Abstract
Mycobacterium avium is widely distributed in the environment, and it is chiefly found in water and soil. M. avium, as well as Mycobacterium smegmatis, has been recognized to produce a biofilm or biofilm-like structure. We screened an M. avium green fluorescent protein (GFP) promoter library in M. smegmatis for genes involved in biofilm formation on polyvinyl chloride (PVC) plates. Clones associated with increased GFP expression ≥2.0-fold over the baseline were sequenced. Seventeen genes, most encoding proteins of the tricarboxylic acid (TCA) cycle and GDP-mannose and fatty acid biosynthesis, were identified. Their regulation in M. avium was confirmed by examining the expression of a set of genes by real-time PCR after incubation on PVC plates. In addition, screening of 2,000 clones of a transposon mutant bank constructed using M. avium strain A5, a mycobacterial strain with the ability to produce large amounts of biofilm, revealed four mutants with an impaired ability to form biofilm. Genes interrupted by transposons were homologues of M. tuberculosis 6-oxodehydrogenase (sucA), enzymes of the TCA cycle, protein synthetase (pstB), enzymes of glycopeptidolipid (GPL) synthesis, and Rv1565c (a hypothetical membrane protein). In conclusion, it appears that GPL biosynthesis, including the GDP-mannose biosynthesis pathway, is the most important pathway involved in the production of M. avium biofilm.
Mycobacterium avium complex is widely distributed in the environment, such as in water and soil, and is a chief component of many natural aquatic biofilms (8). M. avium is also known to cause chronic pulmonary infection in patients with predisposing lung disease, such as previous tuberculosis and chronic obstructive pulmonary disease (28). Urban water systems contain organisms of the M. avium complex in biofilm or a biofilm-like structure, and individuals can potentially be exposed to the bacterium, either by inhalation of aerosol particles or ingestion of contaminated water. Studies have established an association between M. avium in urban water and the development of disseminated disease in individuals with AIDS (36).
Mycobacterium smegmatis, as well as M. avium, has been shown to produce a biofilm or a biofilm-like structure (6, 19). The outermost layers of the M. smegmatis and M. avium cell walls contain glycopeptidolipid (GPL), whereas the outermost layer of M. tuberculosis is made of phenolic glycolipids, dimycocerosate, and lipo-oligosaccharides (24). Recent studies suggest that the M. smegmatis biofilm is associated with a GPL present on the cell wall, and indirect evidence indicates a similar role in M. avium (6). Aspects of biofilm formation have begun to be examined with M. smegmatis. Transposon inactivation of the GPL gene clusters in M. smegmatis decreased the production of biofilm, and the deletion of the genes tmtp and mps revealed their involvement in biofilm formation upon seeding of the bacterium on polyvinyl chloride (PVC) plates (19, 26). The tmtp gene is highly conserved between M. smegmatis and M. avium, with both organisms having genes encoding one small (tmtpA) and two large (tmtpB and tmtpC) putative transmembrane transport proteins in the same operon. The proposed function involves the transport of the precursor of GPL from the inner membrane. The mps genes are identified as pstA, -B, and -C, constituting the GPL gene clusters in M. avium (GenBank accession no. AF143772). The peptide synthetase (mps, Mps protein) has a role in the initial step of GPL synthesis, i.e., in the assembly of the lipopeptide core and acceptor of acyl-Phe, which is modified by sequential addition of threonine, alanine, and alaninol (4). This lipopeptide core may subsequently be glycosylated with rhamnose and 6-deoxytalose, resulting in the nonspecific GPL (nsGPL). The acetyltransferase (atf1) acetylates on 6-deoxytalose in the cell wall, and the putative tmtpC (Tmtpc) protein transports it to the outermost layer of the cell wall (4, 19, 26). However, the roles of GPLs in biofilm formation are still not well defined.
The genetic determinant of biofilm formation in M. avium has not been clearly identified. It was reported that M. avium A5 produced increased amounts of biofilm compared with the M. avium 101, M. avium 104, and M. avium 109 strains (6). Furthermore, M. avium strains produced more biofilm when inoculated in water than in 7H9 broth on a PVC surface. During biofilm formation, microorganisms rarely come into contact with a clean surface and normally colonize a surface that has been modified following the absorption of molecules from the environment, such as water and proteins, etc. The M. avium 101 and 104 strains belong to serotype 1, while M. avium A5 and strain 109 belong to serotype 4. Krzywinska and Schorey (17) described the genomic differences, especially in GPL gene clusters, between M. avium 104 (the strain from which the genome sequence is available) and M. avium A5. Those authors identified that the GPL was highly conserved upstream of the GPL clusters methyl transferase B (mtf B), glycosyl transferase A (gtf A), rhamnosyl transferase A (rtf A), mtf C, mtf D, and dehydrogenase A (dhg A). But downstream of these gene clusters, the GPL is quite different between M. avium 104 and M. avium A5. In addition, it was shown that M. avium A5 has the GDP-d-mannose dehydratase mdht gene (GenBank accession no. AAD20373) and the GDP-6-deoxy-4-keto-d-mannose 3-5-epimerase-4-reductase mer gene (AAD20374) (17). These two enzymes catalyze the transformation of GDP-mannose to GDP-fucose (34). Since GDP-fucose is an important substrate for the GPL, especially for serovar-specific GPL, it is supposed to reflect the different means of biofilm formation between the M. avium 104 and M. avium A5 strains.
