Abstract
Functional and comparative genomic studies have previously shown that the essential protein lysyl-tRNA synthetase (LysRS) exists in two unrelated forms. Most prokaryotes and all eukaryotes contain a class II LysRS, whereas most archaea and a few bacteria contain a less common class I LysRS. In bacteria the class I LysRS is only found in the α-proteobacteria and a scattering of other groups, including the spirochetes, while the class I protein is by far the most common form of LysRS in archaea. To investigate this unusual distribution we functionally annotated a representative phylogenetic sampling of LysRS proteins. Class I LysRS proteins from a variety of bacteria and archaea were characterized in vitro by their ability to recognize Escherichia coli tRNALys anticodon mutants. Class I LysRS proteins were found to fall into two distinct groups, those that preferentially recognize the third anticodon nucleotide of tRNALys (U36) and those that recognize both the second and third positions (U35 and U36). Strong recognition of U35 and U36 was confined to the pyrococcus-spirochete grouping within the archaeal branch of the class I LysRS phylogenetic tree, while U36 recognition was seen in other archaea and an example from the α-proteobacteria. Together with the corresponding phylogenetic relationships, these results suggest that despite its comparative rarity the distribution of class I LysRS conforms to the canonical archaeal-bacterial division. The only exception, suggested from both functional and phylogenetic data, appears to be the horizontal transfer of class I LysRS from a pyrococcal progenitor to a limited number of bacteria.
Aminoacyl-tRNA (aa-tRNA) synthesis is the process whereby an amino acid is esterified to the 3′ end of a tRNA containing the corresponding anticodon sequence (15). After their synthesis, aa-tRNAs are screened and selected by elongation factor Tu (16) and then taken to the ribosomal decoding site, where they are paired with the complementary mRNA codons (19). Consequently, the availability of a cellular pool of correctly aminoacylated tRNAs is essential for the faithful translation of the genetic information encoded in mRNA into the corresponding polypeptide sequence (14). Aminoacyl-tRNAs are primarily made by the aminoacyl-tRNA synthetase (aaRS) protein family (9). The accuracy of aa-tRNA synthesis by the aaRSs is ensured by their extremely high substrate specificity, which is further enhanced in some cases by the existence of proofreading mechanisms. The 20 aaRS proteins, as for example found in Escherichia coli (6), are divided into two mutually exclusive structural groups of 10 members each, termed class I and class II (reviewed in references 1 and 5). The assignment of an aaRS specific for a particular amino acid to one of the two structural classes is almost completely conserved, reflecting the ancient evolutionary origin of this family of proteins (21).
The 20 canonical aaRS proteins do not represent the sole means of cellular aa-tRNA synthesis. It was first observed in Bacillus megaterium that Gln-tRNAGln could be synthesized via Glu-tRNAGln without the use of glutaminyl-tRNA synthetase (GlnRS) (32), and subsequent studies showed that comparable indirect pathways exist for the production of Asn-tRNAAsn and selenocysteinyl-tRNASec (reviewed in reference 11). While the use of these pathways was initially assumed to be idiosyncratic to a small minority of organisms, recent analyses of completed microbial genome sequences has shown them to be widespread in prokaryotes with the corresponding direct pathways absent. GlnRS is absent from all characterized archaea and the majority of bacteria, while asparaginyl-tRNA synthetase (AsnRS) is absent from most archaea and a significant number of bacteria (31). Initial analyses of the complete genome sequences of the archaea Methanococcus jannaschii (3) and Methanobacterium thermoautotrophicum (24) suggested that in addition to AsnRS and GlnRS, genes encoding the cysteinyl-tRNA synthetase (CysRS) and lysyl-tRNA synthetase (LysRS) might also be absent from some organisms. Experimental studies showed that while both CysRS (17, 26) and LysRS (13) are present in archaea, they are encoded by novel or reassigned genes, making the original annotation of the corresponding open reading frames difficult. In the case of LysRS experimental approaches identified a gene encoding a class I aaRS (lysK) in the archaeon Methanococcus maripaludis, in contrast to all previously characterized LysRS proteins that belonged to class II. Comparative genomic analyses showed class II LysRS-encoding genes (lysS) to be absent from the majority of archaea and some bacteria, with lysK being found instead in all cases (e.g., see reference 10). Functional (12) and structural (29) characterizations have shown that the class I and II LysRS proteins are functionally equivalent but structurally unrelated.
