Abstract
3′-Phosphoadenosine-5′-phosphatase (PAPase) is required for the removal of toxic 3′-phosphoadenosine-5′-phosphate (PAP) produced during sulfur assimilation in various eukaryotic organisms. This enzyme is a well-known target of lithium and sodium toxicity and has been used for the production of salt-resistant transgenic plants. In addition, PAPase has also been proposed as a target in the treatment of manic-depressive patients. One gene, halA, which could encode a protein closely related to the PAPases of yeasts and plants, was identified from the cyanobacterium Arthrospira (Spirulina) platensis. Phylogenic analysis indicated that proteins related to PAPases from several cyanobacteria were found in different clades, suggesting multiple origins of PAPases in cyanobacteria. The HalA polypeptide from A. platensis was overproduced in Escherichia coli and used for the characterization of its biochemical properties. HalA was dependent on Mg2+ for its activity and could use PAP or 3′-phosphoadenosine-5′-phosphosulfate as a substrate. HalA is sensitive to Li+ (50% inhibitory concentration [IC50] = 3.6 mM) but only slightly sensitive to Na+ (IC50 = 600 mM). The salt sensitivity of HalA was thus different from that of most of its eukaryotic counterparts, which are much more sensitive to both Li+ and Na+, but was comparable to the PAPase AtAHL (Hal2p-like protein) from Arabidopsis thaliana. The properties of HalA could help us to understand the structure-function relationship underlying the salt sensitivity of PAPases. The expression of halA improved the Li+ tolerance of E. coli, suggesting that the sulfur-assimilating pathway is a likely target of salt toxicity in bacteria as well.
The enzyme 3′-phosphoadenosine-5′-phosphatases (PAPases) have drawn extensive attention during the last few years as a target of sodium and lithium toxicity (4, 8, 22). In plants and yeasts, these enzymes are involved in salt resistance and have been used for the production of salt-resistant transgenic crops (3). These enzymes have also been proposed as a possible target in lithium therapy in the treatment of manic-depressive patients (22). Their mechanism of action in salt tolerance is well known in the yeast Saccharomyces cerevisiae (8, 16, 17). The substrate of PAPases, 3′-phosphoadenosine-5′-phosphate (PAP), is a toxic metabolite and a side product of sulfate assimilation. Sulfur is an essential element for life in all organisms. It is necessary for the synthesis of the amino acids methionine and cysteine, which are used in turn for the synthesis of other sulfur-containing molecules (25). Sulfate, which is the most common form of sulfur under the oxidizing conditions of the atmosphere, is poorly reactive. In general, sulfate is converted into a more reactive form, sulfide, through several reactions in order to synthesize cysteine. PAP is produced during the reduction of 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to sulfite, and this reaction is catalyzed by PAPS reductase (23, 25). Under the action of PAPase, PAP is converted into AMP and phosphate. Since PAPases are sensitive to sodium or lithium ions, the presence of these ions at toxic levels leads to the accumulation of PAP within cells. High levels of PAP become toxic, since it can inhibit the activity of PAPS reductase as well as that of RNA-processing enzymes (8). An increase in the PAPase levels within cells counteracts the inhibitory effect of lithium or sodium.
The family of PAPases belongs to a superfamily of metal-dependent phosphatases with various substrates. These enzymes contain a fructose-1,6-biphosphate 1-phosphatase (FBPase)/inositol monophosphatase (IMPase)/GlpX-like domain and are sensitive to lithium but dependent on magnesium. Two families have been defined for the FBPase/IMPase/GlpX-like domain-containing protein superfamily, the FBPase family, and the IMPase family. FBPases hydrolyze fructose-1,6-biphosphate and are involved in gluconeogenesis (4, 14, 28). The IMPase family includes PAPases, inositol polyphosphate 1-phosphatases (IPPases), IMPases, and PAP/inositol-1,4-bisphosphate phosphatases (PIPases). These enzymes share a common structural core within the active sites and critical residues required for metal binding. Among the well-characterized enzymes of this superfamily, Hal2p from Saccharomyces cerevisiae and AtAHL (Hal2p-like protein) from Arabidopsis thaliana are PAPases (9, 16, 17) and Tol1 from Schizosaccharomyces pombe and Sal1 from Arabidopsis thaliana are PIPases (15, 21).
