Abstract
This study examined the ability of (i) pure nisin, (ii) nisin-producing Lactococcus lactis strain CHCC5826, and (iii) the non-nisin-producing L. lactis strain CHCH2862 to affect the composition of the intestinal microbiota of human flora-associated rats. The presence of both the nisin-producing and the non-nisin-producing L. lactis strains significantly increased the number of Bifidobacterium cells in fecal samples during the first 8 days but decreased the number of enterococci/streptococci in duodenum, ileum, cecum, and colon samples as detected by selective cultivation. No significant changes in the rat fecal microbiota were observed after dosage with nisin. Pearson cluster analysis of denaturing gradient gel electrophoresis profiles of the 16S rRNA genes present in the fecal microbial population revealed that the microbiota of animals dosed with either of the two L. lactis strains were different from that of control animals dosed with saline. However, profiles of the microbiota from animals dosed with nisin did not differ from the controls. The concentrations of nisin estimated by competitive enzyme-linked immunosorbent assay (ELISA) were approximately 10-fold higher in the small intestine and 200-fold higher in feces than the corresponding concentrations estimated by a biological assay. This indicates that nisin was degraded or inactivated in the gastrointestinal tract, since fragments of this bacteriocin are detected by ELISA while an intact molecule is needed to retain biological activity.
There is an increasing consumer demand for gently processed and lightly preserved food products containing fewer chemical preservatives. Such new types of food represent a challenge with respect to microbiological food safety and shelf life. One answer to this challenge is the use of biopreservatives, i.e., live bacteria or products of bacteria, which inhibit the growth of pathogens and spoilage bacteria. Bioprotective organisms primarily include lactic acid bacteria that produce antimicrobial substances such as organic acids, hydrogen peroxide, specific enzymes, small metabolites (reuterin, diacetyl, and fatty acids) and bacteriocins (8). The ability of either bacteriocins or bacteriocin-producing strains to restrict the growth of pathogenic bacteria such as Listeria monocytogenes in various food products is well described. The majority of the investigations are carried out on ready-to-eat foods including cheese (16) and meat products (17, 19); however, fish (23) and vegetables (1) can also be preserved by bioprotection.
Nisin A is a 34-amino-acid peptide (3,500 Da) produced by several Lactococcus lactis strains (13). This antimicrobial peptide can inhibit the growth of a wide range of gram-positive vegetative cells and prevent germination of spores (9). Nisin was the first bacteriocin to be granted “generally recognized as safe” status for use in cheese production in the United States (13). In 1969, nisin was given international acceptance as a food additive by the World Health Organization and is now permitted in a variety of food products around the world (30). From the European Parliament and Council Directive of 1995, nisin is regulated as a preservative with use restricted to specific food products.
While the inhibitory effects of nisin and nisin-producing L. lactis cells in food matrices are well described, no documentation exists on the effect of ingested nisin or nisin-producing L. lactis on the commensal gastrointestinal microbiota. It is not known whether nisin production is induced in the L. lactis strains present in the gut or whether ingested nisin is degraded in the intestinal environment. The most commonly used analytical tools for the determination of nisin concentration are biological assays (agar diffusion tests), but the drawbacks of these methods are lack of specificity and limited sensitivity. Highly specific immunochemistry-based methods are therefore gaining attention (28).
The objective of the present study was to provide information about the unintended effects of nisin and nisin-producing bacteria used for biopreservation. This type of information is highly relevant for the risk assessment of biopreservative products. A gnotobiotic rat model associated with human fecal microbiota (7) was used to investigate the effects of consumption of nisin and nisin-producing lactococci on the indigenous gut bacteria. Issues of (i) degradation of ingested pure nisin and (ii) nisin production by L. lactis present in the gastronintestinal (GI) tract were addressed using a biological as well as a competitive immunoassay.
MATERIALS AND METHODS
Bacterial strains.