Primarily as a result of the sequencing of numerous microbial genomes, over 30 lysK gene sequences are now known. The distribution of organisms containing lysK, which in all but the archaeal Methanosarcina group is to the exclusion of lysS, encompasses most of the archaea and a scattering of bacteria but no eukaryotes. Previous studies have attributed this unusual evolutionary pattern to either horizontal gene transfer (12, 33, 34) or selective retention from the last common ancestor in particular lineages (22). To further investigate the evolution of the lysK gene family, we have now extended our previous preliminary studies and attempted to differentiate a range of class I LysRS proteins based upon their mode of recognition of tRNALys. Comparison of these results with a revised phylogenetic tree of class I LysRS sequences suggests that selective retention was the major factor in the evolution of this protein family with horizontal gene transfer playing a relatively minor role.
MATERIALS AND METHODS
General.
In vivo overproduced E. coli tRNALys variants, in which the anticodon loop nucleotides 34 and 37 have previously been shown to be unmodified (Fig. 1) (4), were prepared as described before (4), as was Borrelia burgdorferi class I LysRS protein (10). Aminoacylation assays were performed as previously described (12). Methanosarcina barkeri fusaro genomic DNA was a gift from Kevin Sowers (University of Maryland Biotechnology Institute, Baltimore), Magnetospirillum magnetotacticum genomic DNA was a gift from Elizabeth Bertani (Division of Biology, California Institute of Technology, Pasadena), Sphingomonas aromaticivorans cells were a gift from Margaret Romine (Pacific Northwest National Lab, Richland, Wash.) and Ferroplasma acidarmanus genomic DNA was a gift from Paul F. Predki (DOE Joint Genome Institute, Walnut Creek, Calif.). S. aromaticivorans cells were grown in 50% Luria broth for 36 h at 30°C and genomic DNA was then prepared according to standard procedures (23).
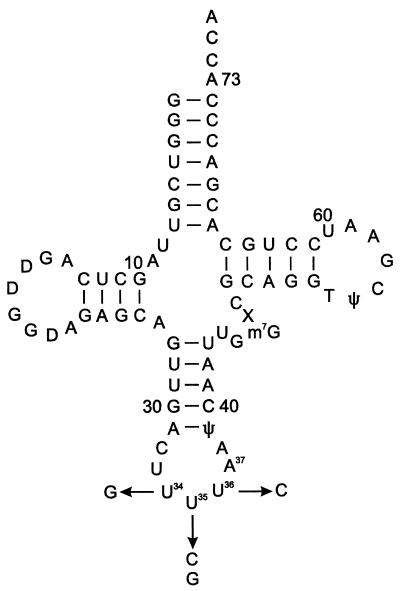
FIG. 1.
Secondary structure of undermodified E. coli tRNALys. Abbreviations: D, dihydrouridine; ψ, pseudouridine; m7G, 7-methylguanosine; X, putative 3-(3-amino-3-carboxypropyl) uridine modification. The undermodified variants used in this study contain uridine in place of 5-methylaminomethyl-2-thiouridine at position 34 and adenosine in place of N6-threonylcarbamoyladenosine at position 37. Changes in the anticodon nucleotides (nucleotides 34 to 36) are indicated.
Cloning of lysK and lysS genes.
LysRS encoding genes were amplified from genomic DNAs as indicated under standard PCR conditions using the following primers (engineered restriction enzyme recognition sites are indicated and underlined for each primer): for S. aromaticivorans lysK, Forward: (NdeI) 5′-GCATATGACAGACGAGATCCGCACC, and Reverse: (BamHI) 5′-GGATCCTCAGTCGAGCGCCTCGGC; for M. barkeri lysK, Forward: (XhoI) 5′-GCTCGAGATGGCTGACACAATTCACTGG, and Reverse: (BlpI) 5′-GCTGAGCTTATGCCTGTTTTTCGGGGC; for M. barkeri lysS, Forward: (NdeI) 5′-GCATATGAGCATGGAAATTAACAATGAAAAAATGTC, and Reverse: (BamHI) 5′-GGATCCTCAGTCTTCCCTTTTCATCTGTGG; for F. acidarmanus lysK, Forward: (XhoI): 5′-GCTCGAGATGTACTGGGCAGATGCACTG, and Reverse: (BamHI): 5′-GGATCCTTACTGTGCAGAATTATTCTGGATAAG; for M. magnetotacticum lysK, Forward: (NdeI) 5′-GCATATGTCCTCCGATTCCCGC, and Reverse: (BamHI) 5′-GGATCCCTACATATCCTCGCCCGC).
