Abstract
The filamentous cyanobacteria Planktothrix spp. occur in the temperate region of the Northern hemisphere. The red-pigmented Planktothrix rubescens bacteria occur in deep, physically stratified, and less eutrophic lakes. Planktothrix is a known producer of the toxic heptapeptide microcystin (MC), which is produced nonribosomally by a large enzyme complex consisting of peptide synthetases and polyketide synthases encoded by a total of nine genes (mcy genes). Planktothrix spp. differ in their cellular MC contents as well as the production of MC variants; however, the mechanisms favoring this diversity are not understood. Recently, the occurrence of Planktothrix strains containing all mcy genes but lacking MC has been reported. In this study, 29 such strains were analyzed to find out if mutations of the mcy genes lead to the inability to synthesize MC. Two deletions, spanning 400 bp (in mcyB; one strain) and 1,869 bp (in mcyHA; three strains), and three insertions (IS), spanning 1,429 bp (in mcyD; eight strains), 1,433 bp (in mcyEG; one strain), and 1,433 bp (in mcyA; one strain), were identified. Though found in different genes and different isolates and transcribed in opposite directions, IS were found to be identical and contained conserved domains assigned to transposable elements. Using mutation-specific primers, two insertions (in mcyD and mcyA) and one deletion (in mcyHA) were found regularly in populations of P. rubescens in different lakes. The results demonstrate for the first time that different mutations resulting in inactivation of MC synthesis do occur frequently and make up a stable proportion of the mcy gene pool in Planktothrix populations over several years.
Microcystins (MC) are cyclic heptapetides regularly produced by cyanobacteria of the genera Anabaena, Microcystis, and Planktothrix. MCs are known to be toxic to aquatic biota, livestock, and humans. They are synthesized nonribosomally by multifunctional enzyme complexes via the so-called thiotemplate mechanism (4). The above-mentioned organisms show an impressive diversity in the production of small bioactive peptides (700 to 1,400 Da) other than microcystins, e.g., anabaenopeptins, aeruginosins, microviridins, cyanopeptolines, etc. (6). Typically, isolates differing in the production of these small peptides cannot be discriminated under the microscope or by traditional molecular taxonomic approaches. Our understanding of the mechanisms and recombination processes affecting existing pathways of secondary metabolite synthesis is rather poor. Recently, progress has been made in the elucidation of the genetic basis of MC synthesis for all three main MC producers occurring in freshwater, i.e., Anabaena, Microcystis, and Planktothrix (2, 27, 36). Three gene clusters responsible for the biosynthesis of microcystins, containing 9 or 10 genes (depending on the genus) and spanning >55 kb, have been sequenced. Using information obtained from nonribosomal peptide synthetases and polyketide synthases in general (17, 32) and from tracer feeding experiments (22), the main pathways of MC biosynthesis could be elucidated (36).
Because of high sequence similarities between the mcy gene clusters in the different genera and the coevolution of 16S rRNA genes and mcy genes, a common ancestor for MC synthesis has been suggested (25). According to this theory, the patchy distribution of mcy genes among strains of one species is understood in terms of repeated-loss processes of the mcy gene cluster during cyanobacterial evolution, albeit the maintenance of mcy in some genera points towards an important but so far unknown function. Our recent report on the occurrence of inactive mcy genotypes (i.e., genotypes possessing the mcy genes but lacking MC production) of Planktothrix spp. in nature (13) might be understood as support for the mcy gene loss hypothesis. The inactivation of MC synthesis in an increasing number of strains might be seen as a first step in evolution towards losing the complete mcy gene cluster. Also, a few inactive mcy genotypes have been described for Microcystis. The loss of MC synthesis has been attributed to the accumulation of point mutations (9, 23). However, so far mutations have not been found among genes of the mcy gene cluster.
The aim of this study was to find out whether the inactivation of MC synthesis can be explained by mutations within the mcy gene cluster, and if so, which mutation types can be found in nature and how frequently they occur or if they should rather be considered exceptional.
(The field data reported in this study partly originated from the M.Sc. thesis submitted by L.Q. to the International Institute for Infrastructural Hydraulic and Environmental Engineering (IHE, Delft, The Netherlands) in the course of the International Postgraduate of Limnology Course at the Limnological Institute in Mondsee, Austria.)
MATERIALS AND METHODS
Experimental organisms and culture conditions.
All nine inactive microcystin genotypes of Planktothrix rubescens (Lake Irrsee strains 12, 62, 65, 87, 94, and 95; Lake Wörthersee strain 67; and Lake Mondsee strain 91/1) and P. agardhii (Lake Gjersjoen strain CCAP1459/36), as reported previously (13), and 20 additional P. rubescens strains (Lake Mondsee strains 40, 110, 120, 130, 135, 137, 154, and 194; Lake Grabensee strains 139, 145, 161, 166, 168, 169, and 170; Lake Wolfgangsee strain 160; Lake Fuschlsee strains 165, 167, and 178; and Lake Obertrumer See strain 197) were isolated and analyzed as described previously (13). All strains gave positive signals in PCRs with a mcyA- and mcyB-specific primer pair but did not contain MCs and were screened for DNA mutations as described below. All strains analyzed contained the internal transcribed spacer region of the phycocyanin operon, detected with primers PcPl+ and PcPl−, specific for Planktothrix spp. (13). Based on the current taxonomical classification (34), strains were assigned to either P. agardhii (green pigmented) or P. rubescens (red pigmented). All strains were cultivated in BG11 medium (26) containing 2 mM NaNO3 and 10 mM NaHCO3 at 15°C with continuous light (5 to 10 μmol m−2 s−1; Osram type L30W/77 Fluora light source). Strains were harvested by incubating 2 ml of culture for 1 h on ice, subsequent centrifuging at 13,000 rpm for 10 min, and lyophilizing the pellet in a vacuum centrifuge at 30°C.