L. lactis strains CHCC5826 (Chr. Hansen Culture Collection, Chr. Hansen A/S, Hørsholm, Demark) and CHCC2862 are isogenic with the exception of the nisin A gene cluster. L. lactis CHCC5826 was constructed by conjugative transposition of this cluster from L. lactis CHCC4098Lac− into strain CHCC2862. The lactose-negative derivative of L. lactis CHCC4098 was constructed by plasmid curing at 40°C and screening as described by McKay et al. (21). Conjugative transposition (20) of the nisin A gene cluster from L. lactis CHCC4098Lac− into strain CHCC2862 was conducted, and transconjugants were selected as lactose-positive and nisin-resistant colonies on lactic agar (21) containing 1,600 IU per ml of nisin. One such colony was named CHCC5826.
HFA animals.
Nineteen germ-free Sprague-Dawley rats (aged 6 weeks) bred at the Danish Institute for Food and Veterinary Research from parental animals originally obtained from IFFA Credo, France, were used. The animals were kept and fed, and the germ-free status was confirmed as described previously (26). All animals were caged individually throughout the duration of the study. Human flora-associaetd (HFA) rats were created by oral gavage of the germ-free rats with 1 ml of human fecal suspension prepared as described below. Twenty days after the introduction of human microbiota, the rats were randomly allocated to five groups and dosed with either nisin-producing or non-nisin-producing L. lactis, nisin A (Chr. Hansen, Hørsholm, Denmark), or phosphate-buffered saline (PBS; Oxoid, Hampshire, England) per os twice during 2 days, as shown in Table 1. The nisin-producing and non-nisin-producing L. lactis isolates were freeze-dried and diluted in saline to the concentrations described in Table 1.
TABLE 1.
Dosage of animals
| Group | No. of rats | Inoculum | Dose
|
|
|---|---|---|---|---|
| First (day 1) | Second (day 2) | |||
| A | 5 | L. lactis (nisin- producing) CHCC5826 | 6 × 109 CFU | 4 × 109 CFU |
| B | 4 | L. lactis (non-nisin- producing) CHCC2862 | 3 × 109 CFU | 3 × 109 CFU |
| C | 3 | Nisin | 60 mg | 60 mg |
| D | 3 | Nisin | 0.6 mg | 0.6 mg |
| Control | 4 | PBS | 1 ml | 1 ml |
Preparation of human fecal suspension.
A human fecal sample was obtained from a 32-year-old woman who had consumed a common diet and had no recent history of gastrointestinal disturbances or antibiotic treatment. The sample (kept under anaerobic conditions and below 5°C for less than 12 h) was placed in an anaerobic chamber and diluted 10-fold in 50% reinforced clostridial medium (Oxoid, Hampshire, England) containing 17% glycerol, homogenized, and transferred to −80°C.
Sampling from rats.
Fecal samples were obtained directly from the rectums of 17 rats during a 7-week period. Two rats, dosed with 60 mg pure nisin and saline, respectively, were euthanized 3 h after the first inoculation.
After euthanization, the gastrointestinal tracts were removed and contents from duodenum, ileum, cecum, and colon were placed in sterile tubes to be used either for immediate plating on selective agars or for DNA extraction. The samples for DNA extraction were stored at −21°C. Fecal samples for nisin quantification were frozen at −80°C. All samples were processed within 3 h.
Selective plating.
All fecal and intestinal samples were examined for total anaerobic counts and total aerobic counts on reinforced clostridial agar (Oxoid) for Lactobacillus on Rogosa agar (Oxoid), for total coliforms on MacConkey agar no. 3 (Oxoid), for Bifidobacterium on Wilkins-Chalgren agar (Oxoid) modified with the addition of glacial acetic acid (1 ml/liter) and mupirocin (100 mg/liter) (24), and for total enterococci/streptococci on Slanetz and Bartley agar (Oxoid). The samples were homogenized, and appropriate dilutions were plated on the selective agars.