In this study, we used two strategies, the screening of a transposon mutant library and of a green fluorescent protein (GFP) promoter library to identify M. avium genes associated with biofilm formation on a solid surface that is encountered in water pipes.
MATERIALS AND METHODS
Mycobacterial strains and plasmids.
M. avium A5 (2) and M. avium 104 are virulent strains, isolated from blood of patients with AIDS. Mycobacterium smegmatis mc2155 was kindly provided by William Jacobs, Jr. (Albert Einstein School of Medicine) (32). They were cultured in either Middlebrook 7H11 agar or 7H9 broth containing 10% oleic acid, albumin, dextrose, and catalase, while M. smegmatis and M. avium mutant clones were plated onto 7H11 Middlebrook agar containing 50 μg/ml or 400 μg/ml of kanamycin, respectively.
Escherichia coli strain DH5α (Stratagene, La Jolla, CA) was used as the host for plasmid construction. GFP-mut2 genes were generated by PCR from the pKEN plasmid, obtained from Rafael Valdivia and Stanley Falkow, Stanford University. GFP-mut2 genes were cloned into the promoterless reporter plasmid pMV261 (9). Temperature-sensitive plasmid pTNGJC was constructed based on the pUC18 vector, as described previously (18).
Briefly, the transposon Tn5367 was cut from pYUB285 and cloned in the EcoRI and HindIII restriction sites, making the plasmid pUC18-Tn5367 (Table 1). The transposon contains the kanamycin-resistant gene as a cassette. The temperature-sensitive mycobacterial origin of replication was removed from pCG79 (provided by B. Gicquel, Institute Pasteur, Paris, France) and inserted in pUC19-Tn5367, using the HindIII restriction site, to create pTNGJC. Transformation with pTNGJC in M. avium A5 was carried out according to protocol described previously (12).
TABLE 1.
Plasmids and strains used in this study
Screening of the M. avium GFP promoter library.
The M. avium GFP promoter library was constructed in M. smegmatis as previously described (9) and stored in pools of five in 96-well plates containing Middlebrook 7H9 broth with 50% glycerol at −70°C. Bacterial pools containing 5 × 107 bacteria were suspended in 200 μl of 7H9 broth and placed into 96-well tissue culture plates (Corning Inc., NY) at 37°C for 3 days. Then 100-μl aliquots of bacteria from each well were transferred to 96-well PVC plates (Becton Dickinson Labware, Franklin Lakes, NJ). The PVC plates were kept at room temperature for 5 days and assayed for the expression of GFP daily (CytoFluor II; subsidiary of Millipore Corp, MA). We were interested in gene up-regulation that precedes the formation of a biofilm. The pools with a ratio of the level of GFP at day 5 to that of GFP at day 1 of over 2.0 were subsequently diluted and plated onto 7H11 agar with 50 μg/ml kanamycin (Km) to obtain isolated clones (9). The experiment was then repeated, with individual isolates, and wells containing clones associated with increased GFP expression greater than 3.0-fold over the baseline were sequenced at the Central Service Laboratory, Oregon State University, using the GFP primer 5′-TTGTGCCCATTAACATCACCA-3′. Database search and sequence comparisons were performed using the BLAST network service at the National Center for Biotechnology Information (NCBI).
Construction of the M. avium A5 transposon library.
M. avium A5 competent cells were washed with 10% glycerol three times at 4°C and 3,000 rpm and diluted in 1 ml 10% glycerol. Bacteria were submitted to electroporation using a Gene Pulser Xcell (Bio-Rad, CA) and plated on 7H11 agar with 400 μg/ml of kanamycin (9). A clone containing the plasmid pTNGJC-KAN was grown at 30°C for 3 weeks in 7H9 broth in the presence of 200 μg/ml of kanamycin. After the number of bacteria in suspension reached approximately 1 × 109 CFU, the culture was placed at 41°C for 3 days. Because pTNGJC-KAN contains a temperature-sensitive Myc origin of replication, the shift in temperature eliminated the plasmid, and all surviving kanamycin-resistant cells necessarily contained pTNGJC-KAN in the bacterial chromosome (18). The suspension was then diluted and plated onto 7H11 agar with kanamycin at 37°C. Colonies were harvested and screened by PCR for the presence of the kanamycin gene (Table 1). Primers for the Km gene were 5′-TGTTCAACAGGCCAGCCA-3′ (forward) and 5′-TAATGTCGGGCAATCAGGTG-3′ (reverse). Twenty colonies were selected and tested for the presence of the transposon-Km gene. All 20 contained the transposon.
Selection of clones deficient in biofilm formation.
Biofilm (or biofilm-like) formation was determined as previously described (6). Briefly, 1 × 108 bacteria in 200 μl of Hanks' buffered salt solution (HBSS) were seeded on PVC 96-well microplates. Plates were incubated at 37°C for up to 14 days. To measure the biofilm formation, the supernatant was removed gently from each well, and 25 μl of a 2% crystal violet solution was added to each well (the dye stains bacterial cells but not the PVC material). The plates were incubated at room temperature for 15 min and rinsed three times with HBSS. The crystal violet was dissolved in 95% ethanol, and biofilm formation was analyzed at 570 nm, as previously reported (6, 26). Two thousand mutants were screened. The experiments were repeated at least five times.