Sampling.
P. rubescens was sampled from June to October in the years 2001-2004 by pulling a plankton net (30-μm mesh size) from a depth of 20 m to the surface at the center of a lake. In parallel, quantitative integrated samples were obtained by collecting 1 liter every meter from the surface to a depth of 20 m. Lakes Irrsee, Mondsee, Offensee, Schwarzensee, Wolfgangssee (Upper Austria, Austria), and Wörthersee (Carinthia, Austria) are generally deep and stratified lakes, and except for the oligotrophic Lake Schwarzensee, are classified as mesotrophic (7). Filaments were assigned to the genus Planktothrix according to previously described morphological criteria (1) and counted from Lugol's solution-fixed integrated samples. The enumeration of cells was done by means of an inverted microscope, using the methods of Utermöhl (37) as described previously (13). Aliquots (a few ml from net samples and 3 to 4 liters from integrated samples) were filtered onto glass fiber filters (BM/C; Ederol, Vienna, Austria) under vacuum pressure and stored frozen (−20°C) until DNA extraction.
Genetic analysis.
DNA extraction from strains and field samples was performed by a standard phenol-chloroform procedure as described previously (12). PCR amplifications were performed in reaction mixtures of 20 μl containing 2 μl of QIAGEN PCR buffer (QIAGEN, VWR International, Austria), 1.2 μl MgCl2 (25 mM; QIAGEN), 0.6 μl deoxynucleoside triphosphates (10 mM [each]; MBI Fermentas, St. Leon-Rot, Germany), 1 μl of each primer (10 pmol μl−1), 0.1 μl Taq DNA polymerase (QIAGEN), 13.1 μl sterile Millipore water, and 1.0 μl of genomic DNA (diluted 100-fold). In order to screen the complete Planktothrix mcy gene cluster (2), 28 primer pairs covering the whole mcy gene cluster were designed and used to amplify fragments of 2 kb without interruption (Table 1). DNA mutations were detected via the difference in PCR product sizes in agarose compared to the corresponding PCR products obtained from strain CYA126/8, whose the mcy gene cluster has been sequenced (AJ441056) (2). The PCR thermal cycling protocol included an initial denaturation step at 94°C for 3 min, followed by 35 cycles at 94°C for 30 s, an annealing temperature of 60°C for 30 s, and an elongation temperature of 72°C for 2 min. In the case of nonsuccessful PCRs, a long-range Taq polymerase (Clontech, BD Biosciences, Palo Alto, CA) was used to amplify PCR products (>4 kb) using adjacent primer pairs following the manufacturer's instructions.
TABLE 1.
Oligonucleotide primers used for the detection of mutations within the mcy gene clustera
| Primer no. | Base pair no. | Forward sequence | Tm (°C) | Reverse sequence | Tm (°C) |
|---|---|---|---|---|---|
| 1 | 452-2022 | ATTGATCCCCTGATCAATGATCA | 62.5 | GCTAAACTGGGGCCATTGAGA | 63.5 |
| 2 | 2002-4050 | TCTCAATGGCCCCAGTTTAGC | 63.5 | CGCTGGCTAATACTCCCTCG | 62.7 |
| 3 | 4031-6021 | CGAGGGAGTATTAGCCAGCG | 62.7 | TTTGCATGGAAAGGGATCAATC | 63.1 |
| 4 | 6000-8002 | GATTGATCCCTTTCCATGCAAA | 63.1 | GATCGCTTCCCTGGGAATAATG | 63.9 |
| 5 | 7981-10011 | CATTATTCCCAGGGAAGCGATC | 63.9 | TGACCGATGGGTTTACCTGTG | 62.9 |
| 6 | 9991-12009 | CACAGGTAAACCCATCGGTCA | 62.9 | CCCAGTTTTCCCAAACCTCC | 62.7 |
| 7 | 11990-13989 | GGAGGTTTGGGAAAACTGGG | 62.7 | TCTCCACGCCCATAGCCAT | 63.8 |
| 8 | 13971-16000 | ATGGCTATGGGCGTGGAGA | 63.8 | CTGGCGGATTTCGAGTTGAT | 62.3 |
| 9 | 15981-18021 | ATCAACTCGAAATCCGCCAG | 62.3 | GCGGCCAGATTAGACTGCATT | 63.6 |
| 10 | 18001-20076 | AATGCAGTCTAATCTGGCCGC | 63.6 | GCGATGAGTGTCTGTTTAGTCCG | 63.2 |