The amplified fragments were cloned into Topo TA (Invitrogen) vector and subsequently subcloned into expression vector pET15b (Invitrogen) using the restriction sites introduced by PCR. Clones were sequenced prior to transformation into E. coli BL21 C+ strain. This procedure allowed the overproduction of N-terminally His6-tagged derivatives, which have previously been found not to perturb substrate recognition by LysRS (14).
Overexpression and purification of LysRS proteins.
Transformants were grown in Luria broth supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) at 37°C to an optical density at 600 nm of 0.6. The expression of the recombinant protein was then induced by IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration of 1 mM) for 3 h at 37°C. The cells were harvested by centrifugation and washed with phosphate-buffered saline; this and subsequent steps were performed at 4°C. The cell paste was resuspended into lysis buffer (50 mM Na2H2PO4, 300 mM NaCl, 5 mM 2-mercaptoethanol, protease inhibitor [Roche]) and sonicated for 10 cycles of 30 s. The cell extract was obtained by ultracentrifugation at 100,000 × g for 1 h and subsequently applied to an Ni-nitrilotriacetic acid agarose column (Qiagen), according to the standard procedure. The protein was eluted with an elution buffer (containing 750 mM imidazole), dialyzed overnight against storage buffer (100 mM Tris-HCl [pH 8.0], 50 mM KCl, 25 mM MgCl2, 5 mM 2-mercaptoethanol, 50% glycerol), and then stored at −20°C. The purity of all the recombinant enzyme preparations was judged to be 95% or greater by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue.
Cloning and in vitro transcription of tRNALys genes.
tRNA genes for in vitro T7 RNA polymerase transcription were cloned by annealing of two cDNA oligonucleotides and direct ligation into pUC18 plasmid using BamHI and HindIII restriction sites. The clones were transformed into E. coli DH5α, sequenced, and amplified. The 3′ end of the transcription template was digested overnight either with BstNI at 60°C or NsiI at 37°C. In vitro T7 RNA polymerase runoff transcription was conducted according to standard procedure (18). tRNALys transcripts were purified on a denaturing 12% polyacrylamide gel and recovered by electroelution (Elutrap; Schleicher and Schuell), followed by desalting on a Nap-5 Column (Pharmacia Biotech). Plasmid T7-911 for preparation of the His6-tagged recombinant T7 RNA polymerase was a gift from T. Shrader (Department of Biochemistry, Albert Einstein College of Medicine, New York, N.Y.). Nucleoside triphosphates were from Sigma.
Genomic and phylogenetic analyses.
All predicted amino acid sequences were obtained from publicly available databases. Sequences were initially aligned using the program CLUSTALX (30). The alignment was then used to generate a neighbor-joining phylogeny using the PHYLIP 3.57c package (distributed by the author [J. Felsenstein]; Department of Genetics, University of Washington, Seattle) with parameters previously used for aaRS phylogenies (20). Maximum-likelihood phylogeny was generated using the program Tree-Puzzle 5.0 (www.tree-puzzle.de) (28) (10,000 puzzle steps, JTT substitution matrix).
RESULTS
Aminoacylation activity and tRNA specificity of recombinant LysRS proteins.
The recombinant LysRS proteins were tested for in vitro activity as determined by their ability to aminoacylate transcripts of their own (cognate) tRNALys genes or other noncognate tRNALys species (Table 1). Of the proteins tested, only the class I LysRS of S. aromaticivorans was able to aminoacylate its own in vitro-transcribed tRNALys. To investigate whether lack of aminoacylation in the other cases resulted from either inactive transcripts and/or inactive enzymes, cross-charging experiments were performed. B. burgdorferi class I LysRS, which has previously been shown to aminoacylate a variety of tRNALys species, was found to aminoacylate in vitro-transcribed F. acidarmanus tRNALys, indicating that this tRNA was active. In addition, F. acidarmanus LysRS was able to charge E. coli tRNALys, indicating that the enzyme was active. Similar tests with the two M. barkeri LysRS proteins and in vitro-transcribed M. barkeri tRNALys showed the class I LysRS and tRNA to be inactive but the class II enzyme to be active. Investigation of M. magnetotacticum showed the class I LysRS to be active, albeit at a significantly lower level than the other active proteins tested, and revealed that in vitro-transcribed M. magnetotacticum tRNALys could be charged by S. aromaticivorans LysRS.
TABLE 1.
