to the unexpected construction of a water drainage system in this area, the sampling positions were moved 3 m south of the initial site after the first sampling time (2 October 2003).
In order to exclude external E. coli inputs to soils, from animals or water, protection boxes (soil exclosures) were used to cover soils at the KS14 site on 16 October 2004. Exclosures were constructed from 121-liter (42-cm-diameter) plastic garbage containers (Rubbermaid, Fairlawn, Ohio) cut to a final height of 30 cm. Four mesh-covered windows were cut into the sides of the exclosures to facilitate air transfer. The open ends of exclosures were buried 10 cm into the soil, and samples were taken at a 10-cm depth from the protected soils on 22 November 2004. Disturbance of the exclosures was not observed during this approximately 1-month period.
Isolation of E. coli strains.
Soil samples from the 0- to 10-cm depth were taken using ethanol-disinfected core tubes and stored in Whirl-Pak bags at 4°C until processed, usually within 24 h. Moist soil samples (10 to 20 g) were diluted with 95 ml of 0.1 M gelatin-ammonium phosphate extraction solution (22) in screw-cap 200-ml bottles and shaken for 30 min at 280 rpm on a wrist-action shaker. Aliquots (20 and 2 ml, separately) of the upper, soil-free phase were filtered, in duplicate, through a 0.45-μm membrane filter (Millipore, Billerica, MA). Membrane filters were placed onto the surface of mFC agar medium (Difco, Detroit, MI) and incubated at 35°C for 2 h, followed by 44.5°C for 22 h. A 10-ml aliquot of the upper soil-free phase was serially diluted in phosphate-buffered saline (pH 7) containing 0.01% gelatin (36). A 200-μl aliquot from each dilution was spread, in duplicate, onto mFC agar plates and incubated at 35°C for 2 h, followed by incubation at 44.5°C for 22 h. Incubations done at 35°C for 2 h were used to resuscitate weakened coliform bacteria (15). Colonies were counted, and results are reported as CFU per g oven-dried soil (39).
Water samples were taken in sterile Whirl-Pak bags, stored at 4°C until processed, usually within 6 h, and analyzed using standard methods as described previously (12). Two volumes of water, 100 ml and 10 ml, were separately filtered through 0.45-μm membrane filters, placed onto the surface of mFC agar, and incubated as described above. Bacterial counts are reported as CFU per 100 ml water. Water samples were taken in April, September, and October for the KS site and in August, September, and October for the NW and SC sites.
Well-isolated, blue colonies appearing on mFC agar were restreaked onto the same medium. After 24 h of incubation at 44.5°C, single colonies on mFC agar were spot inoculated onto MacConkey (Difco) and CHROMagar ECC (CHROMagar Microbiology, Paris, France) agar plates and incubated for 24 h at 37°C. Up to 24 colonies from each sampling site position were transferred to the new medium. Colonies that appeared pink to red on MacConkey agar and were either blue or white on CHROMagar ECC (1) were individually streaked onto plate count agar (Difco) and incubated at 37°C for 24 h. Cells from plate count agar plates were suspended in sterile 50% glycerol (vol/vol), transferred to cryovials and 96-well cell culture plates, and stored at −70°C until used for analyses.
Isolates were confirmed as E. coli by using a series of biochemical tests (14), including indole and methyl red tests, the inability to grow on citrate agar, and the presence of β-d-glucuronide activity using EC-MUG broth (Difco). Strains showing atypical responses to any of these tests were examined by using API20E strips (bioMérieux, Paris, France); only strains identified as E. coli by API20E and with good, very good, or excellent identification (ID) scores were used in subsequent studies. E. coli strain ATCC 25922 and Klebsiella pneumoniae ATCC 35657 served as positive and negative controls, respectively, for all tests.
HFERP DNA fingerprinting.
Horizontal, fluorophore-enhanced, repetitive PCR (HFERP) DNA fingerprinting was performed using the BOXA1R primer as described by Johnson et al. (20). Gel images were normalized and analyzed using Bionumerics (version 2.1) (Applied Maths, Kortrijk, Belgium) as previously described (20). The genetic relatedness of the isolated soil E. coli strains to each other and to those in a library of HFERP DNA fingerprints produced from E. coli obtained from fecal samples of local, wild animals was determined as previously described (19, 20). The Duluth library contained unique HFERP DNA fingerprints from 16, 64, 80, 38, and 148 E. coli isolates from geese, gulls, terns, beaver, and deer, respectively, obtained near Duluth, MN (19). In addition, we examined the relatedness of HFERP DNA fingerprints of the soil E. coli strains to those in a DNA fingerprint library consisting of 1,535 unique E. coli strains isolated from humans and 12 animal sources (dogs, cats, horses, deer, geese, ducks, chickens, turkeys, cows, pigs, goats, and sheep) obtained throughout Minnesota (20).