Sulfate assimilation pathways are well conserved in plants, yeasts, and bacteria (23). PAPS reductase encoded by cysH is also found in a variety of bacterial genomes sequenced so far. However, to our knowledge, the enzymes involved in PAP hydrolysis are poorly defined for bacteria. The most likely candidate of PAPase is the gene product of cysQ from Escherichia coli, since this gene was shown to be required for sulfate assimilation, and its mutation could be complemented by a HAL2-like gene from rice (18, 20). However, the deduced amino acid sequence from cysQ is only distantly related to PAPases from plants and yeasts, and its enzyme activity has never been determined. We have initiated a study on the properties of a putative Hal2p homolog, called HalA, from the cyanobacterium Arthrospira (Spirulina) platensis. Although Arthrospira platensis is found in freshwater environments, this strain is highly resistant to salt and can even be grown in marine water (13, 26). The mechanism of salt resistance of this cyanobacterium is not known. Studies on PAPases from bacteria may help us to understand the structure-function relationship of salt sensitivity of these enzymes and provide information for the engineering of salt-resistant crops.
MATERIALS AND METHODS
Reagents, media, and culture conditions.
All molecular biological reagents were purchased from New England Biolabs, TaKaRa, or Promega. PAP, PAPS, 5′-ADP, 3′-AMP, ATP, NADP, and d-myo-inositol-1,4-bis-phophatase (IBP) were from Sigma. E. coli strain BL21(DE3) was cultured at 37°C in Luria-Bertani (LB) medium or minimal medium M9 (24). Ampicillin (100 μg ml−1) and kanamycin (50 μg ml−1) were added when necessary. Arthrospira platensis came from the culture collection of the Institute of Botany, Chinese Academy of Sciences, Beijing, China. It was grown at 30°C in a medium described by Aiba and Ogawa (1).
Cloning and overexpression of halA in E. coli.
The coding region of the putative open reading frame of halA was amplified by PCR, with one primer containing an NdeI site at the 5′ end and the other primer containing an EcoRI site right after the stop codon. The sequences of the two primers were as follows: 5′-CAAGGATCCCATATGCCCTACGATCGCGAAA-3′ and 5′-CTTGAATTCGGCGGGTTGATGATTCTT-3′. The PCR product was cloned into the expression vector pET28a(+) after digestion with NdeI and EcoRI, and the resulting clone was named pET28a-ArHAL. In this construct, the open reading frame of halA was translationally fused to a sequence encoding a His tag which facilitates the purification of the recombinant protein. The accuracy of the cloned DNA fragment was confirmed by DNA sequencing. The construct was then transformed into E. coli strain BL21(DE3).
Protein purification and enzyme assays.
After the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were cultured for 4 h and then collected by centrifugation and resuspended in lysis buffer (30% sucrose, 0.25 M Tris [pH 8.0], 0.5 M KCl, 25 mM EDTA, 5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride). After sonication and treatment with DNase I, the supernatant was separated from cell debris by centrifugation. The expression of the recombinant protein was monitored by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis.
The recombinant fusion protein His tag-HalA was purified by nickel affinity chromatography using a HisTrap kit (Amersham Inc.) under nondenaturing conditions following the manufacturer's instructions. The optimal concentrations of imidazole were 100 mM in the binding buffer and 200 mM in the elution buffer.
Phosphatase assays were performed as described previously with minor modifications (16). Briefly, a standard assay was conducted in a 100-μl reaction mixture containing 50 mM Tris (pH 8.0), 2 mM MgCl2, and the indicated amounts of purified protein and substrate. After 40 min of incubation at 30°C, the inorganic phosphate released during the reaction was quantified using the malachite green method as described by Baykov et al. (5). Under these conditions, the enzyme activity was linear with protein quantity (up to 300 ng) and reaction time (up to 1 h). The Km for PAP was determined by measuring reaction rates at substrate concentrations of 0.1, 0.2, 0.4, 0.7, and 1 mM. The Km for inositol-1,4-bisphosphate was determined by measuring reaction rates at substrate concentrations of 0.1, 0.2, 0.3, 0.4, and 0.5 mM. The Km for PAPS was determined by measuring reaction rates at substrate concentrations of 0.2, 0.4, 0.6, 0.8, and 1 mM. Specific activity was estimated by normalizing to the protein concentration determined by the Bradford method (Bio-Rad), which was consistent with the relative band intensity on the SDS-polyacrylamide gel, using bovine serum albumin as the standard.