In vitro lysylation of tRNALys substrates by LysRS proteins
| LysRSa protein source | Formation of Lys-tRNALyse
|
||
|---|---|---|---|
| Cognate transcriptb | Noncognate transcriptc | E. coli tRNALysUUUd | |
| F. acidarmanus | − | − | + |
| M. magnetotacticum | − | − | + |
| M. barkeri | − | − | − |
| M. barkeri class II | − | − | + |
| S. aromaticivorans | + | + (M. magnetotacticum) | + |
| B. burgdorferi | +f | + (F. acidarmanus) | +f |
All LysRS proteins are class I except as indicated for M. barkeri.
In vitro-transcribed tRNALys genes (UUU anticodon sequence) derived from the same organism as the LysRS.
In vitro-transcribed tRNALys genes (UUU anticodon sequence) derived from an organism different from that from which the LysRS was derived, as indicated.
Synthesized in vivo; see text for details.
Lysylation of tRNA was performed at 37°C for 15 min with 1 μM enzyme, 100 μM lysine, and 4 μM tRNA. Symbols: −, no detectable formation of Lys-tRNALys compared to an enzyme-free control; +, detectable formation of Lys-tRNALys.
Shown for comparison, taken from reference 13.
Anticodon recognition by class I LysRSs.
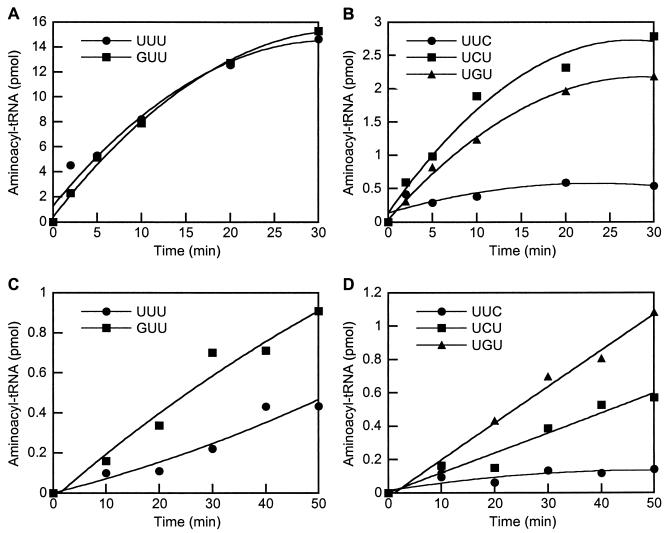
Anticodon recognition was investigated using undermodified E. coli tRNALys variants generated in vivo, which have previously been shown to be efficient substrates for a variety of both class I and class II LysRSs (4, 25). Initial tests using M. magnetotacticum LysRS showed that while it was able to charge the wild-type tRNA and nucleotide 34 mutant (UUU and GUU anticodons, respectively), the level of activity for the other mutants was insufficient to detect charging above background. Since the M. barkeri class I LysRS had been shown to be inactive, the pattern of anticodon recognition was only investigated in more detail for the F. acidarmanus and S. aromaticivorans proteins (Fig. 2). Mutation of U34 to G in tRNALys was found to have no effect on recognition by S. aromaticivorans LysRS (Fig. 2A), changes at U35 resulted in reduced charging and the most-pronounced effect was observed for replacement of U36, which almost completely eliminated activity (Fig. 2B). With F. acidarmanus LysRS replacements of either U34 or U35 by G both led to improvements in recognition compared to wild-type tRNA (Fig. 2C), while mutation of U35 to C did not result in any change in activity (Fig. 2D). The only significant reduction in activity with F. acidarmanus LysRS was observed with tRNALys where U36 was replaced with C, a change that virtually eliminated recognition.
FIG. 2.
Aminoacylation of E. coli tRNALys variants by class I His6-LysRS proteins. Aminoacylation reactions were performed as described in the text (20-μl samples) in the presence of 4 μM tRNA and 100 μM [14C]lysine. Comparison of charging of wild-type and mutant tRNALys using class I LysRS enzymes from S. aromaticivorans (200 nm of enzyme) (A and B) and F. acidarmanus (2 μM enzyme) (C and D).
Anticodon recognition by M. barkeri class II lysyl-tRNA synthetase.