Dendrograms were constructed using the curve-based, Pearson's product-moment correlation coefficient and the unweighted pair group method with arithmetic means (UPGMA) clustering method (20). Multivariate analysis of variance (MANOVA) was performed to cluster E. coli strains from each source group (9, 14). ID bootstrap analysis (at P = 0.9), done using a Bionumerics script (http://www.applied-maths.com/bn/scripts/bnscripts.htm), was performed to identify the potential source(s) of the E. coli isolates from soil samples (9).
Spontaneously occurring antibiotic-resistant E. coli mutants.
E. coli strains obtained from soils at the KS, NW, and SC field sites were evaluated for resistance to nalidixic acid (Nal) and rifampin (Rif). Strains were grown on antibiotic 3 (A3) agar medium (Difco) containing 10, 20, 40, 60, or 80 μg Nal (Fluka, Milwaukee, WI) per ml or 5, 10, 20, 40, 60, or 80 μg Rif (Sigma-Aldrich, St. Louis, MO) per ml. Antibiotic concentrations were gradually increased from 5 μg to 80 μg per ml to obtain doubly marked, antibiotic-resistant strains that were resistant to 30 μg Nal and 60 μg Rif per ml. Strain identity of Nalr Rifr mutants was verified using HFERP DNA fingerprinting studies, growth on laboratory media, and API20E (bioMérieux, Hazelwood, MO) and biochemical analyses. Sequence analysis of the 16S rRNA gene (9, 24) from each strain was also performed to verify that the spontaneously occurring antibiotic-resistant strains were E. coli.
Growth of E. coli strains in soils.
Soils from the KS14, NW5, and SC2 site positions were used for E. coli growth and persistence studies under laboratory conditions. Spontaneously occurring, Nalr and Rifr E. coli strains obtained from each site were grown in M9 minimal medium supplemented with 0.2% glucose (26) at 25°C until an optical density (A600) of 1.0 was reached (approximately 109 cells/ml). Cells were centrifuged at 10,000 × g for 5 min at 4°C, and the pellets were resuspended and washed, three times, in 0.85% NaCl. E. coli cell suspensions were diluted with 0.85% NaCl to obtain final cell concentrations of about 105 cells/ml.
A 1,800- to 2,000-g aliquot of KS14, NW5, and SC2 soil was inoculated with Nalr and Rifr E. coli strains originating from these soils to a final density of about 103 cells/g soil and mixed well. The concentration of E. coli cells used approximated densities found in the tested soils throughout the growing season. Fifteen-gram aliquots of each soil were divided into sterile Whirl-Pak bags and incubated at five different temperatures: 4, 15, 25, 30, and 37°C. Three replicates were used for each soil and temperature condition. The water content of each soil was kept at the field conditions found at each site: 17.2, 9.3, and 76.3% moisture for the KS14, NW5, and SC2 soils, respectively.
E. coli isolates from each soil were extracted and enumerated 0, 1, 2, 3, 4, 8, 16, and 32 days after inoculation using A3 agar medium supplemented with Nal (20 μg/ml) and Rif (40 μg/ml) as described above. The A3 medium containing both antibiotics was incubated at 37°C for 24 h. The identity of 20 randomly selected E. coli colonies recovered from soils 0, 2, and 32 days after inoculation was ascertained by HFERP DNA fingerprinting, growth on laboratory media, and the biochemical reactions described above. Uninoculated soils served as negative controls. Experiments were also conducted with the same strains and soil combinations to examine the growth response of E. coli to shifts in temperature. E. coli strains KS-7NR, NW-13NR, and SC-20NR were inoculated into the three soils and incubated at 15°C for the first 4 days, and the temperature was then increased to 37°C. The population densities of E. coli in soils were monitored daily, by plate count analysis, until 8 days after inoculation.
To determine if the growth evidenced in soils was due to inoculant carryover, washed antibiotic-resistant E. coli soil inoculant strains were added to M9 minimal medium without C and N sources to an initial optical density of 0.05 at 600 nm, and cultures were incubated at 4, 15, 25, 30, or 37°C. Cell growth was monitored spectrophotometrically at 600 nm 0, 1, 2, 3, 4, 8, 16, and 32 days after inoculation.
Mean CFU and standard errors of the means were calculated from replicate cell counts on agar media. The ANOVA subroutine of R project software (version 2.0.1) (http://www.r-project.org/), at α = 0.05, was used to assess the statistical significance of increases or decreases in cell numbers within soils over time.
RESULTS
Isolation of E. coli from soils and water.
A total of 759 E. coli strains were isolated from all sites from 2 October 2003 to 16 October 2004 (Fig. 1). Among these, 586 strains were isolated from soil (252, 157, and 177 from the KS, NW, and SC sites, respectively) and 173 strains were isolated from water (63, 59, and 51 from the KS, NW, and SC sites, respectively). All isolates were confirmed as E. coli by growth and reaction on selective and differential growth media, biochemical tests, and API20E test strips.