Identification of the transposon-inactivated gene.
The genes inactivated were identified by using a nonspecific, nested suppression PCR (34). The primer used was 5′-CCATCATCGGAAGACCTC-3′. PCR cycling was as follows: 35 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 4 min. Prior to the first cycle, a temperature of 94°C was held for 5 min, and at the end of the last cycle, a temperature of 72°C was maintained for 7 min. The primer used for the second PCR was 6 nucleotides (GACCCC) longer at the 3′ end. The PCR cycling was the same as the first PCR, except the annealing cycle was for 30 s at 56°C using Pfu DNA polymerase (Stratagene). The PCR products were run in 1% agarose gel, and each PCR band that appeared on the gel was cut and extracted using a gel extraction kit (QIAGEN). The PCR amplifications were cloned into the pCR2.0 TOPO vector (Invitrogen, Carlsbad, CA) and submitted for sequencing (18).
Complementation of the mutants.
The sequences of the isogenic mutants of 5G4 (MA1565c), 6H9 (sucA), and 9B5 (pcd) were obtained using the M. avium 104 genome BLAST network service of the NCBI. The primers for selected genes were 5G4 forward (5′-GAG AAT TCG CGG GTT TTC GGT AAA TTA GC-3′), 5G4 reverse (′-GTA AGC TTT TTC GAG GCG GCA GAG CCG AT-3′ [2,232 bp]), 6H9 forward (5′-GAG AAT TCA TGT ACC GCA AGT TCC GCG AC-3′), 6H9 reverse (5′-GTA AGC TTT CGG GCA GCT CCA GGC CGA AT-3′ [3,573 bp]), 9B5 forward (5′-GAG AAT TCG AGC ACG CGA TAA CCC AAG CA-3′), and 9B5 reverse 5′-GTA AGC TTA ATC GCG TCG TCC AGC CGG TC-3′ [1,091 bp]). The genes were amplified by PCR, and then the product was digested with both EcoRI and HindIII restriction enzymes. The genes were inserted into EcoRI and HindIII restriction sites of the pMV261-AprII plasmid. The plasmid was transformed into E. coli competent cells (DH5a-T1 chemical competent cell [Invitrogen]) for replication. The plasmid was then purified using a plasmid extraction kit (Stratagene). Plasmids containing the functional genes were electroporated into 5G4, 6H9, and 9B5 competent cells, as described above. The bacteria were then plated onto 7H11 agar containing 400 μg/ml apramycin (Apr). The PCR production was cloned into the pMV261-AprII plasmid containing the hsp60 promoter upstream of the genes and an Apr-resistant gene to create pMV261-5G4, pMV261-6H9, and pMV261-9B5. To confirm transformation, the Apr gene was identified by PCR application; the primers for the Apr gene were 5′-GCATCGCATTCTTCGCATCC-3′ (forward) and 5′-GGCCCACTTGGACTGATCGA-3′ (reverse).
RNA extraction and RT.
M. avium strains were grown in 7H9 broth with 10% oleic acid, albumin, dextrose, and catalase for 5 days (1 × 109/ml) and then pelleted at 3,000 rpm for 15 min at room temperature, resuspended in HBSS, and inoculated on PVC plates for 7 days. Bacteria grown in 7H9 broth at 37°C were used as the control. The bacterial pellets recovered from PVC plates were submitted to RNA extraction. Total RNA was isolated by rapid mechanical cell lysis in a guanidine thiocyanate-based buffer (Trisol) (Invitrogen) in the following manner. The supernatant was removed and added to a 2-ml tube containing the Heavy Phase Lock Gel (Eppendorf, Westbury, NY) and 300 μl chloroform-isoamyl alcohol (24:1). Inverting rapidly, aliquots were centrifuged for 10 min at 4°C, and the aqueous layer was collected and precipitated with isopropanol. Then the pellet was washed with 75% ethanol and dried at room temperature. RNA samples were treated with DNase I (Clontech, Palo Alto, CA) and incubated for 30 min at 30°C. The RNA quantity was determined on 1% denaturing agarose gel, the concentration was calculated, and the quality was determined spectrophotometrically by determining absorption (optical density at 260 or 280 nm). Total RNA was reverse transcribed, and the resulting cDNA was amplified by the SuperScript First Strand synthesis system for reverse transcription (RT)-PCR (Invitrogen) in the following manner. Briefly, total RNA (3 μg) was incubated with 1 μl of a 10 mM concentration of a deoxynucleoside triphosphate mix, 1 μl of random hexamers, and diethyl pyrocarbonate-treated water at 65°C for 5 min and then mixed with 2 μl of 10× RT buffer, 4 μl 25 mM MgCl2, 2 μl 0.1 M dithiothreitol, and 1 μl RNase OUT recombinant RNase inhibitor at 42°C for 2 min. One microliter of SuperScript II reverse transcriptase was added to each tube, and the tube was incubated at 42°C for 50 min. The reaction was terminated at 70°C for 15 min, and the tube was chilled on ice.
Quantitative real-time RT-PCR assay.