Total anaerobes, lactobacilli, and Bifidobacterium were incubated anaerobically for 3 days, whereas coliforms, enterococci/streptococci, and total aerobes were incubated aerobically for 1, 2, or 3 days, respectively. All incubations were carried out at 37°C.
DNA extraction and PCR amplification with universal primers.
DNA was extracted from 200 mg of feces content as previously described by Leser et al. (15). The DNA was purified using the QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) and was finally dissolved in 30 μl autoclaved water and stored at −21°C.
Universal eubacterial primers 1 (5′-CCTACGGGAGGCAGCAG-3′) and 2 (5′-ATTACCGCGGCTGCTGG-3′), described by Muyzer et al. (22), were used for PCR amplification. Forward primer 1 carried a GC clamp (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′) at the 5′ end (27). Aliquots (10 μl) of the extracted DNA were added to the following MasterMix to give a 50-μl PCR mixture: 20 μl of Eppendorf MasterMix (2.5×; Eppendorf, Hamburg, Germany) and 40 pmol of each primer. Amplification was carried out in a PTC-225 thermal cycler (MJ Research, Waltham, Mass.) as a touchdown PCR. Initial denaturation was done at 96°C for 5 min, amplification was carried out using 20 cycles each including denaturation at 94°C for 1 min, annealing for 1 min starting at 65°C and decreasing by 0.5°C for each cycle, and extension at 72°C for 1 min. This was followed by five additional cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72° for 1 min, and final extension at 72°C for 5 min. A final product length of approximately 200 bp was checked by electrophoresis on 2.0% agarose gels (Seakem GTG agarose; Cambrex, Rockland, Maine).
DGGE.
Denaturing gradient gel electrophoresis (DGGE) was carried out using a Dcode Universal Mutation Detection System instrument according to the manufacturer′s instructions (Bio-Rad Laboratories, Hercules, California). The acrylamide concentration in the gel was 9%, and the linear denaturing gradient was 25% to 65% (100% denaturant corresponds to 7 M urea and 40% deionized formamide). PCR products (13 μl) were mixed with 3 μl loading dye and loaded to the bottom of the well. Gels were run in 1× Tris-acetate-EDTA (TAE) at 60°C for 16 h at 36 V and 28 mA. Gels were stained with the fluorescent dye Gelstar (Biowhittaker, Walkersville, Maine) for 45 min.
Indirect competitive immunoassay for detection of nisin in intestinal and fecal samples.
Pure nisin A (Chr. Hansen A/s) was conjugated to keyhole limpet hemocyanin by use of glutaraldehyde as previously described (18). The nisin A-keyhold limpet hemocyanin conjugate was dialyzed against PBS (Oxoid) and mixed (1:1, vol/vol) with Freund's complete adjuvant for the first immunization and Freund's incomplete adjuvant for the following immunizations. Rabbits were immunized each third week and bleedings collected 9 days after each immunization. The blood was centrifuged and the serum analyzed by Western blotting for nisin A-specific antibodies. The serum collected after the third immunization was used for the purification of antibodies against nisin A by affinity chromatography. The specificity of the antibodies was verified against media and fecal extracts.