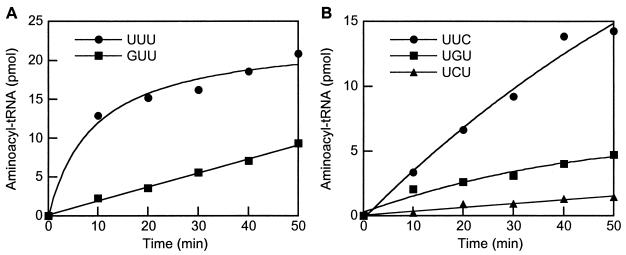
Recognition of the tRNALys anticodon by the class II LysRS of M. barkeri was investigated as described above for class I LysRS proteins. Mutation of the nucleotide U34 to G (Fig. 3A) led to a significant reduction in recognition by M. barkeri class II LysRS, whereas the replacement of U36 with C (Fig. 3B) caused only a slight change, indicating the relative importance of these positions. The strongest effects on aminoacylation were observed for changes in U35 of tRNALys, the G35 and C35 variants both showing significant reductions in activity compared to wild-type (Fig. 3B).
FIG. 3.
Aminoacylation of E. coli tRNALys variants by M. barkeri class II His6-LysRS. Aminoacylation reactions were performed as described in the text (20-μl samples) in the presence of 4 μM tRNA, 100 μM [14C]lysine, and 2 μM enzyme. (A) Position 34 variants; (B) position 35 and 36 variants.
Molecular phylogenies of class I LysRS.
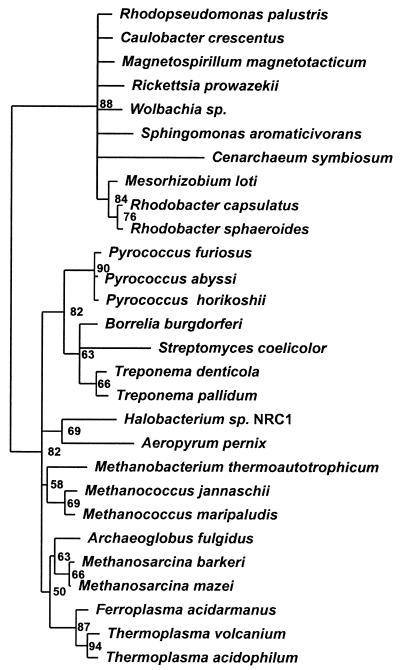
Phylogenetic analyses of the class I LysRS protein family were performed using 28 of the 31 sequences currently available in public databases. The three excluded sequences are all from Borrelia genospecies and show more than 75% amino acid identity to B. burgdorferi (N. Mejlhede, A. Monthán, M. Thesien, and M. Ibba, submitted for publication). As extensive tRNA anticodon recognition data are available for the class I LysRS of B. burgdorferi (25), this sequence was chosen for inclusion in this study. Structural studies have revealed a high degree of similarity between the two subclass Ib aaRSs LysRS and glutamyl-tRNA synthetase (GluRS) (29), suggesting that GluRS sequences may be appropriate for the rooting of LysRS phylogenetic trees. We therefore generated separate sequence alignments using CLUSTALX of 28 class I LysRS and 11 GluRS proteins (chosen as a representative sampling based upon existing phylogenies [33]) and then manually merged these into a single alignment using a structure-based sequence alignment of Pyrococcus horikoshii LysRS and Thermus thermophilus GluRS (29). Positions containing gaps in any of the sequences were then removed, leaving a total of 287 aligned residues present in all sequences. This sequence alignment was then used to generate neighbor-joining and maximum-likelihood phylogenies. In both cases the GluRS sequences formed a single distinct outgroup with high confidence for the corresponding node (84.3% by neighbor joining, 93% by maximum likelihood). A maximum-likelihood tree rooted by GluRS was then generated for the class I LysRS sequences (Fig. 4). The overall class I LysRS phylogeny resembles a somewhat-reduced version of the universal phylogenetic tree with distinct bacterial and archaeal groupings. The only exceptions are the placement of the crenarchaeon Cenarchaeum symbiosum within the major bacterial grouping and a subgroup of several bacteria close to the pyrococcal proteins in the archaeal group.
FIG. 4.
Maximum-likelihood phylogenetic tree of class I LysRS sequences. The tree has been rooted using GluRS sequences. Numbers indicate the percentage occurrence of nodes after 10,000 puzzle steps.
DISCUSSION
Comparative genomics of tRNALys recognition.