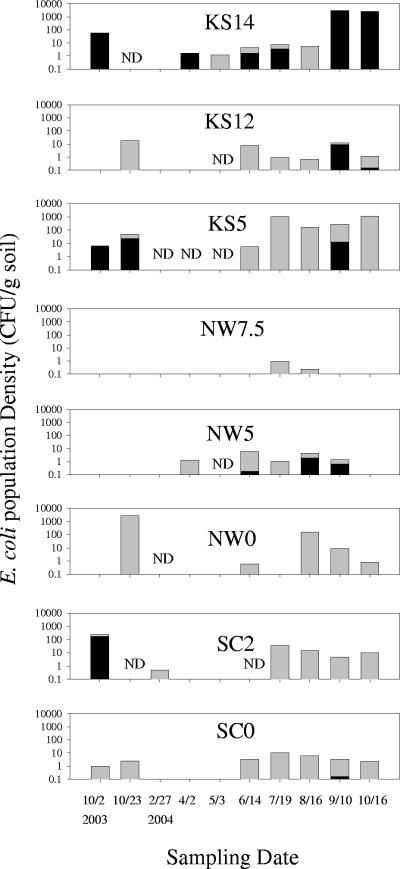
Great variation was observed in the population density of soilborne E. coli at all three sites over the sampling period. In general, E. coli counts as measured as CFU on selective medium were greater in the summer and fall (June to October) and lower during winter and spring (February to May) (Fig. 3). E. coli isolates from the KS14 soil having identical HFERP DNA fingerprints, at a similarity value of greater than 92%, appeared in October 2003 and then again in April 2004 (Fig. 3), indicating that some soilborne E. coli strains can survive over the winter months and grow during the summer months.
FIG. 3.
Seasonal shifts in the population density of E. coli at each sample site. Black bars, naturalized E. coli strains; gray bars, other E. coli strains. ND, not detected.
The greatest numbers of E. coli bacteria were recovered from the KS14 and NW0 soils, with a maximum density of 3 × 103 CFU/g soil. Overall, the KS5 organic soil had the greatest number of E. coli bacteria over all sampling dates, ranging from 5 to 1,150 CFU/g soil. The greatest number of E. coli bacteria recovered from the KS12, NW0, and SC2 sites occurred during October 2003, while the least number of E. coli bacteria (about 1 CFU/g soil) was found at the NW7.5 site, but only during the July-August sampling dates. These differences may, in part, be related to nutrient availability in soils (Table 1). The KS5 soil had the highest organic matter and total P contents of all soils tested, while NW7.5 soil was low in organic matter and P content (Table 1).
Genetic relatedness of E. coli in Minnesota soils.
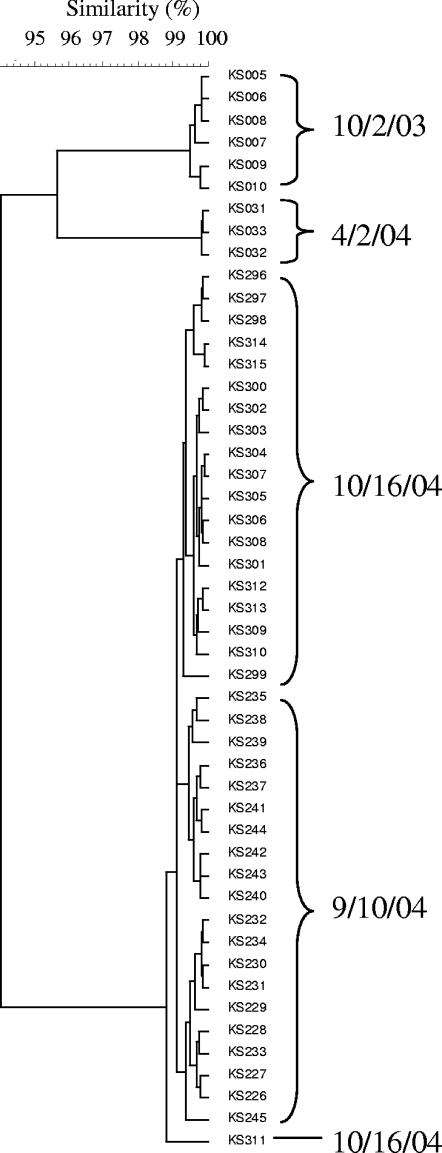
HFERP DNA fingerprint analyses of E. coli strains isolated from soils indicated that the E. coli isolates from the KS, NW, and SC soils were relatively genetically diverse with respect to soil, landscape position, and sampling date (data not shown due to number and excessive length of dendrograms). Relative similarity values of E. coli isolates recovered from the KS, NW, and SC soils ranged from 5 to 100%, 18 to 100%, and 10 to 100%, respectively. However, some of the E. coli strains recovered from each soil were nearly genetically identical with similarity values of 92 to 100%. Johnson et al. (20) reported that repeated HFERP DNA fingerprint analysis of a single E. coli strain produced an average similarity of 92%. Consequently, in this study, isolates having genetic similarity values of ≥92% were defined as being the same strain. According to this scheme, there were 32, 84, and 49 unique E. coli strains recovered from the KS, NW, and SC sites, respectively. Moreover, 89, 46, and 85% of the soil isolates from the KS, NW, and SC sites, respectively, were genetically related at >95% similarity levels and can thus be viewed as the same genotype (Fig. 4).