Pure nisin antigen was used to coat enzyme-linked immunosorbent assay (ELISA) plates for competitive immunoassays. One hundred percent pure nisin was dissolved in 100% dimethyl sulfoxide (DMSO; 1 mg/ml) and diluted 10,000-fold to a final concentration of 100 ng/ml in 0.1 M sodium carbonate-bicarbonate buffer (pH 9.0). Fifty microliters of the solution was used for coating each well of the ELISA plates (Maxisorp; Nunc, Roskilde, Denmark) overnight at 4°C. After washing with washing buffer (1× Tris-buffered saline [TBS] with 0.05% Tween 20), the plate was treated with blocking solution (1× TBS, 1% polyvinylidenepyrrolidone 40, 2% Tween 20). The coated plate could be stored empty and frozen. Purified rabbit antiserum (50 μl/well diluted 1:16,000 in washing buffer) was added. Subsequently, triplicates of 50 μl intestinal or fecal samples or standard solution of pure nisin all in multiple twofold dilutions were additionally added, followed by incubation either overnight at 4°C or at room temperature with shaking for 2 to 3 h. After removal of unbound material (five repeated washes), 150 μl secondary alkaline phosphatase-conjugated anti-rabbit immunoglobulin (Dako-Cytomation, Glostrup, Denmark) diluted 1:30,000 in washing buffer was added to each well and incubated for 2 h at room temperature. After seven repeated washes, 50 μl ELISA alkaline phosphatase amplifier NADPH substrate (GibcoBRL, Paisley, United Kingdom) was added to each well and the plates were incubated for 20 min at room temperature. Subsequently, 50 μl ELISA amplifier solution (GibcoBRL, Paisley, United Kingdom) was added to each well and incubated for an additional 20 min, followed finally by the addition of 50 μl stop solution (0.2 N sulfuric acid) per well. The optical density was measured at 495 nm on an ELISA reader. A detection limit (three times the standard deviation) of 5 ng/ml was obtained for this antigen-competitive ELISA.
Biological assay for the detection of nisin in intestinal and fecal samples.
The biological assay for the determination of active nisin in fecal samples was based on the methods described by Tramer and Fowler (29) and Fowler et al. (5). A straight-line response was obtained for the (log) nisin concentrations ranging from 0.06 to 0.25 μg/g of nisin dissolved in either 0.02 N HCl or 100% DMSO when plotted against the diameters of the inhibition zone. Below concentrations of 0.05 μg/g, no inhibition zone was obtained. Fecal samples were suspended in either 0.02 N HCl or 100% DMSO. Assay plates (Q-tray; BouleNordic, Sweden) contained 200 ml brain-heart infusion agar (Oxoid) inoculated with 106 CFU/ml of the indicator organism Micrococcus flavus strain NCIB 8166 (National Collection of Industrial Bacteria, Aberdeen, Scotland).
Data analysis.
Statistical data analysis of bacterial counts (t tests and one-way analysis of variance) was performed by using SAS software (SAS Institute, Cary, NC). Probability levels (P) of <0.05 were considered to be significant. Standard errors of the means were calculated as standard deviations divided by the square root of the number of observations. Bionumerics software (version 3; Applied Maths, Sint-Martens-Latem, Belgium) was used for the identification of bands and normalization of band patterns from DGGE gels. Subsequently, comparison of band patterns was done by Pearson cluster analysis (the unweighted-pair group method using average linkages) in this software.
RESULTS
Changes in the intestinal microbiota as determined by plating on selective agars.
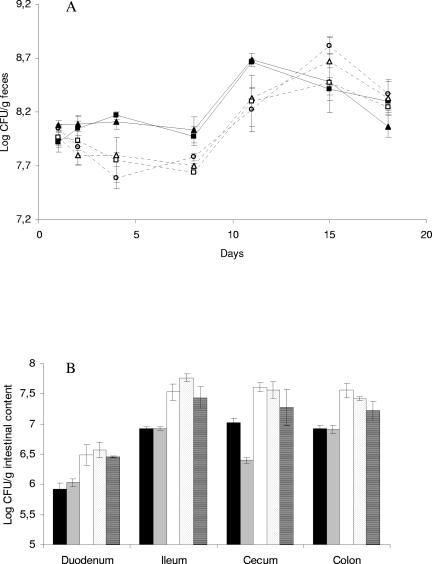
Between the first day and the tenth day after the first dosage, numbers of Bifidobacterium cells per gram of feces detected by selective plating were significantly higher in samples from rats inoculated with the L. lactis strains than in fecal samples from the rats dosed with saline (PBS), 0.6 mg nisin, or 60 mg nisin, respectively. No significant difference was found between the Bifidobacterium concentrations in samples from rats inoculated with the nisin-producing L. lactis CHCC5826 and theisogenic non-nisin-producing L. lactis CHCC2862. Similarly, Bifidobacterium concentrations in fecal samples of rats inoculated with nisin and saline did not differ significantly (Fig. 1A). Total anaerobic and aerobic concentrations, as well as concentrations of lactobacilli, coliforms, and enterococci/streptococci in fecal samples, were similar in all groups of HFA rats, independent of the nature of the dosage given.