Quantitative fluorogenic amplification of cDNA was performed using the iCycler real-time detection system (Bio-Rad) and SYBR green technology (Bio-Rad), according to the method previously described (9). The relative abundances from standard curves were determined from a serially diluted standard pool of cDNAs and normalized to the 16S rRNA mRNA levels. The following primers were designed based on a BLAST search of the NCBI database: guaB2 (1,009 bp; forward, 5′-TCA CCT GCC GCC CCG ACA ACA CGC TGC CCC-3′; reverse, GGC ACC CGG CCC TCG ATG CCC TCG GGC ACC-3′), pmmB (787 bp; forward, 5′-TCC CGA CCC CCG CAC GGC CGC-3′; reverse, 5′-GTC CAC ATC GGC GGC CAG GGT-3′), gtf (374 bp; forward, 5′-ATG GAC GGC GCC GAC CTG CCC-3′; reverse, 5′-AGG ATC GCG GTG ATG CTG CCC-3′), accA2 (1,157 bp; forward, 5′-CGG TGG ATG CGG TGC GCG CGA TGG GCT-3′; reverse, 5′-GTT CCG CCA GCC GCT GGG GAT-3′), pks10 (106 bp; forward, 5′-ATG AGC GTC ATC GCC GGC GTG-3′; reverse, 5′-TCA GTG CCA ACG CAA CAA CAC-3′), and 16S rRNA (934 bp; forward, CGA ACG GGT GAG TAA CAC G-3′; reverse, 5′-TGC ACA CAG GCC ACA AGG GA-3′). The cDNA was denatured for 5 min at 95°C, followed by 30 cycles of amplification. Each cycle consisted of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and primer extension at 72°C for 2 min. The threshold cycle (Ct), which is defined as the fractional cycle number at which the fluorescence reaches 10 times the standard deviation of the baseline, was quantitated as described in User Bulletin no. 2 for the ABI PRISM 7700 sequence detection system. Changes in (Δ) gene expression (n-fold) were calculated as follows: ΔCttarget = CtM. avium A5 mRNA − Ct16S rRNA; ΔCtcontrol = CtM. avium 104 mRNA − Ct16S rRNA; and Δ(ΔCt) = ΔCttarget − ΔCtcontrol; change (n-fold) = 2−Δ(ΔCt).
Statistical analysis.
Analysis of variance was used for the comparisons among three or more groups. Student's t test was used to compare means between the two groups. The P values that were <0.05 were considered statistically significant.
RESULTS
Biofilm-associated promoters identified using a GFP promoter library.
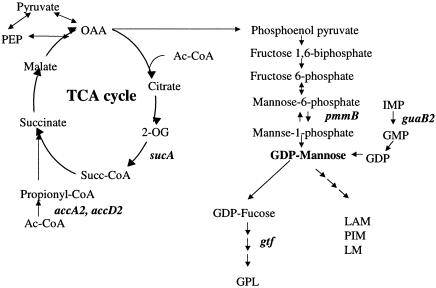
To identify M. avium genes regulated upon the formation of a biofilm or biofilm-like structure on PVC, an M. avium promoter library containing 10,000 clones of M. smegmatis was screened. Because we were interested in genes expressed during the initial phase of biofilm formation, GFP expression was monitored during the first 5 days of M. avium exposure to PVC. Twelve clones were identified with increased expression of GFP. Sequencing of the 12 genes showed that guaB2 (IMP dehydrogenase), AccA2 and AccD2 (alpha and beta subunits of acetyl/propionyl coenzyme A [CoA] carboxylase, respectively), pks10 (polyketide synthase family), pmmB (mannose-1-phosphatase), Itp3 (lipid carrier protein or keto acyl-CoA thiolase LTP3), ccsA (cytochrome c-type biogenesis protein), and gtf (glycosyl transferase, a homologue of M. tuberculosis CDC1551) were up-regulated. Four genes out of the 12 encode hypothetical proteins (Table 2). The AccA2 and AccD2 genes encode enzymes which are part of the TCA cycle (Fig. 1). The guaB2 and gtf genes take part in the biosynthesis of GDP-mannose and GPLs, respectively, as shown in Fig. 1.
TABLE 2.
M. avium genes identified as up-regulated upon incubation on PVC plates using the GFP promoter library
| Clone(s) | Gene | Protein | Homologue of M. tuberculosis H37Rv or CDC1551 | Fold increase in GFP expression |
|---|---|---|---|---|
| 4A11 | gtf | Glycosyltransferase | MT0564 | 3.6 |
| 4D7, 4H5, 5E12, 12B7, 1C9 | guaB2 | IMP dehydrogenase | Rv3411 | 3.8 |
| 4H11 | ccsA | Cytochrome c-type biogenesis protein | Rv0529 | 3.2 |
| 10H6 | accD2 | Acetyl/propionyl-CoA carboxylase (β subunit) | Rv0974 | 4.0 |
| 19H7 | pks10 | Polyketide synthase family | Rv1660 | 3.5 |
| 22A2, 22H3 | pmmB | Mannose-1-phosphatase | Rv3308 | 3.8 |
| 22H5 | accA2 | Acetyl/propionyl-CoA carboxylase (α subunit) | Rv0973 | 3.4 |
| 22B8 | Itp3 | Lipid carrier protein or keto acyl-CoA thiolase | Rv3523 | 3.9 |
| 3A12 | Integral membrane proteina | Rv0359 | 4.1 | |
| 20B5 | Acid phosphatasea | Rv3310 | 3.2 | |
| 5B10 | Oxidoreductasea | Rv3526 | 3.8 | |
| 22H7 | Integral membrane transport proteina | Rv1258c | 3.7 |
Probable hypothetical protein.