Phylogenic analysis and structural modeling.
Phylogenetic construction and illustration were performed using MEGA3.1 and the neighbor-joining method (11). Bootstrap analysis was performed with 3,000 resampling replicates.
The alignment between HalA and Hal2p was produced with ClustalX1.8 (27) and manually optimized based on their secondary structures. This alignment was submitted as the input to the SWISS-MODEL server (http://swissmodel.expasy.org/SWISS-MODEL.html), with the high-resolution X-ray structure of Hal2p (PDB identification, 1ka1A) as a template to build the three-dimensional model of HalA. Swiss Pdb-Viewer was used to view the structure and the Ramachandran plot as well as to produce the stereo figures. The Ramachandran plot of the HalA model showed that 283 residues (88.7%) had their dihedral angles clustered in the most favored regions, 23 residues (7.2%; proline and glycine residues were not counted) in the additional allowed regions, and 9 residues (2.8%; proline and glycine residues were not counted) in the disallowed regions. All the residues having disallowed dihedral angles were located at the coil regions (on the surface of the structure) and far from the active center.
Nucleotide sequence accession number.
The sequence of halA has been deposited into the National Center for Biotechnology Information databank, and the accession number is DQ185137.
RESULTS
Determination of a gene, halA, from Arthrospira platensis encoding a putative plant-like PAPase.
The genome of Arthrospira platensis is currently being sequenced. Preliminary analysis identified several open reading frames whose products were similar to proteins from eukaryotes. One of the open reading frames, halA, is 957 bp long in size and could encode a protein of 319 amino acid residues and a molecular mass of 34.2 kDa. The predicted gene product showed significant sequence identity to Hal2p of Saccharomyces cerevisiae, as well as the PAPases Sal1 and Sal2 of Arabidopsis thaliana (Fig. 1). The three conserved motifs (motifs A, B, and C) of the protein family of magnesium-dependent, lithium-sensitive phosphatases were present (4, 14, 28). These motifs contained all critical residues involved in substrate and metal binding (data not shown), as defined for Hal2p of yeast (2, 19).
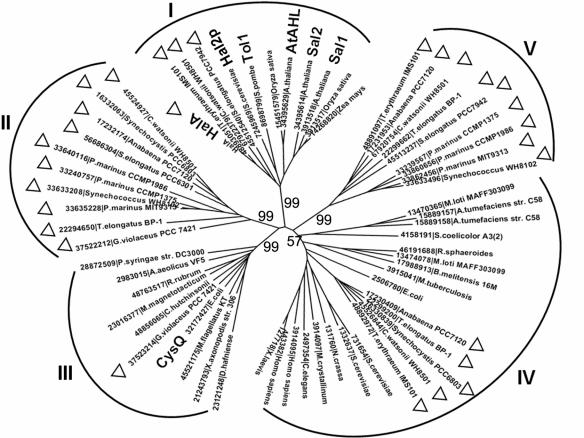
FIG. 1.
Phylogenic analysis of various IMPases. For each protein, the corresponding organisms and the accession number from the databanks are shown. The full names of the species under study, except those already mentioned in the text or the figure, were Agrobacterium tumefaciens C58, Aquifex aeolicus VF5, Brucella melitensis 16M, Caenorhabditis elegans, Cytophaga hutchinsonii, Desulfitobacterium hafniense, Gloeobacter violaceus PCC7421, Magnetospirillum magnetotacticum, Mesembryanthemum crystallinum, Mesorhizobium loti MAFF303099, Methylobacillus flagellatus KT, Mycobacterium tuberculosis, Neurospora crassa, Prochlorococcus marinus MIT9313, Prochlorococcus marinus CCMP1375, Prochlorococcus marinus CCMP1986, Pseudomonas syringae DC3000, Rhodobacter sphaeroides, Rhodospirillum rubrum, Streptomyces coelicolor A3(2), Synechococcus elongatus PCC6301, Thermosynechococcus elongatus BP-1, Xanthomonas axonopodis 306, and Xenopus laevis. Based on this analysis, five different clusters can be clearly defined (clades I through V). Proteins from cyanobacteria are marked with a triangle. Proteins of particular relevance to this work are shown in boldface. Bootstrap values are labeled on the major branches.