FIG. 1.
(A) Bifidobacterial populations in feces of rats dosed with nisin-producing L. lactis (closed triangles), non-nisin-producing L. lactis (closed squares), 60 mg nisin (open trangles), 0.6 mg nisin (open squares), and PBS (open circles). Each point represents the mean of data from animals in one dose group. Error bars designate standard errors of the means. Animals were dosed on day 1 and day 2. Statistical analysis confirmed that, between day 2 and day 11, numbers of bifidobacteria in the feces of animals inoculated with either of the L. lactis strains (closed symbols) were higher than in the other three groups (open symbols). (B) Densities of enterococci/streptococci in intestinal samples from rats 25 days after dosage with nisin-producing L. lactis (black), non-nisin-producing L. lactis (gray), 60 mg nisin (white), 0.6 mg nisin (dots), and PBS (stripes). Each point represents the mean of data from animals in one dose group. Error bars designate standard errors of the means. Statistical analysis confirmed that the densities of enterococci/streptococci in all sections of the gut were lower in animals dosed with either of the L. lactis strains (black and gray shaded bars) than in the other three groups.
Rats dosed with either of the L. lactis strains had generally lower level of enterococci/streptococci in the duodenum, ileum, cecum, and colon at the time of euthanization (25 days after dosage) than rats dosed with saline or nisin (Fig. 1B). The observed concentrations of total anaerobes, aerobes, lactobacilli, coliforms, and Bifidobacterium in intestinal and fecal samples at the day of euthanization were not significantly different between the five groups of rats (data not shown).
Changes in intestinal microbiota analyzed by DGGE.
DGGE profiles of PCR-amplified 16S rRNA genes were created from DNA extracted from fecal samples from HFA rats dosed as described in Table 1. The profiles contained between 10 and 24 bands (data not shown).
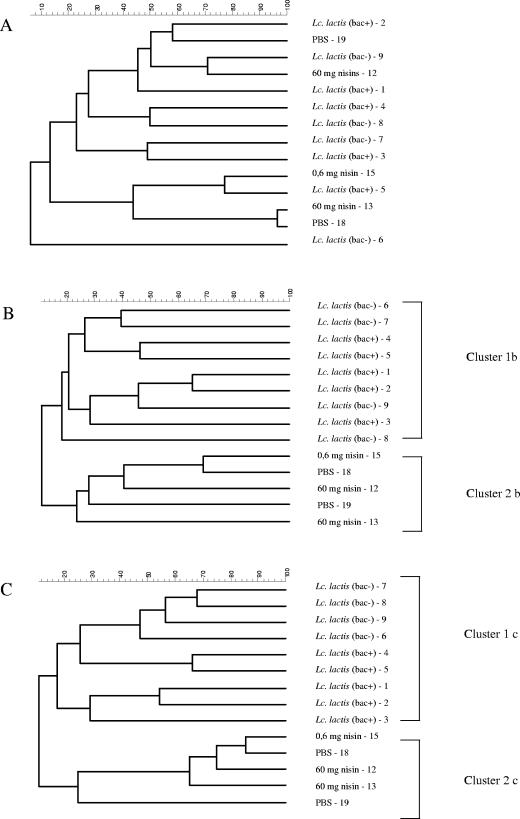
Generation of dendrograms for the comparison of profiles obtained the day after the second dosage with either the lactococcal strain, nisin, or saline resulted in two clusters. One cluster contained the profiles from rats dosed with the nisin-producing L. lactis CHCC5826 or non-nisin-producing L. lactis CHCC2862, while another cluster contained profiles obtained from rats dosed with either pure nisin or saline (Fig. 2B). Similar clustering patterns were obtained 3 days after cessation of dosage (Fig. 2C), while a completely different pattern prevailed prior to dosage (Fig. 2A).