FIG. 1.
Genes of biofilm formation associated with the TCA cycle and GDP-mannose and glycopeptidolipid biosynthesis. The TCA cycle provides phosphoenolpyruvate, converted from oxaloacetate (OAA). The pmmB2 and guaB2 genes encode the enzymes to accelerate the biosynthesis of GDP-mannose. The gtf gene encodes the enzyme to biosynthesize GPL. Ac, acetyl; PEP, phosphoenolpyruvate; 2-OG, alpha-ketoglutarate; LAM, lipoarabinomannan; PIM, phosphatidylinositol mannoside; LM, lipomannan.
Detection of M. avium mRNA expression using real-time PCR.
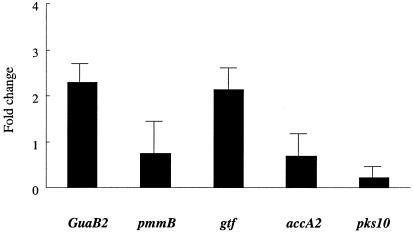
To confirm the findings obtained by screening the GFP promoter library, five genes were selected and real-time PCR was performed using M. avium strains seeded on PVC plates. The genes guaB2 and gtf were chosen because they represent different pathways, while pks10 was selected due to the importance of polyketides in mycobacteria. The M. avium 104 and M. avium A5 strain mRNAs were obtained from bacteria growing on PVC plates for 5 days. All five genes showed a significant increase (P < 0.05 using Student's t test) of expression upon biofilm formation, and the levels of expression of two genes were comparable between the strains A5 and 104, although the level of expression sometimes varied. The levels of expression of guaB2 and gtf in the M. avium A5 strain were increased, respectively, 2.28 ± 0.37- and 2.12 ± 0.46-fold above the levels of expression of similar genes in the M. avium strain, while expression of the pks10 gene of M. avium A5 decreased by 0.21 ± 0.25-fold compared with that of M. avium strain 104 (Fig. 2). These data confirmed the relevance of the promoter assay.
FIG. 2.
Changes in mRNA expression (n-fold) in M. avium A5 using real-time PCR. The expression of guaB2, gtf, pmmbB, accA2, and pks10 in M. avium A5. The expression of guaB and gtf was greater in M. avium A5 than in 104, while the expression of pks mRNA in M. avium A5 was lower than in M. avium 104. Values are means ± standard deviations (n = 3). The levels of expression of pmmB and accA2 were comparable in both strains. A P of <0.05 was considered significant for the expression of all genes.
M. avium A5 transposon mutants attenuated on biofilm formation.
As shown in Table 3, the screening of the transposon library led to the identification of five clones with an impaired ability to form biofilm. The possibility that the mutants would bind more to the crystal violet was ruled out by preliminary analysis with a similar number of mutant and wild-type bacteria stained with crystal violet. Four sequences were obtained out of the five mutants. The 2F1 and 6H9 mutants have the transposon inserted in the gene homologous to M. tuberculosis H37Rv sucA (Rv1248c), which encodes 2-oxoglutarate dehydrogenase. The 5G4 mutant had inactivation of a gene encoding a hypothetical membrane protein (Rv1565c), while the 9B5 mutant had the transposon interrupting a pcd (Rv3293) homologue of piperideine-6-carboxylic acid dehydrogenase (P6CDH). The 4B2 mutant had inactivation of a gene homologous to the pstB gene of the M. avium 2151 genome (GenBank accession no. AF143772).
TABLE 3.
M. avium genes identified using the transposon mutant system
| Clone(s) | Gene(s) | Protein | Homologue of M. tuberculosis H37Rv or CDC1551 |
|---|---|---|---|
| 4B2 | nrp (pstB)a | Protein synthetase | Rv0101 |
| 5G4 | Hypothetical membrane protein | Rv1565c | |
| 2F1, 6H9 | sucA | 2-Oxoglutarate dehydrogenase | Rv1248 |
| 9B5 | pcd | Piperideine-6-carboxylic acid dehydrogenase | Rv3293 |
Homologue of the M. avium 2151 genome (GenBank accession no. AF143772).
Biofilm formation by the wild type, mutants, and complemented strains.
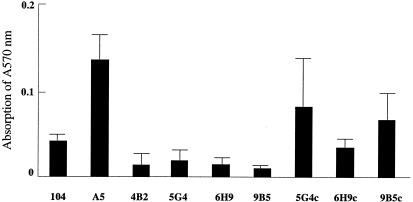
The ability to form biofilm was evaluated comparatively among the wild-type, mutant, and complemented strains. M. avium 104 and M. avium A5 had a spectrophotometer reading of 0.041 ± 0.001 and 0.136 ± 0.031. Biofilm formation of mutants 5G4 (0.019 ± 0.012), 6H9 (0.014 ± 0.009), and 9B5 (0.010 ± 0.002) was significantly impaired compared with that of M. avium A5. The biofilm formation of complemented strains 5G4c (0.082 ± 0.064), 6H9c (0.034 ± 0.009), and 9B5c (0.067 ± 0.041) was significantly increased compared with that of the 5G4, 6H9, and 9B5 mutants (Fig. 3), although no complete complementation was achieved.