A phylogenic analysis was performed with various proteins similar to Hal2p of S. cerevisiae, CysQ of E. coli, and representatives of IMPases or PIPases (Fig. 1). Also included in this analysis were putative orthologues from various cyanobacterial strains available in databanks. As shown in Fig. 1, five distinct clades (clades I through V) could be defined based on protein sequence similarity. Clusters II and V consist exclusively of cyanobacterial proteins, and their catalytic domains are more closely related to IMPases according to protein domains defined in the Pfam database. Cluster I includes HalA, Hal2p, and Sal1, as well as three additional cyanobacterial members from, respectively, the marine filamentous diazotrophic strain Trichodesmium erythraeum, the marine unicellular diazotrophic strain Crocosphaera watsonii (strain WH8501), and the freshwater unicellular strain Synechococcus elongatus PCC 7942. All members of clade III are of bacterial origin, whereas those from clades I and IV are from both prokaryotic and eukaryotic organisms. In some cases, members of this protein family from the same organism were found in different clades. In Anabaena strain PCC 7120, for example, three proteins belonging to IMPases were found in clades II, IV, and V, respectively.
HalA is a magnesium-dependent PAPase.
The coding region of halA was cloned in an expression vector, and its corresponding protein was produced in E. coli as a fusion product with the His tag. After induction with IPTG and protein separation on an SDS-12% polyacrylamide gel, a polypeptide with an apparent molecular mass of 41.8 kDa could be identified (Fig. 2). The size of this polypeptide was slightly higher than the theoretical molecular mass of the recombinant protein (36.4 kDa) predicted from the corresponding DNA sequence of halA together with the His tag sequence. As expected, the BL21(DE3) strain transformed with the vector pET28a(+) without insert did not give rise to the corresponding polypeptide (Fig. 2). Purification using a nickel affinity chromatography column (see Materials and Methods) yielded the recombinant protein with good homogeneity (Fig. 2).
FIG. 2.
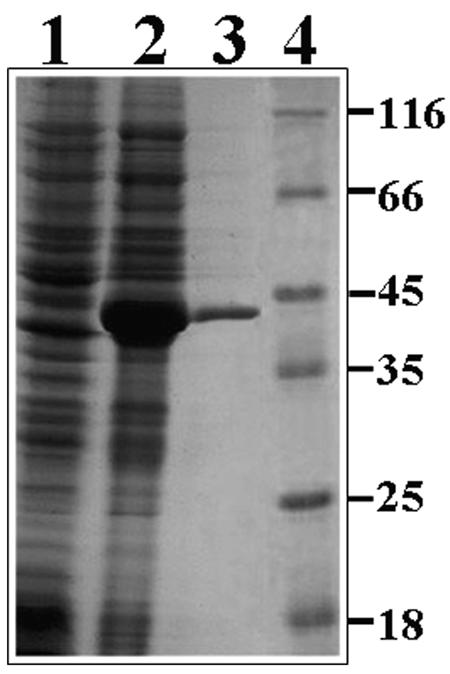
Analysis of the purified HalA on an SDS-12% polyacrylamide electrophoresis gel stained with Coomassie brilliant blue. Lane 1, protein extract from E. coli transformed with plasmid pET28a(+) (control); lane 2, protein extract from E. coli transformed with plasmid pET28a-ArHAL (containing the coding region of the halA gene); lane 3, HalA after purification in a HisTrap affinity column; lane 4, molecular mass standards in kilodaltons.
Protein sequence comparison suggested that HalA could be a PAPase or a PIPase by using both IBP and PAP as substrates (Fig. 1). A variety of phosphorylated compounds were tested as substrates for purified HalA (Table 1). The results showed that HalA had a narrow substrate specificity, with PAP serving as the best substrate, followed by PAPS. The activity by using PAPS as substrate represented 42% of that obtained using PAP as substrate. A weak enzymatic activity was detected with ADP, but no activity was detectable with IBP, 3′-AMP, ATP, or NADP. Therefore, the HalA protein is a 3′,5′-bisphosphate nucleotidase with PAP as the best substrate.
TABLE 1.