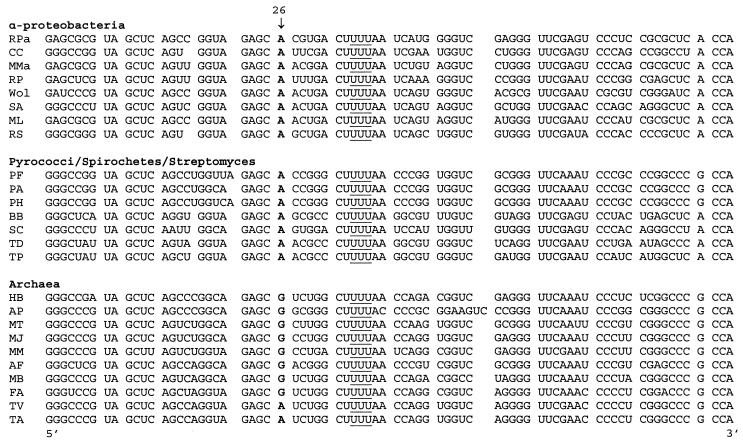
While all of the LysRSs, with the exception of the class I protein from M. barkeri, were found to be active when heterologously produced, only one was found to recognize its cognate in vitro-transcribed tRNA substrate. Previous studies showed that the B. burgdorferi (12) and P. horikoshii (29) class I LysRSs both recognized cognate in vitro-transcribed tRNALys, while the M. maripaludis enzyme did not (12). These differences in tRNA gene transcript recognition may result from the absence of essential RNA modifications necessary for recognition or folding, as reported for human mitochondrial tRNALys (8). We attempted to correlate sequence differences between tRNALys genes (UUU anticodon) that gave rise to active transcripts in vitro and those that did not with respect to potential nucleotide modification sites. This was done by comparing known positions of nucleotide modifications described for tRNALys from E. coli (Fig. 1) and Haloferax volcanii (7) with positions conserved in either the “active” or “inactive” tRNALys gene sets, but not in both (Fig. 5). These comparisons identified a notable difference at position 26, which is found as N2,N2-dimethylguanosine in H. volcanii and as adenosine in E. coli. All tRNALys genes whose transcripts are recognized by their cognate class I LysRSs contain A26, with the exception of M. magnetotacticum, while those that are not recognized contain G26. While the possible role of a modified G26 in tRNALys folding and/or recognition now requires a more extensive cataloguing of in vivo RNA modification patterns, the strong correlation between experimental and comparative genomic approaches provides a testable hypothesis for the observed differences in the activity of in vitro transcribed tRNALys genes. This is further supported by the observation that the distribution of A versus G at position 26 broadly correlates with the molecular phylogeny of class I LysRSs (Fig. 4 and 5).
FIG. 5.
Alignment of tRNALys sequence (UUU anticodon) from organisms encoding class I LysRSs. Nucleotide 26 is indicated and shown in boldface type, and the anticodon sequence is underlined. Abbreviations: Rpa, Rhodopseudomonas palustris; CC, Caulobacter crescentus; MMa, M. magnetotacticum; RP, R. prowazekii; Wol, Wolbachia sp.; SA, S. aromaticivorans; ML, Mesorhizobium loti; RS, Rhodobacter sphaeroides; PF, Pyrococcus furiosus; PA, Pyrococcus abysii; PH, Pyrococcus horikoshii; BB, B. burgdorferi; SC, Streptomyces coelicolor; TD, Treponema denticola; TP, Treponema pallidum; HB, Halobacterium sp.; AP, Aeropyrum pernix; MT, M. thermoautotrophicum; MJ, M. jannaschii; MM, M. maripaludis; AF, Archaeoglobus fulgidus; MB, M. barkeri; FA, F. acidarmanus; TV, Thermoplasma volcanium; TA, Thermoplasma acidophilum. Sequences of genes encoding tRNALys (UUU) were obtained from The Institute for Genomic Research (http://www.tigr.org), the Sanger Center (http://www.sanger.ac.uk/Projects/), and public databases (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table_cgi).
Evolutionary divergence of anticodon recognition among class I LysRSs.