FIG. 3.
Biofilm formation by the M. avium wild-type strain, transposon mutants, and strains with complementations of the inactivated genes. The bacterial strains (6 × 108 CFU/ml) were inoculated on PVC 96-well plates with HBSS for 14 days. The biofilm was evaluated using crystal violet stain as described in Materials and Methods. Values are means ± standard deviations. The A570 readings from three experiments are shown. 5G4, 6H9, and 9B5 were the transposon mutants of M. avium A5 strain. 5G4c, 6H9c, and 9B5c were the complemented strains of transposon mutants. A P of <0.005 was used for the comparisons between 4B2, 5G4, 6H9, 9B5, and 104 or A5. A P of >0.005 was used for the comparisons between 5G4 and 5G4c, 6H9 and 6H9c, and 9B5 and 9B5c.
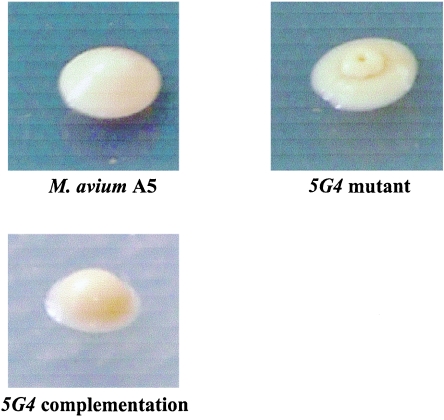
Colony morphology of M. avium A5 strain and mutants and between mutant and complemented strain.
The colony morphology of M. avium A5 appears like a white dome on 7H11 agar for 30 days (Fig. 4). On the other hand, the colonies of the depleted transposon mutants 5G4, 6H9, and 9B5 were white, flat, and round with a central small dome on 7H11 agar with Km at 400 μg/ml. The complemented transposon mutants of 5G4, 6H9, and 9B5 were all white and formed a dome on 7H11 agar, with Km at 400 μg/ml and Apr at 400 μg/ml, which were almost the same shape as the M. avium A5 strain. All mutant strains grew in agar in a fashion similar to that of the wild-type A5 strain (data not shown) at 37°C.
FIG. 4.
Colony morphologies of M. avium A5, its transposon mutants, and its complemented strains. M. avium A5 was cultured and observed for 30 days on 7H11 agar. The transposon mutant, 5G4, was grown on 7H11 with 400 μg/ml of kanamycin, and its complementation, 5G4c, was grown on 7H11 with 400 μg/ml of kanamycin and 400 μg/ml of apramycin.
DISCUSSION
M. avium is an environmental bacterium that can infect humans. M. avium is known to form biofilm or biofilm-like structures, and it is commonly recovered from sauna walls, swimming pools, and urban PVC water pipes. The ability to form biofilm has been associated with chronic bacterial infection. Persistent M. avium infection is frequently seen in individuals with chronic lung pathology, such as emphysema and cystic fibrosis. Since it is plausible that the ability to establish a biofilm can be associated with the difficulty to eliminate the infection, we attempted, as the first stage, to identify bacterial genes involved in biofilm formation. We screened an M. avium GFP promoter library of M. smegmatis and an M. avium transposon library. The use of an M. avium library with M. smegmatis is a very effective and rapid strategy that can have the results confirmed by investigating the transcription of identified genes in M. avium (9).
The GFP assay revealed several genes, up-regulated concomitantly with the formation of the biofilm. Two of the genes (AccA2 and AccD2) were members of the TCA cycle and have homology to the alpha and beta subunits of acetyl/propionyl-CoA carboxylase, respectively. Three genes (guaB2, pmmB, and gtf) are associated with GDP-mannose and GPL biosynthesis. The Itp3 and pks10 genes encode proteins that participate in fatty acid biosynthesis, while the other identified genes encode hypothetical proteins.
Biofilm formation occurs by phases in which the levels of gene regulation may differ. In our screen, we were interested in genes up-regulated during the establishment of the biofilm and not later phases. Synthesis of GPL, therefore, must be important for the adherence and initial establishment of biofilms.
In the synthesis of GPL, phosphoenolpyruvate is synthesized from oxaloacetate in the TCA cycle (22, 35). The pathway leads to GDP-mannose through fructose 1,6-biphosphate, fructose 6-phosphate, mannose-6-phosphate, and mannose-1-phosphate. The GDP-mannose is provided by two different biosynthetic pathways (Fig. 1), which suggests the importance of the pathway. The gene pmmB, which was identified by a GFP promoter library in the present study, is homologous to mannose-1-phosphatase. In an alternate pathway for GPL synthesis, the GDP may be synthesized. GDP is a product of the conversion of GMP and IMP, and the enzyme IMP dehydrogenase (guaB2) converts IMP into GMP. Considering mRNA expression upon biofilm formation, guaB2 expression was greater than that of pmmB, perhaps implying increased importance of the pathway under the conditions used. However, only by creating a null mutation on the genes can this hypothesis be addressed. The reason the synthesis of IMP appears to be of more significance than the synthesis of mannose-6-phosphate for biofilm formation is presently unknown.