FIG. 2.
Dendrograms generated by unweighted-pair group method using average linkages cluster analysis of PCR-DGGE profiles of 16S rRNA obtained from fecal samples of HFA rats inoculated as given in Table 1. Samples were taken an hour before first inoculation (A), 1 day after last inoculation (B), and 3 days after last inoculation (C). Numbers 1 to 19 designate the individual animals.
A general tendency of increased difference between profiles obtained from individual rats was observed the first day after dosage of the animals (Fig. 2B). Three days after the last dosage, this variation between fecal samples from different animals was again similar to the variation observed before dosage (Fig. 2A and C).
Presence of nisin in the GI tract.
Nisin was detected in the duodena and ilea of HFA rats 3 hours after dosage with 60 mg nisin by a biological assay as well as by a competitive ELISA. The concentrations estimated by ELISA were approximately 10-fold higher than the concentrations estimated by the biological assay. Similarly, nisin was detectable by both assays in fecal samples taken 24 h after dosage. However, the concentration estimated by the biological assay was 0.4 μg/g feces while a concentration of 76.8 μg/g feces was estimated by competitive ELISA (Table 2). Nisin was not detectable in intestinal and fecal samples from rats dosed with saline (data not shown).
TABLE 2.
Presence of nisin in the gastrointestinal tract of HFA rats estimated by a biological assay and a competitive immunoassay
| Sampling source | Nisin dose (mg)a | Time after dosage (h) | Nisin concn (μg/g intestinal or fecal content) byb:
|
|
|---|---|---|---|---|
| Biological assay | Competitive immunoassay | |||
| Duodenum | 60 | 3 | 1.8 ± 0.1 | 18.9 ± 5.6 |
| Ileum | 60 | 3 | 6.9 ± 1.5 | 62.4 ± 16.1 |
| Cecum | 60 | 3 | <LD | <LD |
| Colon | 60 | 3 | <LD | <LD |
| Fecesc | 60d | 24 | 0.4 ± 0.5 | 76.8 ± 18.2 |
| 0.6e | 24 | <LD | 5.5 ± 1.4 | |
The concentrations of nisin were below the LD for intestinal and fecal samples from rats inoculated with PBS (data not shown).
LD, limit of detection. The LD was 50 ng/g for the biological assay and 5 ng/g of intestinal or fecal content for the competitive immunoassay.
Fecal samples other than the intestinal samples were obtained from other rats.
Nisin was detected in the two rats inoculated with 60 mg nisin.
Nisin was detected in only one of three rats inoculated with 0.6 mg nisin, which corresponds to the nisin concentration approved for use in food by the European Union (European Parliament and Council Directive 95/2/EC).
Similarly, nisin was not detectable by biological assay or competitive ELISA on fecal or intestinal samples from rats inoculated with either of the L. lactis strains in samples taken either 3 hours or 25 days after the first dosage (data not shown).
DISCUSSION
Community analysis carried out by PCR-DGGE of 16S ribosomal genes suggests that the presence of the nisin-producing as well as non-nisin-producing L. lactis has an effect on the composition of fecal bacteria but that this effect is not related to the presence of nisin-encoding genes in this strain (Fig. 2). L. lactis is metabolically active in the GI tract (4) and may affect the surrounding intestinal microbiota by competition for specific nutrients and/or adhesion sites (6). However, previous studies have shown that ingested L. lactis remains in the gut for only up to 3 days (12, 14), while in the present study, changes in the fecal densities of Bifidobacteria induced by dosage with L. lactis were detectable by traditional selective plating 10 days after dosage, indicating that the induced changes remained also after clearance of L. lactis (Fig. 1A). Furthermore, the Enterococcus/Streptococcus populations of the intestinal sections investigated 25 days after dosage were still lower in animals dosed with L. lactis than in animals from all groups not dosed with this strain (Fig. 1B).