| 11 | 20054-21990 | CGGACTAAACAGACACTCATCGC | 63.2 | GATTTCCCGGTTTCATTGAGTG | 62.7 |
| 12 | 21969-23983 | CACTCAATGAAACCGGGAAATC | 62.7 | TTAGCAGCATTGGCTAAGACTGC | 62.9 |
| 13 | 23961-25999 | GCAGTCTTAGCCAATGCTGCTAA | 62.9 | GTTGTGAGATTGTTGGCGCC | 63.8 |
| 14 | 25980-28065 | GGCGCCAACAATCTCACAAC | 63.8 | CTGAAATAACGGTATGTTGATCGGT | 62.3 |
| 15 | 28042-30072 | CCGATCAACATACCGTTATTTCAG | 62.3 | GTTGCGCTTGAATGAAACGG | 63.8 |
| 16 | 30053-32040 | CCGTTTCATTCAAGCGCAAC | 63.8 | GTCCTTTGGGGTCTGTTGGATAC | 62.9 |
| 17 | 32010-34020 | GTATCCAACAGACCCCAAAGGAC | 62.9 | AAGGCTCCCGTTGCTAAAAC | 63.1 |
| 18 | 34001-36035 | GTTTTAGCAACGGGAGCCTT | 63.1 | CGGGAATGGGTTCTCCTGTATAA | 63.2 |
| 19 | 36013-38042 | TTATACAGGAGAACCCATTCCCG | 63.2 | AGGAAGCCACACTCCAACCAT | 63.0 |
| 20 | 38022-40022 | ATGGTTGGAGTGTGGCTTCCT | 63.0 | CTGTTAATTCGGGACGATGGAG | 62.8 |
| 21 | 40001-42060 | CTCCATCGTCCCGAATTAACAG | 62.8 | TTGTCATGGATGTGACGAGCA | 63.3 |
| 22 | 42060-44052 | TGCTCGTCACATCCATGACAA | 63.6 | CTGATTGGACAGAACTGCTTGGA | 63.7 |
| 23 | 44030-46036 | TCCAAGCAGTTCTGTCCAATCAG | 63.7 | GGTAAATGAACGCGGGGAAT | 63.0 |
| 24 | 46017-48031 | ATTCCCCGCGTTCATTTACC | 63.0 | AATCGTCCCAAGTCTCCGGTA | 62.9 |
| 25 | 48011-50000 | TACCGGAGACTTGGGACGATT | 62.9 | GACCGCCCATTCTAGCGATT | 63.5 |
| 26 | 49981-51970 | AATCGCTAGAATGGGCGGTC | 63.5 | AAAACCAAGGGCAGGAGGAA | 63.0 |
| 27 | 51951-54058 | TTCCTCCTGCCCTTGGTTTT | 63.0 | GTGAGTGCCATCCTGACAGCTAT | 62.7 |
| 28 | 54036-55470 | ATAGCTGTCAGGATGGCACTCAC | 62.7 | GCACCCTAACTAACTCTGCAATCG | 63.3 |
The primers allow for the amplification of the whole mcy synthetase gene cluster of Planktothrix (2) and were designed during this study.
In order to detect mutant genotypes directly in the field, primers specific for the detected mutation events were designed (Table 2). The deletion-specific primer pairs produced distinctly shorter amplification products than the amplicons resulting from unmutated gene regions (mcyHA deletion in strain 62, 1,869 bp; mcyB deletion in strain CCAP1459/36, 707 bp). In contrast, the insertion (IS)-specific primer pairs (mcyDIS1, strain 110; mcyAIS, strain 40) consisted of the forward primer binding to a locus within mcyD or mcyA and the reverse primer binding to the inserted gene region or vice versa. All five primer pairs (Table 2) were found to be specific for the corresponding genotype and did not give PCR products for other typically co-occurring Planktothrix strains and other cyanobacteria (Aphanizomenon spp., Microcystis sp., Nostoc sp., and Synechococcus spp.). For field analysis, the PCR thermal cycling protocol included an initial denaturation step at 94°C for 3 min, followed by 35 cycles at 94°C for 30 s, an annealing temperature of 60°C for 30 s, and an elongation temperature of 72°C for 30 s. To control for the occurrence of Planktothrix and the mcy gene cluster, each sample was analyzed with the PcPl+/− primers, amplifying the intergenic spacer region within the phycocyanin operon (PC-IGS), and the peamso+/− primers, amplifying a region within the mcyA gene, as described previously (13). Pilot experiments showed that PCRs using the primer pair PcPl+/− and the mutation-specific primers mcyDIS1 (3′ end) and mcy HA (deletion) all had a detection limit of 10 cells per assay. PCR products (4 μl of the reaction mix) were visualized by electrophoresis on 1.0% (strains) or 1.5% (field samples) agarose in 0.5× TBE (Tris-borate-EDTA buffer), with ethidium bromide staining.
TABLE 2.