The data presented above (Fig. 2) support earlier proposals (e.g., those in reference 25) that the anticodon is a crucial site in tRNALys for recognition by class I LysRSs. In agreement with studies using class I LysRSs from the bacterium B. burgdorferi and the archaeon M. maripaludis, the first nucleotide of the anticodon (position 34) was not found to play a significant role during recognition. Nucleotides U35 and especially U36 were both shown to be important during recognition by the class I LysRS from S. aromaticivorans, while only U36 was found to be important for the F. acidarmanus enzyme. This predominant role for U36 in tRNALys recognition was also observed for the class I LysRS of M. maripaludis but differs from the B. burgdorferi (25) and P. horikoshii (29) enzymes, for which both U35 and U36 were found to be critical recognition elements. Taken together with previous studies, the data presented here suggest that the class I LysRSs can be divided into two broad groups defined by whether either U36, or U35 and U36 are the preferred anticodon nucleotides for recognition. This functional divergence among the class I LysRSs can also be correlated with the overall phylogeny of this family (Fig. 4, discussed below), in contrast to the class II LysRS family where the pattern of anticodon recognition is more highly conserved.
Phylogenetic conservation of anticodon recognition by class II LysRSs.
Previous studies on the activity of tRNALys anticodon variants with bacterial (E. coli [4] and Helicobacter pylori [25]) class II LysRSs have shown that all three positions of the anticodon are recognized, with U35 being the predominant recognition site. A similar pattern was also observed during studies of the human class II LysRS (27), with U35 and U36 both recognized and the former being the main recognition element. The data presented here (Fig. 3) indicate that an archaeal class II LysRS recognizes the anticodon of tRNALys in the same way as its bacterial and eukaryotic counterparts do, with U35 being the most important position. This suggests that, in contrast to their class I counterparts, class II LysRSs share a pattern of tRNA anticodon recognition which is conserved in the three primary domains of the universal phylogenetic tree. This difference in the evolutionary divergence of a function duplicated in the two LysRS classes supports earlier evidence (12) suggesting that these two families are functionally but not mechanistically equivalent.
Evolution of the class I LysRS family.
The phylogenetic tree for class I LysRS sequences contains two major branches broadly corresponding to the bacteria and the archaea (Fig. 4). The inclusion of GluRS sequences places the root of the tree between the bacterial and archaeal branches, indicating that the evolutionary pattern of class I LysRS sequences is in agreement with that of the universal phylogenetic tree derived from rRNA sequences (see, for example, reference 2). While the class I LysRS tree appears canonical it is somewhat restricted, lacking any known eukaryotic examples and being mainly limited to α-proteobacteria in the bacterial branch and euryarchaeota in the archaeal branch. This observation of a limited canonical phylogeny among the class I LysRSs suggests that selective retention of the lysK gene in certain lineages has contributed substantially to their present distribution, as previously suggested (22).
Closer examination of the class I LysRS phylogeny, however, suggests that limited gene transfer has also contributed to the phylogenetic distribution of class I LysRSs. One possible exception to the canonical universal phylogeny among the class I LysRSs is seen for the C. symbiosum protein, which is positioned within the main bacterial branch. Earlier analyses based upon smaller data sets placed C. symbiosum as a branch preceding the α-proteobacteria (33, 34), possibly indicating horizontal transfer from an ancestral crenarchaeote. While our present analysis suggests horizontal transfer from rather than to the α-proteobacteria, a more reliable interpretation of the exact positioning of the C. symbiosum protein within the class I LysRS tree, and of possible gene transfer events, is now dependent on the availability of other closely related archaeal sequences. Within the archaeal branch of the class I LysRS tree, the group containing Pyrococcus species also contains several bacterial examples of LysRS. Functional analyses have shown that bacterial (B. burgdorferi) and archaeal (P. horikoshii) enzymes from within this group share common tRNALys anticodon recognition properties (see above) not seen for class I LysRSs from elsewhere in the phylogenetic tree, supporting a possible common origin. The positioning of this group within the canonical archaeal branch suggests that the corresponding bacterial LysRSs (Borrelia, Treponema, and Streptomyces) originated from a gene transfer event from the pyrococcal progenitor into an ancestral bacterium. Thus, while selective retention appears to have been the main determinant of the distribution of class I LysRSs, horizontal gene transfer has also played a significant but less widespread role.
Acknowledgments
We thank Sylvain Blanquet and Pierre Plateau for the gift of tRNAs; Kevin Sowers, Elizabeth Bertani, and Paul Predki for genomic DNAs; Margaret Romine for cells; and Dieter Söll for his encouragement and support.
Parts of this work were supported by funds from The Ohio State University (M.I.) and the National Institutes of Health (GM-22854 to Dieter Söll).