Nisin was not detectable in feces from rats inoculated with the nisin-producing L. lactis. This may be due to the lack of induction of nisin expression in the intestinal environment but may also be explained by the continuous degradation of nisin by proteases present in the intestine or by the hydrophobic binding of nisin to the cell walls of the producer strain (2) or to intestinal surfaces (11).
Estimations of nisin concentrations in intestinal and fecal samples from rats dosed with pure nisin were significantly higher when based on a competitive ELISA than when based on biological activity. In the upper part of the intestinal tract (duodenum and ileum), the nisin concentrations estimated by ELISA were approximately 10-fold higher than those estimated by the bioassay, while in fecal samples, the difference was around 200-fold (Table 2). This indicates that degradation or inactivation of nisin occurs in the GI tract, since proteolytic fragments of nisin will be detected in the competitive ELISA (3), while a nearly intact nisin molecule is needed to retain biological activity (25). Nisin is reported to undergo proteolytic degradation mediated by the pancreas enzyme α-chymotrypsin present in the GI tract (10).
In agreement with previous observations from our lab, some variation between DGGE profiles of the individual HFA rats was seen prior to dosage (Fig. 2A). The first day after dosage, a temporary increase of this variation among individual rats was observed (Fig. 2B). We speculate that this could be caused by stress of the animals associated with the procedure of oral gavage.
Under the experimental conditions used in the present study, no nisin-mediated disturbance of the commensal microbiota of HFA rats resulting from dosing with nisin or a nisin-producing strain could be identified. Dosage of HFA rats with either of the L. lactis strains increased the number of bifidobacteria detected in feces. This effect is generally considered beneficial. None of the results obtained give reason to assume that the use of nisin as a biopreservative represents a risk for human consumers by unintended changes of the intestinal microbiota.
Acknowledgments
This work was financed by the Danish FØTEK 3 program (3401-44-03-691) and the Danish Bacon and Meat Council. The work was conducted in collaboration with Chr. Hansen A/S, Danisco A/S, and the Danish Toxicology Centre.
We thank Pia Bjerring for constructing Lactococcus lactis strain CHCC5826.
REFERENCES
- 1.Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim. Biophys. Acta 1373:47-58. [DOI] [PubMed] [Google Scholar]
- 2.Bouksaim, M., C. Lacroix, R. Bazin, and R. E. Simard. 1999. Production and utilization of polyclonal antibodies against nisin in an ELISA and for immuno-location of nisin in producing and sensitive bacterial strains. J. Appl. Microbiol. 87:500-510. [DOI] [PubMed] [Google Scholar]
- 3.Daoudi, L., C. Turcotte, C. Lacroix, and I. Fliss. 2001. Production and characterization of anti-nisin Z monoclonal antibodies: suitability for distinguishing active from inactive forms through a competitive enzyme immunoassay. Appl. Microbiol. Biotechnol. 56:114-119. [DOI] [PubMed] [Google Scholar]
- 4.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler, G. G., B. Jarvis, and J. Tramer. 1975. The assay of nisin in foods, p.91-105. In R. G. Board and D. G. Lovelock (ed.), Some methods for microbiological assay. Academic Press, London, United Kingdom.
- 6.Freter, R. 1983. Mechanisms that control the microflora of the large intestine, p. 33-54. In D. Hentges (ed.), Human intestinal flora in health and disease. Academic Press, New York, N.Y.