Substrate specificity of the phosphatase activity of HalA
| Substrate | Relative activity | Km (mM) | Vmax (mM min−1 mg−1) |
|---|---|---|---|
| PAP | 100 | 1.03 | 3.98 |
| PAPS | 42 | 0.61 | 1.37 |
| 5′-ADP | 5.6 | 41.5 | 0.26 |
| IBP | <1 | NDa | ND |
| ATP | <1 | ND | ND |
| 3′-AMP | <1 | ND | ND |
| NADP | <1 | ND | ND |
ND, not determined.
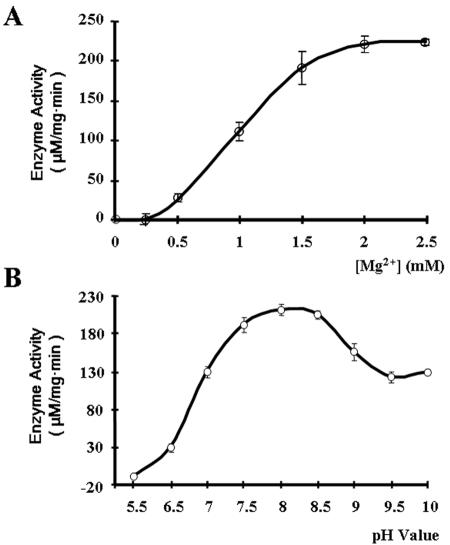
HalA, similar to all other characterized PAPases (28), had an absolute requirement for Mg2+ for its activity. No activity was detectable in the absence of Mg2+, and the activity increased over increasing concentrations of Mg2+ to reach its optimal values in the presence of 2 to 2.5 mM Mg2+ (Fig. 3). The PAPase activity of HalA was favored at slightly basic pH values, with the optimal pH at 8; it was strongly inhibited at an acidic pH but remained at a level corresponding to 60% of its maximal activity when the pH of the reaction sample increased to 10 (Fig. 3).
FIG. 3.
Effects of Mg2+ (A) and pH (B) on the activity of HalA. Error bars indicate standard deviations.
In the presence of 2 mM Mg2+, at pH 8, the specific activity of HalA displayed a hyperbolic dependence on the PAP concentrations. Using PAP as the substrate, the Vmax was determined to be 3.98 mM Pi min−1 mg of protein−1 and the Km to be 1.03 mM (Table 1). When PAPS was used as the substrate, the Vmax and Km were estimated to be 1.37 mM Pi min−1 mg of protein−1 and 0.61 mM, respectively.
Li+ inhibition.
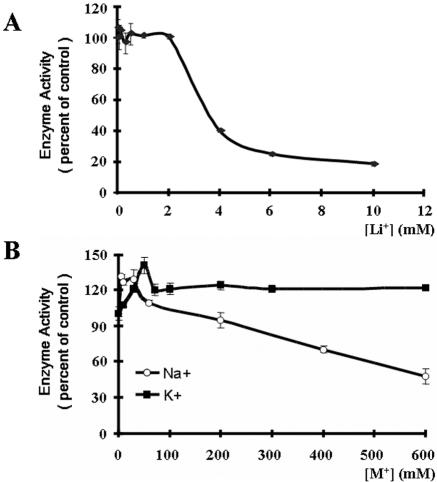
Most PAPases from yeasts and plants are strongly inhibited by Li+ ions and to a lesser extent by Na+ ions (4, 8, 22). The effects of several ions on the activity of HalA were thus tested. When the concentration of Li+ was 2 mM or less, little inhibitory effect on the PAPase activity of HalA was observed. However, once the concentration of Li+ was increased to more than 2 mM, a drastic inhibitory effect on the enzyme activity was obtained. IC50, an index measuring the concentration of an inhibitor to reduce the enzyme activity by 50%, was estimated to be 3.6 mM (Fig. 4). To determine if the effect of Li+ on the activity of HalA was specific, LiCl was replaced with NaCl or KCl. The results showed that K+ at a concentration as high as 600 mM had no inhibitory effect on the activity of HalA. When the concentration of K+ was at 50 mM, a stimulatory effect was observed (Fig. 4). Low concentrations of Na+ stimulated the activity of HalA as K+, and as its concentrations increased, a weak inhibitory effect was found (IC50 = 600 mM).
FIG. 4.
Effects of various ions on the enzyme activity of HalA with 0.5 mM PAP as substrate. The activity was measured in a buffer containing 50 mM Tris, pH 8.0, and 2 mM MgCl2 at 30°C for 40 min. The activity obtained at optimal conditions was considered 100%, and that obtained in the presence of ions was normalized to the optimal activity. Error bars indicate standard deviations.