REFERENCES
- 1.Arnez, J. G., and D. Moras. 1997. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22:211-216. [DOI] [PubMed] [Google Scholar]
- 2.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon. Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 4.Commans, S., M. Lazard, F. Delort, S. Blanquet, and P. Plateau. 1998. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J. Mol. Biol. 278:801-813. [DOI] [PubMed] [Google Scholar]
- 5.Cusack, S. 1997. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 7:881-889. [DOI] [PubMed] [Google Scholar]
- 6.Eriani, G., G. Dirheimer, and J. Gangloff. 1991. Cysteinyl-tRNA synthetase: determination of the last E. coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 19:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta, R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 259:9461-9471. [PubMed] [Google Scholar]
- 8.Helm, M., R. Giegé, and C. Florentz. 1999. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38:13338-13346. [DOI] [PubMed] [Google Scholar]
- 9.Ibba, M., H. D. Becker, C. Stathopoulos, D. L. Tumbula, and D. Söll. 2000. The adaptor hypothesis revisited. Trends Biochem. Sci. 25:311-316. [DOI] [PubMed] [Google Scholar]
- 10.Ibba, M., J. L. Bono, P. A. Rosa, and D. Söll. 1997. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 94:14383-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibba, M., A. W. Curnow, and D. Söll. 1997. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem. Sci. 22:39-42. [DOI] [PubMed] [Google Scholar]
- 12.Ibba, M., H. C. Losey, Y. Kawarabayasi, H. Kikuchi, S. Bunjun, and D. Söll. 1999. Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc. Natl. Acad. Sci. USA 96:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibba, M., S. Morgan, A. W. Curnow, D. R. Pridmore, U. C. Vothknecht, W. Gardner, W. Lin, C. R. Woese, and D. Söll. 1997. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science 278:1119-1122. [DOI] [PubMed] [Google Scholar]
- 14.Ibba, M., and D. Söll. 1999. Quality control mechanisms during translation. Science 286:1893-1897. [DOI] [PubMed] [Google Scholar]
- 15.Ibba, M., and D. Söll. 2000. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69:617-650. [DOI] [PubMed] [Google Scholar]
- 16.LaRiviere, F. J., A. D. Wolfson, and O. C. Uhlenbeck. 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294:165-168. [DOI] [PubMed] [Google Scholar]
- 17.Lipman, R. S., K. R. Sowers, and Y. M. Hou. 2000. Synthesis of cysteinyl-tRNACys by a genome that lacks the normal cysteine-tRNA synthetase. Biochemistry 39:7792-7798. [DOI] [PubMed] [Google Scholar]
- 18.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 20.Ribas de Pouplana, L., J. R. Brown, and P. Schimmel. 2001. Structure-based phylogeny of class IIa tRNA synthetases in relation to an unusual biochemistry. J. Mol. Evol. 53:261-268. [DOI] [PubMed] [Google Scholar]
- 21.Ribas de Pouplana, L., and P. Schimmel. 2001. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem. Sci. 26:591-596. [DOI] [PubMed] [Google Scholar]
- 22.Ribas de Pouplana, L., R. J. Turner, B. A. Steer, and P. Schimmel. 1998. Genetic code origins: tRNAs older than their synthetases? Proc. Natl. Acad. Sci. USA 95:11295-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: A laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Smith, D. R., L. A. Doucette-Stamm, C. DeLoughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Söll, D., H. D. Becker, P. Plateau, S. Blanquet, and M. Ibba. 2000. Context-dependent anticodon recognition by class I lysyl-tRNA synthetases. Proc. Natl. Acad. Sci. USA 97:14224-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stathopoulos, C., T. Li, R. Longman, U. C. Vothknecht, H. D. Becker, M. Ibba, and D. Söll. 2000. One polypeptide with two aminoacyl-tRNA synthetase activities. Science 287:479-482. [DOI] [PubMed] [Google Scholar]
- 27.Stello, T., M. Hong, and K. Musier-Forsyth. 1999. Efficient aminoacylation of tRNALys,3 by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res. 27:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terada, T., O. Nureki, R. Ishitani, A. Ambrogelly, M. Ibba, D. Söll, and S. Yokoyama. 2002. Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nature Struct. Biol. 9:257-262. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tumbula, D. L., H. D. Becker, W. Z. Chang, and D. Söll. 2000. Domain-specific recruitment of amide amino acids for protein synthesis. Nature 407:106-110. [DOI] [PubMed] [Google Scholar]
- 32.Wilcox, M., and M. Nirenberg. 1968. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl. Acad. Sci. USA 61:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese, C. R., G. J. Olsen, M. Ibba, and D. Söll. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64:202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689-710. [PubMed] [Google Scholar]