Planktothrix strains sequenced for mutations (red-pigmented strains [P. rubescens] and green-pigmented strains [P. agardhii])
| Strain | Yr of isolation | Origin | Type of mutation | Sequence accession no. | Position | Forward primer name, 5′-3′ sequencea | Temp (°C) | Reverse primer name, 5′-3′ sequencea | °C | Product size (bp)b |
|---|---|---|---|---|---|---|---|---|---|---|
| 110 (P. rubescens) | 2001 | Mondsee, Austria | Insertion (mcyDIS1) (1,424 bp) at 11,918 | AM039940 | 11,745 (fwd primer) | mcyDIS1 5′ end F, AAGAAT ATCCCCAAATGCGTTG | 59 | mcyDIS1 5′ end R, GGGAATGTA AGTCTTACTCCATAGGG | 59 | 351 |
| 12,047 (rev primer) | mcyDIS1 3′ end F, CCTTTT TCCCTGATTTTTTCGG | 56 | mcyDIS1 3′ end R, TCGCCCCCA TTTTCGATAA | 55 | 300 | |||||
| 2001 | Mondsee, Austria | Insertion (mcyEGIS) (1,433 bp) at 23,822 | AM055629 | |||||||
| 139, 145, 161, 166, 169, 170 (P. rubescens) | 2001 | Grabensee, Austria | Insertion (mcyDIS2) at 11,918 (1,435 bp) | AM039941- AM039946 | ||||||
| 178 (P. rubescens) | 2001 | Fuschlsee, Austria | Insertion (mcyDIS2) at 11,918 | AM039947 | ||||||
| 40 (P. rubescens) | 2001 | Mondsee, Austria | Insertion (mcyA) (1,433 bp) at 41,274 | AM039939 | 41,167 (fwd primer) | mcyAIS 5′ end F, TCATCT CGTCGTAGATGGG | 58 | mcyAIS 5′ end R, CACTGCTCA ACACCCTAGGATTG | 60 | 455 |
| 41,454 (rev primer) | mcyAIS 3′ end F, CTTTTT CCCTGATTTTTTCGGA | 58 | mcyAIS 3′ end R, TTGAGTGTT ACCGTGATCTCTGC | 58 | 351 | |||||
| 62 (P. rubescens) | 2001 | Irrsee, Austria | Deletion (mcyHA) (1,869 bp) at 32,938 | AM039937 | 32,791 (fwd primer) | Magnetic 1fwd, CCATTTATC GTCTTCGCTCCC | 60 | Magnetic 1rev, CGGCAAATA GACGAACAACATG | 59 | 300 (2223) |
| CCAP1459/36 (P. agardhii) | 1968 | Lake Gjersjoen, Norway | Deletion (mcyB) (400 bp) at 43,423 | AM039938 | 43,226 (fwd primer) | 1459/36 1F, AGAAATCCA AAGGCTGAGTGATG | 58 | 1459/36 301R, AAAGATTGC TGGGTCAGCAAA | 58 | 301 (707) |
The primers for the specific detection of mcyAIS and mcyDIS1 genotypes (on both ends) and mcyHA/mcyB deletion genotypes in field samples and corresponding base pair numbers of the mcy gene cluster (AJ441056) from strain CYA126/8 (2) are given.
Sizes of PCR products obtained without deletions are given in parentheses.
Sequence alignment and analysis.
The PCR products obtained in field samples were sequenced directly from PCR products by standard automated fluorescence techniques (Applied Biosystems, Weiterstadt, Germany). Sequences were aligned using multiple sequence alignment (Clustal W 1.8). Similarity values between nucleotide sequences were calculated using the program DNADIST of the PHYLIP software package (version 3.6[alpha3]) (5).
Nucleotide sequence accession numbers.
The sequence data obtained in this study have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AM039937 to AM039947, AM040461 to AM040485, and AM055629 (Tables 2 and 3).
TABLE 3.
Sequence accession numbers of mutant mcy genotypes obtained from field samples by sequencing of PCR products obtained with mutation-specific primers (Table 2)
| Sampling site | Date | Sequence accession no.
|
||||
|---|---|---|---|---|---|---|
| mcyDIS1 5′ end | mcyDIS1 3′ end | mcyAIS 5′ end | mcyAIS 3′ end | mcyHA deletion | ||
| Irrsee | 1 December 03 | AM040466 | ||||
| Mondsee | 15 April 03 | AM040471 | AM040478 | |||
| Mondsee | 29 April 03 | AM040472 | AM040476 | |||
| Mondsee | 14 October 03 | AM040473 | AM040479 | AM040482 | ||
| Mondsee | 11 November 03 | AM040468 | AM040461 | AM040474 | AM040477 | AM040483 |
| Offensee | 1 July 03 | AM040462, AM040467 | ||||
| Offensee | 10 October 02 | AM040465 | ||||
| Wörthersee | 26 September 01 | AM040469 | AM040463 | AM040480 | AM040484 | |
| Wörthersee | 28 August 03 | AM040470 | AM040464 | AM040475 | AM040481 | AM040485 |
RESULTS
Identification of mutations linked to the inactivation of MC synthesis.