Expression of halA improves Li+ tolerance of E. coli.
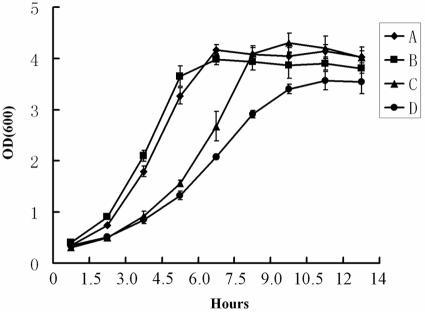
No reliable genetic transformation system is available for Arthrospira platensis. Since key components of sulfur metabolism seem to be conserved in various organisms (23, 25), we sought to determine whether the expression of halA in E. coli could have any effect on Li+ tolerance (Fig. 5). E. coli strain BL21 was transformed by the pET28a(+) vector alone or pET28a(+) with the coding region of halA under the control of the IPTG-inducible promoter (plasmid pET28a-ArHAL). E. coli was grown in either the rich LB medium containing readily available amino acids or minimal medium M9 supplemented with 2 mM MgSO4 as the sulfur source. In both media, the synthesis of HalA could be induced with 0.04 mM IPTG, as visualized by SDS-polyacrylamide gel electrophoresis (data not shown). When cells were grown in the LB medium, the induction of HalA synthesis by IPTG made no difference to the growth rate in the presence or absence of lithium (data not shown). When grown in the M9 medium with 0.04 mM IPTG, both transformed strains grew similarly (Fig. 5). In M9 medium supplemented with 0.4 M LiCl and 0.04 mM IPTG, the strain transformed by pET28a(+) bearing halA showed a better growth rate than the control strain under the same conditions. When sulfite was used as the only sulfur source, the two strains grew better than those in the presence of sulfate, and no difference was observed with or without HalA in the presence of LiCl (data not shown). These results suggest that the expression of halA conferred a better resistance to LiCl in E. coli and that the reduction steps of sulfate to sulfite were the likely target of Li+ inhibition, a situation similar to that found in plants and yeasts (4, 8, 16, 17).
FIG. 5.
Effects of the expression of halA in E. coli on the tolerance toward LiCl. E. coli strain BL21 (DE3) was transformed with either pET28a-ArHAL (A and C) or pET28a(+) (B and D). The two transformed strains were grown in M9 medium supplemented with MgSO4 as the sulfur source and 0.04 mM IPTG as the inducer. For A and B, no LiCl was added; for C and D, 0.4 M LiCl was added. The growth rate was followed by measuring the optical density (OD) by spectroscopy at 600 nm. Error bars show the results of three independent experiments.
DISCUSSION
In the present study, we showed that proteins similar to IMPases are widespread among cyanobacteria. Phylogenic analysis indicated that these proteins were in different clades; two clades consist exclusively of cyanobacterial members, while others have either only prokaryotic members or both prokaryotic and eukaryotic ones. These results suggest that IMPases have multiple evolutionary origins in cyanobacteria. Some species have more than one member distributed among different clades, and such enzymes might use different substrates, such as inositol monophosphate or inositol-1,4-bisphosphate.
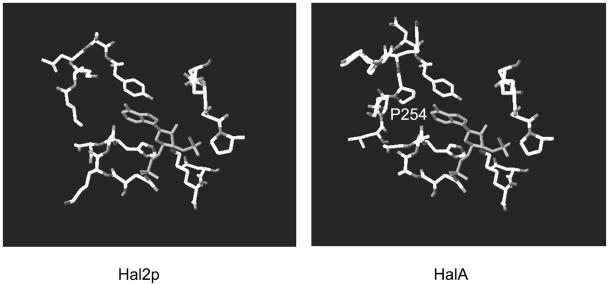
Protein sequence alignment suggested that HalA could have a PAPase activity. This was confirmed by biochemical analysis indicating that HalA could use PAP as the best substrate tested. These data were comparable to those obtained with Hal2p-like protein (AtAHL) from Arabidopsis thaliana (9). The activity of AtAHL by using PAPS as the substrate was 52% of that obtained using PAP. By comparison with other PAPases from yeasts and plants, HalA had a high Km value and reaction velocity (Table 2). Structural modeling using the structure of Hal2p from yeast as a template suggested that a very similar substrate-binding pocket was present in HalA (Fig. 6). However, the putative substrate-binding motif of HalA has a proline residue (P254) which might lower its affinity with PAP.