FIG. 4.
Partial dendrogram of naturalized E. coli strains from the KS site from October 2003 to October 2004. The dendrogram was generated from HFERP DNA fingerprints using Pearson's product-moment correlation coefficient and the UPGMA clustering method.
The DNA fingerprints of E. coli recovered from soils were compared with those in the Duluth fingerprint library by using ID bootstrap analyses at a P value of >0.90. Results of this analysis indicated that 7 and 12% of the E. coli isolates from the KS site most likely originated from beaver and terns, respectively. Likewise, 5, 2, 5, and 1% of the E. coli isolates from the NW site presumably originated from beaver, deer, geese, and terns, respectively, and 2, 2, and 11% of E. coli isolates at the SC site most likely originated from deer, terns, and beaver, respectively. The potential sources of the remainder of the recovered soil E. coli isolates (81, 87, and 85% of the KS, NW, and SC isolates, respectively) could not be determined using this fingerprint library at the probability level adopted or using the more comprehensive Minnesota fingerprint library containing over 1,500 unique E. coli fingerprints. This indicated that the majority of soilborne E. coli isolates may be unique to soil and were not present in the two DNA fingerprint libraries examined.
Naturalized E. coli in Minnesota soils.
Further examination of the dendrograms indicated that some of the same E. coli strains with similarity values of ≥92% appeared over time at each site, suggesting that they were part of the autochthonous soil bacterial community (Fig. 4). For example, the same E. coli strain was repeatedly isolated from the KS14 site from October 2003 to October 2004. Similar results were obtained from the NW and SC sites (data not shown). These strains represented 28, 17, and 14% of all E. coli strains isolated from the KS, NW, and SC soils, respectively, and their sources could not be determined by ID bootstrap analysis using the Duluth or Minnesota fingerprint libraries. We propose to use the term naturalized E. coli to describe these isolates. In this study naturalized E. coli isolates were defined by the following criteria: (i) they had unique HFERP DNA fingerprints that were not similar to E. coli strains from known source DNA fingerprint libraries at a P value of 0.9, (ii) their HFERP DNA fingerprints clustered together on the dendrogram at a similarity value of ≥92%, and (iii) the same E. coli strain could be repeatedly isolated from each site over time.
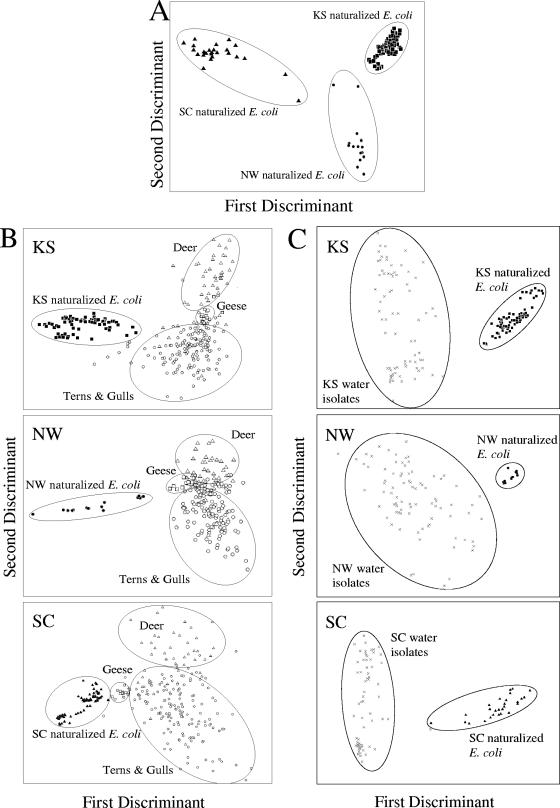
The relatedness of the naturalized E. coli strains recovered from each site was examined using MANOVA. Results presented in Fig. 5A show that the strains from each site clustered together, indicating that different populations became independently naturalized at each site. The first and second discriminants accounted for 100% of the variation, indicating that the strains are tightly clustered by site.
FIG. 5.