GDP-fucose synthetase converts GDP-mannose to GDP-fucose. Fucose is found widely distributed in complex carbohydrates as a component of glycoconjugates, such as glycoproteins and glycolipids, in a wide variety of bacteria (15, 20). Fucose is added to glycoconjugates by specific transferases that utilize GDP-fucose as the sugar donor. In gram-negative bacteria, fucose is present as a component of the capsular polysaccharides and lipopolysaccharides which function in antigenic determination and participate in biofilm formation. On the other hand, GDP-mannose is the precursor of phosphatidylinositol and phosphatidylinositol mannosides in M. tuberculosis. GDP-mannose also provides the lipid anchor of two lipoglycans, lipomannan and lipoarabinomannan, the latter being an important modulator of the immune response in the course of tuberculosis and leprosy (7, 23), as well as a key ligand in the interactions between M. tuberculosis and phagocytic cells (13, 30). However, the roles of phosphatidylinositol mannosides and lipoarabinomannan in M. avium have not been extensively studied, and their role in biofilm formation could be novel.
The genes which were identified in the present study have shed light on the metabolic regulation of biofilm formation. Martinez et al. (19) reported the inactivation of genes after use of the mariner transposon system and identified the mps and tmtpC genes as being involved in biofilm formation. The mps is homologue of a peptide synthetase (GenBank MAC 104 genome; pstA, pstB, and pstC) (4). The roles of peptide synthetase may be modified by an N-acylated Phe acceptor by sequential addition of Thr, Ala, and alaninol residues by peptidosynthetase, encoded by mps. This lipopeptide core may subsequently be glycosylated with rhamnose and 6-deoxy talose, resulting in the nonspecific core GPL that is found in all members of M. avium as well as M. smegmatis (3). The nsGPLs are further elaborated with oligosaccharide structures to produce the antigenically important serovar-specific GPLs (5). M. smegmatis, which produces only nsGPL, has been used to identify the genes for the glycosyltransferase and methyltransferase involved in elaborating the nsGPL with the hepatenic oligosaccharides of M. avium serovar 2 (11, 21). Furthermore, the M. avium A5 transposon mutant of 4B2 obtained in the present study had the pstB gene inactivated, which was encoded in the sequences of the GPL biosynthesis gene cluster and daunorubicine gene in M. avium 2151 (GenBank accession no. AF143772) (4).
The role that mycolic acid plays in mycobacterial biofilm formation is unclear. Polyketide synthetase is responsible for the synthesis of methyl-branched fatty acids in M. tuberculosis. Analysis of the M. tuberculosis genome sequence has revealed the existence of several polyketide synthases (pks) (29). In an attempt to determinate the function of mycobacterial pks, mutants deficient in the expression of the pks10 and ppsB/ppsC, pks2, pks3/4, pks10, and pks15/1 genes were constructed by allelic replacement in M. tuberculosis or M. bovis BCG. The pks gene, disruption of which in M. tuberculosis H37Rv also caused dimycocerosate deficiency without affecting the ability of the mutant strain to synthesize mycocerosic acids, is thought to be involved in the production of phthiocerol derivatives (14, 31). The precise function of pks10 is currently unclear. Recent work suggests that polyketides are important in the ability of M. tuberculosis to prevent activation of the innate immunoresponse (27), but its role in the formation of biofilm will need further investigation.
The transposon library of M. avium strain A5, screened for the identification of clones with an impaired ability to produce biofilm on PVC, led to the identification of five clones (four genes). The transposon mutant clone, 6H9, has the gene homologue of 2-oxoglutarate dehydrogenase (OX) in the TCA cycle inactivated. The conversion of propionyl-CoA and succinyl-CoA leads to succinate and malate in the TCA cycle (16). It has been reported for Salmonella enteritidis that sucD, the succinyl-CoA synthetase alpha subunit and an enzyme upstream of 2-oxodehydrogenase, has been identified as an important enzyme in the formation of biofilm by the bacterium (33). The mutants 6H9 and 4B2 encode sucA and pstB, which are associated with GPL synthetase. The gene inactivated in the 5G4 mutant was highly homologous to M. tuberculosis genomic Rv1565c, a hypothetical membrane protein. It is located upstream of the trehalose synthetase genes treX, treY, and treZ. These genes encode malto-oligosyltrehalose synthase, malto-oligosyltrehalose synthase, and malto-oligosyltrehalose, respectively, which are well conserved between the M. tuberculosis and M. avium 104 genomes. In M. tuberculosis, trehalose has been known to participate in mycolic acid synthesis; mycolic acids are complemented by glycolipids such as a,a′-trehalose dimycolate and a,a′-trehalose monomycolate (5). The 9B5 clone of M. avium A5 has the pcd gene inactivated. The pcd gene encoding P6CDH (Rv3293) is involved in the biosynthesis of α aminoadipic acid. It is located in the cephamycin C gene cluster of Streptomyces clavuligerus (1, 25). P6CDH, which converts 1-piperideine-6-carboxylic acid into α aminoadipic acid, a precursor of cephamycin C, has recently been purified (10). Since there was no cephamycin C gene cluster around the pcd gene in the mycobacterial genome, it is not easy to explain the roles of P6CDH with biofilm formation or GPL biosynthesis in M. avium. The complemented strains of both 5G4 and 9B5 restored the ability of the strains to form biofilms.
In summary, work with the strain A5 identified several genes associated with biofilm formation. Most of genes are involved in GPL biosynthesis, which indicates that the outer surface of the bacterium is likely to be important for the establishment of biofilm. The future aim of this work is to find a regulator(s) for the formation of biofilm.