TABLE 2.
Comparison of different parameters of various enzymes
| Enzyme | Substrates | Sensitivity (IC50) (mM) to:
|
Km for PAP (μM) | Family | Source or reference | |
|---|---|---|---|---|---|---|
| Na+ | Li+ | |||||
| HalA | PAP, PAPS | 600 | 3.6 | 1,035 | PAPase | This study |
| Hal2p | PAP, PAPS | 20 | 0.1 | <20 | PAPase | 16 |
| AtAHL | PAP, PAPS | 50 | 10 | 160 | PAPase | 9 |
| Tol1 | PAP, PAPS, IBP | SMa | SMMb | >20 | PIPase | 15 |
| Sal1 | PAP, PAPS, IBP | 200 | 0.2 | 2-10 | PIPase | 21 |
| Sal2 | PAP, PAPS, IBP | 200 | 10 | >20 | PIPase | 9 |
SM, submolar concentration.
SMM, submillimolar concentration.
FIG. 6.
Modeling of the substrate-binding site of HalA. The model was based on the high-resolution X-ray structure of Hal2p from yeast (2, 19). A major difference between Hal2p and HalA was found at the substrate-binding pocket with the presence of the ring structure of Pro254 in HalA.
Similar to other PAPases, HalA was dependent on Mg2+ for its activity. Compared to other PAPases, HalA was relatively less sensitive to Li+ and Na+. Hal2p and Tol1 from yeast and Sal1 from plants are all highly sensitive to Li+ and Na+, while HalA and AtAHL are much less sensitive to these ions (Table 2). Structural analysis using X-ray crystallography of Hal2p from yeast indicates that lithium ions bind to the ternary complex formed by Hal2p, magnesium, and PAP and that they exert their inhibitory effect by blocking the release of the products of PAP hydrolysis from the active site (2, 19). The structural basis of lower sensitivity of HalA and AtAHL is unknown, as the residues of site 2 of Hal2p involved in lithium binding are identically conserved in these two proteins. Some subtle differences in the ternary structure of the enzyme-substrate-Mg2+ complex could be responsible.
It has been shown that the expression of Sal1 of Arabidopsis thaliana enhances salt tolerance in yeast (21). Similarly, the expression of halA in E. coli enhances its resistance to lithium, suggesting that lithium tolerance mediated by HalA-like proteins could also take place in bacteria. This phenotype was observed only when sulfate was used as the sole sulfur source for the growth of E. coli (Fig. 5). In the rich LB medium, where amino acids were readily available, no difference in growth rate was observed between the strain expressing halA and the control strain. Similarly, when sulfite replaced sulfate, both strains grew better and no growth difference was observed with or without LiCl. These results correlate well with the inhibition of sulfur metabolism by lithium in bacteria and the fact that PAP is produced during the reduction of sulfate to sulfite (2, 17). The properties of HalA could be helpful for studying the structure-function relationship of PAPases and understanding their molecular basis of salt resistance. Furthermore, the relatively higher resistance of HalA to salt makes it a good candidate for expression in plants in order to improve the salt resistance of crops.
Sulfur metabolism is poorly understood in cyanobacteria. Several genes involved in sulfate transport have been characterized in Synechococcus elongatus strain PCC7942 (10, 12). Sulfur deprivation has a profound impact on the physiology and relocation of cell resources (7). But, in general, how sulfur metabolism is regulated in response to environmental changes remains largely unknown. Mutants of a cyanobacterial strain resistant to LiCl have been isolated, but the targets of these mutations have never been clearly identified (6). Characterization of enzymes involved in sulfur metabolism could be helpful for the understanding of the regulation of sulfur metabolism in cyanobacteria.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 30125003), the National Hi-Tech Program of China (grant number 2001AA223131), and the Cheung Kong Scholar Foundation.