MANOVA of HFERP DNA fingerprints from E. coli strains. The first two discriminants are represented by the distances along the x and y axes. (A) MANOVA of HFERP DNA fingerprints from naturalized E. coli strains from the KS (▪), NW (•), and SC (▴) sites; (B) MANOVA of HFERP DNA fingerprints obtained from naturalized E. coli strains from the KS (▪), NW (•), and SC (▴) sites and E. coli isolated from feces of geese (□), terns and gulls (○), and deer (▵); (C) MANOVA of HFERP DNA fingerprints of naturalized E. coli obtained from soils at the KS (▪), NW (•), and SC (▴) sites and E. coli isolated from water at the same sites (×).
The genetic relatedness of the naturalized E. coli strains from the KS, NW, and SC sites to E. coli strains in the Duluth HFERP DNA fingerprint library was further explored by using MANOVA (Fig. 5B). Naturalized E. coli strains from each sampling site were distinctly clustered and could be readily separated from E. coli strains present in the fingerprint library. The first and second discriminants of the MANOVAs of E. coli from the KS, NW, and SC sites accounted for 84, 79, and 86% of the variation, respectively, indicating that the naturalized strains at each site were distinct and easily distinguishable from strains obtained from local animal feces.
The HFERP DNA fingerprints of naturalized soilborne E. coli strains were also compared to E. coli strains isolated from water at the KS, NW, and SC sites. MANOVAs indicated that the naturalized E. coli strains from each soil had different DNA fingerprints from those in water (Fig. 5C). The first and second discriminants of the MANOVAs of E. coli from the KS, NW, and SC sites accounted for 89, 84, and 97% of the variation, respectively, indicating that the naturalized strains at each site were easily differentiated from E. coli strains obtained from water. Moreover, cluster analyses indicated that the E. coli strains obtained from water samples were more genetically diverse within a site (similarity values ranging from 15 to 99%) than the naturalized E. coli obtained from soils (data not shown). However, within-site diversity of the naturalized soil isolates was relatively low; strains were 92 to 100% similar. These results are consistent with our initial hypothesis that the soilborne E. coli isolates became naturalized to the sites and were not simply reinoculated from water into soils.
Further evidence for the naturalization of E. coli strains comes from studies done with exclosure boxes that protected soils from additional E. coli inputs due to the deposition of fecal materials from animals or from rain-derived runoff. For example, all E. coli strains recovered from exclosure-protected KS14 soil had HFERP DNA fingerprints that were >95% similar to those of naturalized E. coli strains previously isolated from this site. These data suggest that the soilborne, naturalized strains were adapted to this soil and were not continuously added from external sources.
Growth of E. coli in nonsterile soils.
Since the same strains could be repeatedly isolated from the same soil, incubation studies were done to examine the persistence and growth of naturalized E. coli strains in natural, nonsterile soils. Antibiotic-resistant E. coli strains KS-7NR, NW-13NR, and SC-20NR were developed from naturalized E. coli strains obtained from the KS, NW, and SC sites, respectively. All of the spontaneously occurring, antibiotic-resistant strains were identified as E. coli by using API20E test strips, by growth on selective and differential media, and by biochemical reactions. Sequence analysis of the nearly complete 16S rRNA gene confirmed that these strains were bona fide E. coli, with greater than 99.5% nucleotide sequence identity with the E. coli K-12 reference strain MG1655 (accession number U00096). Moreover, HFERP DNA fingerprint analyses indicated that the spontaneously occurring, antibiotic-resistant strains were >98% similar to their respective parent strains.
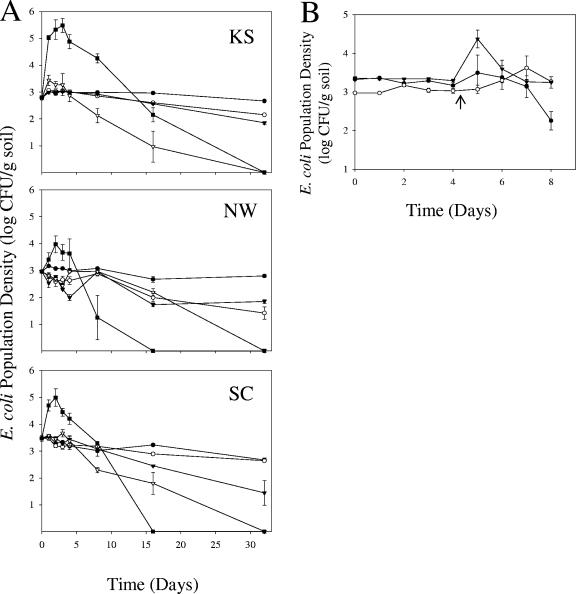
Nonsterile soils from these sites were inoculated with antibiotic-resistant strains originating from the same location at that site to an initial cell density of about 103 CFU/g soil. Representative colonies recovered at 0, 2, and 32 days after inoculation were identified as the inoculant E. coli strains using growth, biochemical, and HFERP DNA fingerprint analyses. Population densities of all strains significantly increased (α = 0.05) in the KS, NW, and SC soils when incubated at 37°C (Fig. 6A). Cell densities increased to a maximum of 4.2 × 105, 1.8 × 104, and 1.7 × 105 CFU/g soil in the KS, NW, and SC soils, respectively, 2 to 3 days after inoculation. This represents a 20- to 655-fold increase relative to initial cell densities added to soils. Thereafter, however, cell densities decreased to a level lower than the initial inoculation amount, and to less than the detection limit (approximately 1 CFU/g soil), by 32 days after inoculation. No colonies appeared on A3 agar medium supplemented with antibiotics from any of the uninoculated, negative-control soils. Moreover, cell densities (measured at 600 nm) did not increase when the KS-7NR, NW-13NR, and SC-20NR strains were inoculated into M9 medium, without C and N sources, and incubated at 37°C. Taken together, these results indicate that the growth of E. coli observed in these soils was not due to nutrient carryover from the inocula or detection of noninoculated E. coli or other bacteria from soils. These results support the contention that at least some resident E. coli strains can utilize nutrients in each soil to replicate at 37°C.