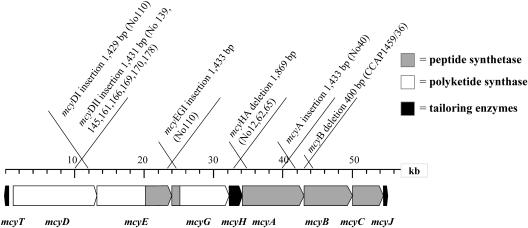
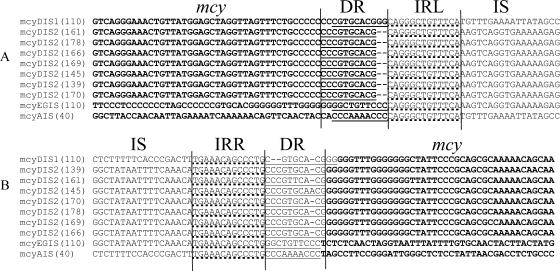
Sequencing of the PCR amplicons that differed in size from the expected 2.0 kb revealed deletions and insertions (Fig. 1; see the supplemental material). Insertions were detected at three different sites by significantly larger-than-expected PCR products, using in one case primer pair 6, binding to mcyD, in the second case primer pair 12, binding to the spacer region between mcyE and mcyG, and in the third case primer pair 21, binding to mcyA. The insertion affecting mcyD (mcyDIS1) was 1,429 bp long and found at position 11,918 of the mcy gene cluster (AJ441056), and it occurred in strain 110. The insertion sequences observed in strains 139, 145, 161, 166, 169, 170, and 178 were located in mcyD at the same position (mcyDIS2). Both mcyDIS1 and mcyDIS2 had a left inverted repeat (IRL; 5′-CAGGGCTGTTTCA-3′) and a right inverted repeat (IRR; 5′-TGAAACAGCCCTG-3′) and were highly similar (99.0 to 99.4% identity), but mcyDIS2 was transcribed in the opposite direction (Fig. 2). The variability among the seven strains containing mcyDIS2 (1,397 bp) was low (0 to 0.6%). Two other insertions with identical IRL and IRR sequences affected the spacer between mcyE and mcyG (1,434 bp; named mcyEGIS) in strain 110 at position 23,822 and affected mcyA (1,433 bp; named mcyAIS) at position 41,274 in strain 40. All mcyDIS and mcyAIS mutants had short directly repeated sequences (DR) of 10 bp in length at the downstream end (mcyDIS1, 5′-CGT GCA CGG G-3′; mcyDIS2, 5′-CCC GTG CAC G-3′; mcyEGIS, 5′-GGC TGT TCC C-3′; and mcyAIS, 5′-CCC AAA ACC C-3′). Consequently, mcyDIS2 transcribed in the opposite direction but inserted at the same position should have originated from a second independent insertion event. All IS showed 98.5 to 99.4% identity and carried a single predicted open reading frame (ORF) (encoding 363 amino acids [aa]) constituting 76% of mcyDIS1 (bp 237 to 1328). The ORF included conserved domains assigned to transposase 11 (E value, 7e − 08), a transposase DDE domain known to be necessary for efficient DNA transposition (3). We concluded that all insertions found among the mcy gene cluster involved this same transposase 11.
FIG. 1.
mcy gene cluster of Planktothrix isolate CYA126/8 (2) and locations of mutations found in inactive mcy genotypes. Strains 110 and 40 were isolated from Lake Mondsee (Austria), strains 139, 145, 161, 166, 169, and 170 were isolated from Lake Grabensee (Austria), strain 178 was isolated from Lake Fuschlsee (Austria), strains 12, 62, and 65 were isolated from Lake Irrsee (Austria), and strain CCAP1459/36 was isolated from Lake Gjersjoen (Norway).
FIG. 2.
Sequence alignment of putative transposases (mcyDIS1 and mcyDIS2, observed in mcyD [at base pair 11,918 of the mcy gene cluster of Planktothrix; AJ441056], mcyEG [at base pair 23,822], and mcyA [at base pair 41,274]). The 5′ end (A) and the 3′ end (B) of each insertion (IS) and part of the mcy genes (bold) are shown. The short directly repeated sequences (DR) of 10 bp in length (straight lines) and the left inverted repeats (IRL) and right inverted repeats (IRR) (dotted lines) are indicated.
Deletions were identified by shorter-than-expected PCR amplicons at two different sites. In one case, PCR amplification constantly failed to give an amplicon with primer pairs 17, 18, and 19. Subsequently, long-range PCR amplification with primers 17fwd and 19rev was performed, yielding an amplicon of only 4 kb instead of the expected 6 kb (data not shown). This deletion (called the mcyHA deletion) was found in strains 12, 62, and 65 and spanned 1,869 bp within mcyH and mcyA, from the Walker motif in mcyH to the core motif A2 of the first adenylation domain (Ad) of mcyA (Ad1). This deletion was only found in genotypes containing mcyA Ad1 without the N-methyltransferase domain and not in genotypes containing mcyA Ad1 with the NMT domain (14). The second deletion (called the mcyB deletion) was identified with primer pair 22 and was located within mcyB Ad1 of strain CCAP1459/36, resulting in the loss of 400 bp located before the core motif A2. For the other 16 strains analyzed, no detectable differences in PCR product size could be identified.