REFERENCES
- 1.Aiba, S., and T. Ogawa. 1977. Assessment of growth yield of a blue-green alga, Spirulina platensis, in axenic and continuous culture. J. Gen. Microbiol. 102:179-182. [Google Scholar]
- 2.Albert, A., L. Yenush, M. R. Gil-Mascarell, P. L. Rodriguez, S. Patel, M. Martinez-Ripoll, T. L. Blundell, and R. Serrano. 2000. X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity. J. Mol. Biol. 295:927-938. [DOI] [PubMed] [Google Scholar]
- 3.Arrillaga, I., R. Gil-Mascarell, C. Gisbert, E. Sales, C. Montesinos, R. Serrano, and V. Moreno. 1980. Expression of the yeast HAL2 gene in tomato increases the in vitro salt tolerance of transgenic progenies. Plant Sci. 136:219-226. [Google Scholar]
- 4.Atack, J. R., H. B. Broughton, and S. J. Pollack. 1995. Structure and mechanism of inositol monophosphatase. FEBS Lett. 361:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Baykov, A. A., O. A. Evtushenko, and S. M. Avaeva. 1988. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171:266-270. [DOI] [PubMed] [Google Scholar]
- 6.Bhargava, S., R. K. Saxena, P. K. Pandey, and P. S. Bisen. 2003. Mutational Engineering of the cyanobacterium Nostoc muscorum for resistance to growth-inhibitory action of LiCl and NaCl. Curr. Microbiol. 47:5-11. [DOI] [PubMed] [Google Scholar]
- 7.Collier, J. L., and A. R. Grossman. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 174:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dichtl, B., A. Stevens, and D. Tollervey. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16:7184-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil-Mascarell, R., J. M. Lopez-Coronado, J. M. Belles, R. Serrano, and P. L. Rodriguez. 1999. The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 17:373-383. [DOI] [PubMed] [Google Scholar]
- 10.Green, L. S., D. E. Laudenbach, and A. R. Grossman. 1989. A region of a cyanobacterial genome required for sulfate transport. Proc. Natl. Acad. Sci. USA 86:1949-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Laudenbach, D. E., and A. R. Grossman. 1991. Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J. Bacteriol. 173:2739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, C., and A. Vonshak. 2002. Effects of salinity stress on photosystem II function in cyanobacterial Spirulina platensis cells. Physiol. Plant. 114:405-413. [DOI] [PubMed] [Google Scholar]
- 14.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto, R., R. Sugiura, S. Kamitani, T. Yada, Y. Lu, S. O. Sio, M. Asakura, A. Matsuhisa, H. Shuntoh, and T. Kuno. 2000. Tol1, a fission yeast phosphomonoesterase, is an in vivo target of lithium, and its deletion leads to sulfite auxotrophy. J. Bacteriol. 182:3619-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murguia, J. R., J. M. Belles, and R. Serrano. 1995. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267:232-234. [DOI] [PubMed] [Google Scholar]
- 17.Murguia, J. R., J. M. Belles, and R. Serrano. 1996. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 271:29029-29033. [DOI] [PubMed] [Google Scholar]
- 18.Neuwald, A. F., B. R. Krishnan, I. Brikun, S. Kulakauskas, K. Suziedelis, T. Tomcsanyi, T. S. Leyh, and D. E. Berg. 1992. CysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 174:415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel, S., M. Martinez-Ripoll, T. L. Blundell, and A. Albert. 2002. Structural enzymology of Li+-sensitive/Mg2+-dependent phosphatases. J. Mol. Biol. 320:1087-1094. [DOI] [PubMed] [Google Scholar]
- 20.Peng, Z., and D. P. Verma. 1995. A rice HAL2-like gene encodes a Ca2+-sensitive 3′(2′),5′-diphosphonucleoside 3′(2′)-phosphohydrolase and complements yeast met22 and Escherichia coli cysQ mutations. J. Biol. Chem. 270:29105-29110. [DOI] [PubMed] [Google Scholar]
- 21.Quintero, F. J., B. Garciadeblas, and A. Rodriguez-Navarro. 1996. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quiroz, J. A., T. D. Gould, and H. K. Manji. 2004. Molecular effects of lithium. Mol. Interv. 4:259-272. [DOI] [PubMed] [Google Scholar]
- 23.Saito, K. 2004. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136:2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2:145-177. [PubMed] [Google Scholar]
- 26.Stanier, R. Y., and G. Cohen-Bazire. 1977. Phototrophic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31:225-274. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.York, J. D., J. W. Ponder, and P. W. Majerus. 1995. Definition of a metal-dependent/Li(+)-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc. Natl. Acad. Sci. USA 92:5149-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]