FIG. 6.
Influence of temperature on survival and growth of naturalized E. coli in Minnesota soils. Nalidixic acid- and rifampin-resistant naturalized E. coli strains KS-7NR, NW-13NR, and SC-20NR were inoculated into KS, NW, and SC soils, respectively. Values presented are means of CFU ± standard errors on A3 agar medium. (A) Soils were incubated at constant temperatures of 4°C (•), 15°C (○), 25°C (▾), 30°C (▿), or 37°C (▪); (B) E. coli strains KS-7NR (•), NW-13NR (○), and SC-20NR (▾) were inoculated into their respective soils of isolation and incubated at 15°C for 4 days and then shifted (↑) to 37°C for an additional 4 days.
Only limited, or no, E. coli growth was seen at the other temperatures examined, and cell densities decreased to a level lower than the initial inoculation level, or the detection limit, by 32 days after inoculation. At 30°C, strain KS-7NR reached a maximum cell density of 3.4 × 103 CFU/g soil in the KS soil 1 day after inoculation, a 5.6-fold increase. While E. coli strains incubated at 4, 15, and 25°C did not show an increase in cell abundance, they were able to survive longer at these temperatures than when incubated at 30°C or 37°C (Fig. 6A). Repeated experiments done in all three soils showed a similar trend: E. coli cell density increased at 37°C after 2 days of incubation, and cells survived up to 32 days of incubation at 15°C (data not shown).
Results in Fig. 6B show that there was no growth response when E. coli strains were incubated at 15°C. However, strain SC-20NR showed a rapid increase in growth after the soil temperature was shifted to 37°C. Similarly, strain NW-13NR showed a significant increase in growth following the temperature shift, but the magnitude (1-log increase) and rate of growth were not as great as those seen with strain SC-20NR. Similar to what was found in the constant-temperature experiments, the population densities of all strains decreased to values less than or equal to the inoculation level 1 to 2 days after the temperature shift. In a separate experiment, strain NW-13NR was inoculated to 9 × 102 CFU/g soil into NW15 soil and incubated at 15°C for 16 days and then shifted to 37°C for 8 days. The cell density of this strain gradually declined to 1.1 × 102 CFU/g soil 16 days after inoculation and then increased, 10-fold, to 1.04 × 103 CFU/g soil 4 days after the temperature increased to 37°C. However, cell numbers of NW-13NR decreased to 7.7 CFU/g soil 8 days after the temperature shift (data not shown).
DISCUSSION
In the current study, we investigated whether E. coli could grow and persist in temperate riverine soils of northern Minnesota, which are subjected to temperature and moisture extremes. While there was great seasonal and site variation in E. coli population densities in soils examined, E. coli could be routinely isolated from these soils. Seasonal and site variation in E. coli counts in these temperate soils is most likely related to temperature and moisture changes occurring in this region and the patch distribution of E. coli within the sampling plots (9). The mean high temperature in Duluth, Minnesota, was 25°C for 2003 to 2004 and occurred during July. The highest daily air temperature during this period near the test sites was 32.2°C. In contrast, the mean low temperature was −20°C in January. Results from our studies also indicated that the same strain (genotype) of E. coli survived over the winter months, through several freeze-thaw cycles, and subsequently grew during summer months. While Byappanahalli et al. (9) recently reported that the population dynamics of E. coli strains from temperate Indiana coastal forest soils were similar to those reported here, extremes in temperature were not as pronounced at the sites they examined. In addition to temperature, variation in the population dynamics of E. coli found in the soils used in this study might also be partially due to nutrient availability. Tate (33) observed that E. coli populations were three times greater in an organic soil than in a sandy soil, suggesting that nutrients in the organic matter support growth.