- 7.Hirayama, K., and K. Itoh. 2005. Hum. flora-associated (HFA) animals as a model for studying the role of intestinal flora in human health and disease. Curr. Issues Intest. Microbiol. 6:69-75. [PubMed] [Google Scholar]
- 8.Holzapfel, W. H., R. Geisen, and U. Schillinger. 1995. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int. J. Food. Microbiol. 24:343-362. [DOI] [PubMed] [Google Scholar]
- 9.Hurst, A. 1981. Nisin. Adv. Appl. Microbiol. 27:85-123. [Google Scholar]
- 10.Jarvis, B., and R. R. Mahoney. 1969. Inactivation of nisin by alpha-chymotrypsin. J. Dairy Sci. 52:1448-1449. [DOI] [PubMed] [Google Scholar]
- 11.Joosten, H. M. L. J., and M. Nunez. 1995. Adsorption of nisin and enterocin 4 to polypropylene and glass surfaces and its prevention by Tween 80. Lett. Appl. Microbiol. 21:389-392. [Google Scholar]
- 12.Kimoto, H., M. Nomura, M. Kobayashi, K. Mizumachi, and T. Okamoto. 2003. Survival of lactococci during passage through mouse digestive tract. Can. J. Microbiol. 49:707-711. [DOI] [PubMed] [Google Scholar]
- 13.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 14.Klijn, N., A. H. Weerkamp, and W. M. De Vos. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61:2771-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Møller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loessner, M., S. Guenther, S. Steffan, and S. Scherer. 2003. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl. Environ. Microbiol. 69:1854-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lungu, B., and M. G. Johnson. 2005. Fate of Listeria monocytogenes inoculated onto the surface of model Turkey frankfurter pieces treated with zein coatings containing nisin, sodium diacetate, and sodium lactate at 4 degrees C. J. Food Prot. 68:855-859. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, J. M., M. I. Martinez, A. M. Suarez, C. Herranz, P. Casaus, L. M. Cintas, J. M. Rodriguez, and P. E. Hernandez. 1998. Generation of polyclonal antibodies of predetermined specificity against pediocin PA-1. Appl. Environ. Microbiol. 64:4536-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattila, K., P. Saris, and S. Työpponen. 2003. Survival of Listeria monocytogenes on sliced cooked sausage after treatment with pediocin AcH. Int. J. Food. Microbiol. 89:281-286. [DOI] [PubMed] [Google Scholar]
- 20.McKay, L. L., K. A. Baldwin, and P. M. Walsh. 1980. Conjugal transfer of genetic information in group N streptococci. Appl. Environ. Microbiol. 40:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKay, L. L., K. A. Baldwin, and E. A. Zottola. 1972. Loss of lactose metabolism in lactic streptococci. Appl. Microbiol. 23:1090-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson, L., H. H. Huss, and L. Gram. 1997. Inhibition of Listeria monocytogenes on cold-smoked salmon by nisin and carbon dioxide atmosphere. Int. J. Food. Microbiol. 38:217-227. [DOI] [PubMed] [Google Scholar]
- 24.Rada, V., and J. Petr. 2000. A new selective medium for the isolation ofglucose non-fermenting bifidobacteria from hen caeca. J. Microbiol. Methods 43:127-132. [DOI] [PubMed] [Google Scholar]
- 25.Rollema, H. S., J. W. Metzger, P. Both, O. P. Kuipers, and R. J. Siezen. 1996. Structure and biological activity of chemically modified nisin A species. Eur. J. Biochem. 241:716-722. [DOI] [PubMed] [Google Scholar]
- 26.Schlundt, J., P. Saadbye, B. Lohmann, B. L. Jacobsen, and E. M. Nielsen. 1994. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microb. Ecol. Health Dis. 7:59-69. [Google Scholar]
- 27.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1989. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suarez, A. M., J. M. Rodriguez, P. E. Hernandez, and J. I. Azcona-Olivera. 1996. Generation of polyclonal antibodies against nisin: immunization strategies and immunoassay development. Appl. Environ. Microbiol. 62:2117-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tramer, J., and G. G. Fowler. 1964. Estimation of nisin in foods. J. Sci. Food Agric. 15:522-528. [Google Scholar]
- 30.Vandenbergh, P. A. 1993. Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol. Rev. 12:221-237. [Google Scholar]