Frequency of occurrence of mutant mcy genotypes in field samples.
In total, 123 samples from six lakes were analyzed, and all samples (except for two integrated samples from Lake Schwarzensee [collected on 15 April 2004 and 8 July 2004]) gave positive PCR signals for PC-IGS and mcyA, implying that Planktothrix and mcy gene-containing genotypes were present (Table 4). The extremely low density (zero filaments in 50 ml of sedimentation volume) in Lake Schwarzensee was responsible for the two negative sampling dates, since the net samples taken in parallel gave PCR products indicative of Planktothrix and mcyA. Otherwise, there was a good correlation between the frequencies of occurrence of PCR results from net samples and integrated samples, indicating that the mcy genotype composition did not differ between plankton net samples (containing a >1,000-fold larger sample of the population in terms of individual numbers) and integrated samples (containing all the individuals sampled from approximately 4 liters of integrated lake water).
TABLE 4.
Occurrence of P. rubescens, biovolume and filament abundance in study lakes, and numbers of integrated and net samples showing specific mutations
| Lake | Sampling time (day.mo.yr) (no. of sampling dates) | Biovolume (mm3 liter−1) (minimum-median-maximum) | No. of filaments per mla (minimum-median-maximum) | Integrated sample (I)/net sample (N) | No. of samples with indicated mutation (relative proportion [%])b
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PC-IGS | mcyA | mcyAIS (insertion) | mcyDIS1 (insertion) | mcyB (deletion) | mcyHA (deletion) | |||||
| Irrsee (Austria) | 9.9.2002-2.8.2004 (18) | 0.001-0.015-0.09 | 0.04-0.7-3.9 | I | 18 (100) | 18 (100) | 0 | 1 (6)* | 0 | 0 |
| 9.9.2002-2.8.2004 (18) | N | 18 (100) | 18 (100) | 0 | 0 | 0 | 0 | |||
| Mondsee (Austria) | 25.9.2001-6.8.2004 (23) | 0.06-0.31-1.81 | 5.9-25.9-227.1 | I | 23 (100) | 23 (100) | 15 (65) | 18 (78) | 0 | 22 (96) |
| 25.9.2001-6.8.2004 (23) | N | 23 (100) | 23 (100) | 14 (61)* | 22 (96)* | 0 | 23 (100)* | |||
| Offensee (Austria) | 5.8.2002-13.7.2004 (5) | 0.02-0.23-0.54 | 1.8-5.7-42.9 | I | 5 (100) | 5 (100) | 0 | 0 | 0 | 0 |
| 5.8.2002-13.7.2004 (6) | N | 6 (100) | 6 (100) | 0 | 2 (33)* | 0 | 0 | |||
| Schwarzensee (Austria) | 10.9.2001-8.7.04 (6) | 0-0-0.01 | 0-0-1.4 | I | 4 (67) | 4 (67) | 0 | 0 | 0 | 0 |
| 10.9.2001-8.7.04 (7) | N | 7 (100) | 7 (100) | 0 | 0 | 0 | 0 | |||
| Wolfgangsee (Austria) | 26.6.2003-21.6.2004 (4) | 0 | 0 | I | 4 (100) | 4 (100) | 0 | 0 | 0 | 0 |
| 26.6.2003-21.6.2004 (4) | N | 4 (100) | 4 (100) | 0 | 0 | 0 | 0 | |||
| Wörthersee (Austria) | 16.10.03-23.8.04 (4) | 1.9-2.2-2.5 | 120-185-250 | I | 4 (100) | 4 (100) | 2 (50) | 4 (100) | 0 | 4 (100) |
| 26.9.2001-23.8.2004 (5) | N | 5 (100) | 5 (100) | 5 (100)* | 5 (100)* | 0 | 5 (100)* | |||
For Lugol counting, the detection limit is one filament in 50 ml sedimentation volume.
*, for sequence accession numbers, see Table 3.
PCR products indicative of insertions (mcyDIS1 and mcyAIS) were found to be ubiquitously distributed, i.e., the mcyDIS1 genotype was found in Lakes Mondsee, Wörthersee, Offensee, and Irrsee and the mcyAIS genotype was found in Lakes Mondsee and Wörthersee (Table 4; see the supplemental material). The mcyHA deletion was detected in Lakes Mondsee and Wörthersee. The mcyB deletion was not detected in field samples. For Lakes Mondsee and Wörthersee, both insertions (mcyDIS1 and mcyAIS) and the mcyHA deletion were found regularly over 4 years. In net samples from Lake Mondsee, the mcyHA deletion and mcyDIS1 were found as frequently as mcyA and PC-IGS. For net samples from Lake Wörthersee, both insertions and the mcyHA deletions occurred with a 100% frequency of detection. For populations showing the lowest abundance of Planktothrix throughout the study period (Lakes Wolfgangsee and Schwarzensee), no mcy mutations were detected.
Similarity of sequences of mcy mutations from field samples.