HFERP DNA fingerprint analyses indicated that identical E. coli strains (similarity values of ≥92%) could be isolated from the same sites over multiple seasons, suggesting that these bacteria can persist in soil over the long term. Further evidence for the autochthonous nature of these E. coli strains was provided by DNA fingerprint analyses showing that unique dominant strains were isolated from particular soils and locations. HFERP DNA fingerprint analyses also showed that these strains were different from E. coli strains obtained from wildlife commonly found in the studied habitats or river water, indicating that these strains were most likely not directly deposited or replaced by E. coli from animal feces or water, similar to what Byappanahalli et al. (9) observed in Indiana soil. In this study, we referred to these bacteria as being naturalized. Furthermore, E. coli strains recovered from the exclosure-protected KS14 soil in November had HFERP DNA fingerprints identical to those of naturalized strains recovered from the KS soil during the previous year. Consequently, the Minnesota soils we examined appear to be a recurrent reservoir for E. coli, whose presence overtime is not likely solely due to the inoculation of new genotypes into soils from local or transient point and nonpoint sources.
Our results also support the contention that naturalized E. coli strains were mobile in soils. For example, a unique, naturalized E. coli population was present at the near-shore KS5 and KS12 sites only when this strain was abundant at the upslope KS14. This observation suggests that the naturalized population may have moved downslope from KS14 towards the river by erosion or runoff events. These results are consistent with those reported by Solo-Gabriele et al. (31), who showed that soil- and sediment-borne E. coli can move due to tidal events.
The soil incubation studies presented here show that naturalized E. coli strains have the ability to grow in nonamended, nonsterile, Minnesota soils. While several previous reports have suggested that E. coli has the potential to grow in soils, these studies were done using gamma-irradiated sterile soil (8), addition of bile salts to inhibit nonfecal coliform bacteria, or addition of small amounts of carbon- and nitrogen-containing compounds as nutrients (4, 7, 8, 35). However, growth in soils was significantly impacted by temperature and occurred only if soil temperatures were at or above 30°C. Moreover, our laboratory studies clearly showed that E. coli grew to highest cell densities at 37°C. Thus, maximum cell growth was most likely not realized at the sites examined, since it is doubtful that soils in Minnesota reach 37°C at any time during the year. However, the highest daily air temperature was 32.2°C in July 2004, most likely providing enough heat in the upper surface of soils to promote moderate E. coli growth.
We speculate that growth of E. coli strains in minimal medium before introduction into soils helped E. coli adjust to the nutrient-limiting conditions found in these soils. This was demonstrated in preliminary soil incubation experiments, where inocula from naturalized E. coli strains prepared in nutrient-rich medium (A3 medium) failed to grow in soils (data not shown). This failure may be one reason why others have previously demonstrated growth of E. coli only in nutrient- and manure-amended soils (4, 7, 8, 35). Moreover, growth and survival of E. coli in water, sediment, and soils have been reported to be strain dependent (2, 17, 35), and the naturalized E. coli strains that we studied here may have a selective advantage to grow in soils.
E. coli cell densities decreased after rapid cell growth at both 30 and 37°C, suggesting that E. coli exhausted the bioavailable nutrients or predation controlled the final E. coli population size. While Topp et al. (35) also observed a decline of E. coli in soils after a rapid cell increase, Berry and Miller (4) reported that E. coli O157:H7 maintained a stable population density after growth in soil. The differences in the observed growth rates of E. coli from the KS, NW, and SC soils may also be due to the nutrient content and availability in the three soils examined. E. coli grew fastest in the KS soil, which had a relatively large amount of organic matter, and more slowly in soils having less organic matter.
While E. coli isolates did not show an increase in cell density in soils at 25°C, they survived longer at this temperature than if incubated above 30°C. These results are similar to those reported by Berry et al. (3), Terzieva and McFeters (34), and Topp et al. (35), who demonstrated that the survival of E. coli in soils was related to soil incubation temperatures. However, Berry and Miller (4) showed the growth of E. coli at lower temperature, around 19°C, in manure-rich soils. In our studies, E. coli incubated in soils at 15°C for 4 days or longer showed growth if the soil temperature was raised to 37°C. These findings suggested that the soilborne E. coli isolates can persist in colder soils and are poised for growth if favorable conditions occur. Jones et al. (21) reported that E. coli cells adopted an elongated morphology when incubated in liquid culture at 4°C, and these cells were ready to replicate when the temperature was raised above 6°C. Consequently, the presence of naturalized E. coli in northern temperate soils, even if frozen for part of the year, may confound the use of E. coli as a reliable indicator of fecal contamination.
Acknowledgments
We thank Chang-Hun Lee for help with soil incubation studies, John Ferguson for help in sampling, Wendy Hieb for donation of the known-source Duluth HFERP DNA fingerprint library, and Nir Shapir and Murulee Byappanahalli for reviewing the manuscript.
This study was supported, in part, by grants from the Minnesota Sea Grant program (to R.E.H. and M.J.S.) and from the University of Minnesota Agricultural Experiment Station (to M.J.S.).