Acknowledgments
We are grateful to Denny Weber for editing the manuscript.
This work was supported by a grant of the National Institute of Allergy and Infectious Diseases (AI-43199).
REFERENCES
- 1.Alexander, D. C., and S. E. Jensen. 1998. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J. Bacteriol. 180:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., J. T. Crawford, and K. D. Eisenach. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 177:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belisle, J. T., and P. J. Brennan. 1989. Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 171:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 6.Carter, G., M. Wu, D. C. Drummond, and L. E. Bermudez. 2003. Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J. Med. Microbiol. 52:747-752. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, D., and K. H. Khoo. 1998. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8:113-120. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danelishvili, L., M. J. Poort, and L. E. Bermudez. 2004. Identification of Mycobacterium avium genes up-regulated in cultured macrophages and in mice. FEMS Microbiol. Lett. 239:41-49. [DOI] [PubMed] [Google Scholar]
- 10.de La Fuente, J. L., A. Rumbero, J. F. Martin, and P. Liras. 1997. Delta-1-piperideine-6-carboxylate dehydrogenase, a new enzyme that forms alpha-aminoadipate in Streptomyces clavuligerus and other cephamycin C-producing actinomycetes. Biochem. J. 327:59-64. [PMC free article] [PubMed] [Google Scholar]
- 11.Eckstein, T. M., J. M. Inamine, M. L. Lambert, and J. T. Belisle. 2000. A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J. Bacteriol. 182:6177-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilhot, C., I. Otal, I. Van Rompaey, C. Martin, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang, B. K., and L. S. Schlesinger. 1998. Characterization of mannose receptor-dependent phagocytosis mediated by Mycobacterium tuberculosis lipoarabinomannan. Infect. Immun. 66:2769-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 15.Kordulakova, J., M. Gilleron, K. Mikusova, G. Puzo, P. J. Brennan, B. Gicquel, and M. Jackson. 2002. Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J. Biol. Chem. 277:31335-31344. [DOI] [PubMed] [Google Scholar]
- 16.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krzywinska, E., and J. S. Schorey. 2003. Characterization of genetic differences between Mycobacterium avium subsp. avium strains of diverse virulence with a focus on the glycopeptidolipid biosynthesis cluster. Vet. Microbiol. 91:249-264. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., E. Miltner, M. Wu, M. Petrofsky, and L. E. Bermudez. 2005. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol. 7:539-548. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, A., S. Torello, and R. Kolter. 1999. Sliding motility in mycobacteria. J. Bacteriol. 181:7331-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon, S., M. Stahl, R. Kumar, G. Y. Xu, and F. Sullivan. 1999. Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from Escherichia coli. J. Biol. Chem. 274:26743-26750. [DOI] [PubMed] [Google Scholar]
- 21.Mills, J. A., M. R. McNeil, J. T. Belisle, W. R. Jacobs, Jr., and P. J. Brennan. 1994. Loci of Mycobacterium avium ser2 gene cluster and their functions. J. Bacteriol. 176:4803-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay, B., E. M. Concar, and R. S. Wolfe. 2001. A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis. J. Biol. Chem. 276:16137-16145. [DOI] [PubMed] [Google Scholar]
- 23.Nigou, J., M. Gilleron, M. Rojas, L. F. Garcia, M. Thurnher, and G. Puzo. 2002. Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes Infect. 4:945-953. [DOI] [PubMed] [Google Scholar]
- 24.Ortalo-Magne, A., A. Lemassu, M. A. Laneelle, F. Bardou, G. Silve, P. Gounon, G. Marchal, and M. Daffe. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178:456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Llarena, F. J., A. Rodriguez-Garcia, F. J. Enguita, J. F. Martin, and P. Liras. 1998. The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of alpha-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J. Bacteriol. 180:4753-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Recht, J., and R. Kolter. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84-87. [DOI] [PubMed] [Google Scholar]
- 28.Rosenzweig, D. Y. 1979. Pulmonary mycobacterial infections due to Mycobacterium intracellulare-avium complex. Clinical features and course in 100 consecutive cases. Chest 75:115-119. [DOI] [PubMed] [Google Scholar]
- 29.Rousseau, C., T. D. Sirakova, V. S. Dubey, Y. Bordat, P. E. Kolattukudy, B. Gicquel, and M. Jackson. 2003. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149:1837-1847. [DOI] [PubMed] [Google Scholar]
- 30.Schlesinger, L. S., S. R. Hull, and T. M. Kaufman. 1994. Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J. Immunol. 152:4070-4079. [PubMed] [Google Scholar]
- 31.Sirakova, T. D., V. S. Dubey, H. J. Kim, M. H. Cynamon, and P. E. Kolattukudy. 2003. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect Immun. 71:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 33.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Dien, S. J., Y. Okubo, M. T. Hough, N. Korotkova, T. Taitano, and M. E. Lidstrom. 2003. Reconstruction of C(3) and C(4) metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 149:601-609. [DOI] [PubMed] [Google Scholar]
- 36.von Reyn, C. F., J. N. Maslow, T. W. Barber, J. O. Falkinham III, and R. D. Arbeit. 1994. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137-1141. [DOI] [PubMed] [Google Scholar]