Twenty-five PCR amplification products of mutant genotypes from field samples were sequenced in order to validate the results of PCR detection and to find out whether the insertion events observed among strains may occur independently in nature. In general, the similarity among sequences of mutations, even if obtained from different populations, was high. Sequence identities for the sequences obtained from the 5′ end and 3′ end of mcyDIS1 were 96.6 to 100% (270 bp; n = 4) and 95.9 to 100% (198 bp; n = 8), respectively. According to the difference in DR sequences of the mcyDIS1 3′ ends, two independently arisen genotypes occurred in lake samples, and one mcyDIS1 3′-end genotype was found in Lake Offensee only (see the supplemental material). In contrast, only one mcyAIS genotype DR sequence was observed, and sequence identities for the 5′ end and 3′ end of mcyAIS were 99.5 to 100% (369 bp; n = 6) and 99 to 100% (298 bp; n = 7), respectively. All mcyHA deletion events occurred at the same position of the mcy gene cluster, and the sequences flanking the mcyHA deletion (266 bp; n = 4) were identical. The results demonstrate that mutations within mcy may arise independently and are detectable for years.
DISCUSSION
Types of naturally occurring mutations.
Of the 29 strains that were inactive in microcystin synthesis, 13 were found to contain mutations within the mcy gene cluster. One strain, no. 110, even contained two mutations (mcyDIS1 and mcyEGIS). Since mcyEGIS was located in the spacer region between mcyE and mcyG, it should not disturb the translation process and result in a nonfunctional protein as mcyDIS1 does. The other 16 strains without detectable MC did not reveal insertions or deletions, and consequently, they may have acquired point mutations within the mcy gene cluster, as suggested by other authors (9, 23). This study is the first showing that mutations do occur frequently within the mcy gene cluster and that a large proportion of mutations are caused by insertion of an IS element at different sites (observed in 9 of 13 strains). A relatively large number of genes encoding putative transposases have been reported for sequenced genomes of Nostoc PCC7120 (10), Nostoc punctiforme (18), and Synechocystis PCC6803 (11). Transposition has been reported to result in different Synechocystis PCC6803 genotypes (24) and to affect gas vesicle genes in Microcystis PCC7806 (20). The IS elements found in this study contained ORFs that putatively encode a transposase of 363 amino acids, including the conserved domains assigned to the DDE motif of the active site, which are involved in DNA cleavage at a specific site followed by a strand transfer reaction (16). It is obvious from the different insertion sites and flanking regions that insertion of this IS element occurred several times. No stop codons were detected within the ORFs, i.e., transposition activity should still be maintained. Notably, characterization of the mcy gene clusters from Anabaena, Microcystis, and Planktothrix and the nda gene cluster from Nodularia revealed associations with transposases (2, 21, 27, 36). Those transposases were members of families distinct from that of the transposase observed in this study.
Frequency of occurrence of mutations.
Although mcyDIS1, mcyAIS, and mcyHA were found ubiquitously distributed, the populations investigated in this study were found to differ in the frequency of occurrence of these mutations. Since PCR assays for the total population (PlPc+/−) and for the mcyDIS1 3′ end and mcyHA were shown to have the same detection limit, the lower abundance in some populations is considered unbiased by the PCR approach used in this study. The results correspond to an earlier quantification of inactive mcy genotypes (13) in Lake Mondsee (21% inactive genotypes) and Lake Irrsee (5% inactive genotypes) and may imply that transposase-mediated mutations do occur more frequently in populations with a larger number of individuals. The relatively high proportion of mcy mutants occurring in populations is surprising since it indicates that MC is not needed for the survival of individual cells in nature. Moreover, the mutants have existed for long periods of time, as indicated by the detection of some of them in different lakes and by point mutations that allow for discrimination between genotypes bearing the same mutation. Indeed, directed mutagenesis of mcy genes in Microcystis PCC7806 revealed no difference in growth between mutant and wild-type cells under different light conditions (8). On the other hand, no population has been found without any MC production, and it may be assumed that MC as a toxin is necessary to keep the ambient density of a specific group of potential antagonists low (15, 28, 29, 31). Thus, it may be sufficient if some of the genotypes in a population produce MC in defense against grazers or other organisms.
Microevolution of mcy genes.
The mutants identified during this study support the idea of a frequent loss of the ability to synthesize MC during evolution (25). On the other hand, the inactivation of the mcy gene cluster by transposable elements as observed in this study might be seen as an intermediate step in reorganization of the mcy gene cluster towards cell types with modified MC synthesis. Typically, organisms that lack an immune system are prolific producers of secondary metabolites (33). Recombination has been recognized as a general feature in the formation of mcy gene clusters for the synthesis of new structural variants of MC (14, 19, 35) and the related toxic peptide nodularin (21). Generally, transposases have been recognized as a major factor in the rearrangement of genes (30). In this study, mcyE was found flanked by two functional IS modules resembling transposon elements shown to mobilize DNA sequences encoding antibiotic resistance. It remains to be established whether there is a connection between the recombination processes observed within mcy genes, resulting in the formation of new MC variants, and the nonrandom mutations induced by transposable elements observed in this study.
Supplementary Material
Acknowledgments
We are grateful to Martin Meixner for DNA sequencing. Johanna Schmidt provided excellent technical assistance. We are grateful to the Federal Agency for Water Management, Scharfling (Upper Austria), Austria, for field sampling (by Günter Bruschek and Karl Mayrhofer). Gerti Roidmayr counted Planktothrix rubescens cell numbers.