REFERENCES
- 1.Alonso, J. L., A. Soriano, O. Carbajo, I. Amoros, and H. Garelick. 1999. Comparison and recovery of Escherichia coli and thermotolerant coliforms in water with a chromogenic medium incubated at 41 and 44.5°C. Appl. Environ. Microbiol. 65:3746-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, C., B. J. Lloyd, and J. S. Colbourne. 1991. Effect of heat shock on recovery of Escherichia coli from drinking water. Water Sci. Technol. 24:85-88. [Google Scholar]
- 4.Berry, E. D., and D. N. Miller. 2005. Cattle feedlot soil moisture and manure content: II. Impact on Escherichia coli O157. J. Environ. Qual. 34:656-663. [DOI] [PubMed] [Google Scholar]
- 5.Bogosian, G., L. E. Sammons, P. J. Morris, J. P. O'Neil, M. A. Heitkamp, and D. B. Weber. 1996. Death of the Escherichia coli K-12 strain W3110 in soil and water. Appl. Environ. Microbiol. 62:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brettar, I., and M. G. Höfle. 1992. Influence of ecosystematic factors on survival of Escherichia coli after large-scale release into lake water mesocosms. Appl. Environ. Microbiol. 58:2201-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byappanahalli, M., and R. Fujioka. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50:27-32. [PubMed] [Google Scholar]
- 8.Byappanahalli, M. N., and R. S. Fujioka. 1998. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 38:171-174. [Google Scholar]
- 9.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8 [Online.] doi: 10.1111/j.i4622920.2005.00916.x. [DOI] [PubMed]
- 10.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao, W. L., and R. L. Feng. 1990. Survival of genetically engineered Escherichia coli in natural soil and river water. J. Appl. Bacteriol. 68:319-325. [DOI] [PubMed] [Google Scholar]
- 12.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 13.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour, A. P., E. R. Strickland, and V. J. Cabelli. 1981. Membrane filter method for enumerating Escherichia coli. Appl. Environ. Microbiol. 41:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujioka, R. S. 2001. Monitoring coastal marine waters for spore-forming bacteria of faecal and soil origin to determine point from non-point source pollution. Water Sci. Technol. 44:181-188. [PubMed] [Google Scholar]
- 17.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 18.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hieb, W. S. 2005. Identifying the sources of fecal coliform bacteria in Lake Superior watersheds. M.S. thesis. University of Minnesota, Duluth.
- 20.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T., C. O. Gill, and L. M. McMullen. 2004. The behavior of log phase Escherichia coli at temperatures that fluctuate about the minimum for growth. Lett. Appl. Microbiol. 39:296-300. [DOI] [PubMed] [Google Scholar]
- 22.Kingsley, M. T., and B. B. Bohlool. 1981. Release of Rhizobium spp. from tropical soils and recovery for immunofluorescence enumeration. Appl. Environ. Microbiol. 42:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 25.Leclerc, H., D. A. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ram, J. L., R. P. Ritchie, J. Fang, F. S. Gonzales, and J. P. Selegean. 2004. Sequence-based source tracking of Escherichia coli based on genetic diversity of β-glucuronidase. J. Environ. Qual. 33:1024-1032. [DOI] [PubMed] [Google Scholar]
- 29.Savageau, M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732-744. [Google Scholar]
- 30.Sherer, B. M., J. R. Miner, J. A. Moore, and J. C. Buckhouse. 1992. Indicator bacterial survival in stream sediment. J. Environ. Qual. 21:591-595. [Google Scholar]
- 31.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tassoula, E. A. 1997. Growth possibilities of E. coli in natural waters. Int. J. Environ. Stud. 52:67-73. [Google Scholar]
- 33.Tate, R. L., III. 1978. Cultural and environmental factors affecting the longevity of Escherichia coli in histosols. Appl. Environ. Microbiol. 35:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terzieva, S. I., and G. A. McFeters. 1991. Survival and injury of Escherichia coli, Campylobacter jejuni, and Yersinia enterocolitica in stream water. Can. J. Microbiol. 37:785-790. [DOI] [PubMed] [Google Scholar]
- 35.Topp, E., M. Welsh, Y.-C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microb. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 36.Turco, R. F. 1994. Coliform bacteria, p. 145-158. In R. W. Weaver, J. S. Angle, and P. J. Bottomley (ed.), Methods of soil analysis. Soil Science Society of America, Madison, Wis.
- 37.United States Environmental Protection Agency. 1986. Ambient water quality criteria for bacteria—1986. United States Environmental Protection Agency, Washington, D.C.
- 38.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuberer, D. 1994. Recovery and enumeration of viable bacteria, p. 119-144. In R. W. Weaver, J. S. Angle, and P. J. Bottomley (ed.), Methods of soil analysis. Soil Science Society of America, Madison, Wis.