This study was supported by grant P15709 from the Austrian Science Fund to R.K. (Linking CYanoTOxin production to GENEtic diversity).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anagnostidis, K., and J. Komarek. 1988. Modern approach to the classification system of cyanophytes, 3-Oscillatoriales. Algol. Stud. 50-53:327-472. [Google Scholar]
- 2.Christiansen, G., J. Fastner, M. Erhard, T. Börner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies, D. R., L. M. Braam, W. S. Reznikoff, and I. Rayment. 1999. The three-dimensional structure of a Tn5 transposase-related protein determined to a 2.9-A resolution. J. Biol. Chem. 274:11904-11913. [DOI] [PubMed] [Google Scholar]
- 4.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle, Wash.
- 6.Fujii, K., K. Sivonen, E. Naganawa, and K. Harada. 2000. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron 56:725-733. [Google Scholar]
- 7.Gassner, H., A. Jagsch, D. Zick, G. Bruschek, and I. Frey. 2002. Die Wassergüte ausgewählter Seen des oberösterreichischen und steirischen Salzkammergutes, Band 15. Schriftenreihe BAW, Vienna, Austria.
- 8.Hesse, K., E. Dittmann, and T. Börner. 2001. Consequences of impaired microcystin production for light-dependent growth and pigmentation of Microcystis aeruginosa PCC 7806. FEMS Microbiol. Ecol. 37:39-43. [Google Scholar]
- 9.Kaebernick, M., T. Rohrlack, K. Christoffersen, and B. A. Neilan. 2001. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 12.Kurmayer, R., G. Christiansen, and I. Chorus. 2003. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis and determines its microcystin net production in Lake Wannsee. Appl. Environ. Microbiol. 69:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurmayer, R., G. Christiansen, J. Fastner, and T. Börner. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831-841. [DOI] [PubMed] [Google Scholar]
- 14.Kurmayer, R., G. Christiansen, M. Gumpenberger, and J. Fastner. 2005. Genetic identification of microcystin ecotypes in toxic cyanobacteria of the genus Planktothrix. Microbiology 151:1525-1533. [DOI] [PubMed] [Google Scholar]
- 15.Kurmayer, R., and F. Jüttner. 1999. Strategies for the co-existence of zooplankton with the toxic cyanobacterium Planktothrix rubescens in Lake Zürich. J. Plankton Res. 21:659-683. [Google Scholar]
- 16.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 18.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Phycol. Res. 70:85-106. [DOI] [PubMed] [Google Scholar]
- 19.Mikalsen, B., G. Boison, O. M. Skulberg, J. Fastner, W. Davies, T. M. Gabrielsen, K. Rudi, and K. S. Jakobsen. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mlouka, A., K. Comte, A.-M. Castets, C. Bouchier, and N. Tandeau de Marsac. 2004. The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J. Bacteriol. 186:2355-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffitt, M. C., and B. A. Neilan. 2004. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Environ. Microbiol. 70:6353-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, R. E., J. L. Chen, B. S. Moore, and G. M. L. Patterson. 1991. Biosynthesis of microcystin-LR. Origin of the carbons in the Adda and Masp units. J. Am. Chem. Soc. 113:5083-5084. [Google Scholar]
- 23.Nishizawa, T., M. Asayama, K. Fujii, K. I. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520-529. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto, S., M. Ikeuchi, and M. Ohmori. 1999. Experimental analysis of recently transposed insertion sequences in the cyanobacterium Synechocystis sp. PCC6803. DNA Res. 6:265-273. [DOI] [PubMed] [Google Scholar]
- 25.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-27. [DOI] [PubMed] [Google Scholar]
- 27.Rouhiainen, L., T. Vakkilainen, B. L. Siemer, W. Buikema, R. Haselkorn, and K. Sivonen. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sedmak, B., and G. Kosi. 1998. The role of microcystins in heavy cyanobacterial bloom formation. J. Plankton Res. 20:691-708. [Google Scholar]
- 29.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro, J. A. 1999. Transposable elements as the key to a 21st century view of evolution. Genetica 107:171-179. [PubMed] [Google Scholar]
- 31.Skulberg, O. M. 2000. Microalgae as a source of bioactive molecules—experiences from cyanophyte research. J. Appl. Phycol. 12:341-348. [Google Scholar]
- 32.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 33.Stone, M. J., and D. H. Williams. 1992. On the evolution of functional secondary metabolites (natural products). Mol. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 34.Suda, S., M. M. Watanabe, S. Otsuka, A. Mahakahant, W. Yongmanitchai, N. Nopartnaraporn, Y. Liu, and J. G. Day. 2002. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 52:1577-1595. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe, Y., K. Kaya, and M. M. Watanabe. 2004. Evidence for recombination in the microcystin synthetase (mcy) genes of toxic cyanobacteria Microcystis spp. J. Mol. Evol. 58:633-641. [DOI] [PubMed] [Google Scholar]
- 36.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 37.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplanktonmethodik. Mitt. Int. Verein. Limnol. 2:1-38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.