Abstract
We present the complete genome sequence of Yersinia pestis KIM, the etiologic agent of bubonic and pneumonic plague. The strain KIM, biovar Mediaevalis, is associated with the second pandemic, including the Black Death. The 4.6-Mb genome encodes 4,198 open reading frames (ORFs). The origin, terminus, and most genes encoding DNA replication proteins are similar to those of Escherichia coli K-12. The KIM genome sequence was compared with that of Y. pestis CO92, biovar Orientalis, revealing homologous sequences but a remarkable amount of genome rearrangement for strains so closely related. The differences appear to result from multiple inversions of genome segments at insertion sequences, in a manner consistent with present knowledge of replication and recombination. There are few differences attributable to horizontal transfer. The KIM and E. coli K-12 genome proteins were also compared, exposing surprising amounts of locally colinear “backbone,” or synteny, that is not discernible at the nucleotide level. Nearly 54% of KIM ORFs are significantly similar to K-12 proteins, with conserved housekeeping functions. However, a number of E. coli pathways and transport systems and at least one global regulator were not found, reflecting differences in lifestyle between them. In KIM-specific islands, new genes encode candidate pathogenicity proteins, including iron transport systems, putative adhesins, toxins, and fimbriae.
Yersinia pestis is the causative agent of bubonic and pneumonic plague, which has caused widespread loss of human life during recurrent pandemics. Over the past several decades, research on Y. pestis has greatly advanced our understanding of this organism, the disease it causes, and its mechanisms (5, 36, 44). Significant progress has been made in a number of areas related to the ability of Y. pestis to cause disease. These include expression of virulence genes and their regulation (43), bacterial iron acquisition (35), and prevention of host immune responses via paralysis of phagocytic cells, as well as suppression and disruption of signal transduction (44). However, many aspects of this organism's biology remain to be studied, and an effective vaccine that induces long-lived immunity against bubonic and pneumonic plague is still lacking. Information about the whole genome sequence and genes will help us to better understand this organism and to identify possible targets for disease treatment and vaccine development.
Y. pestis strains fall into three subtypes, or biovars: Antiqua, Mediaevalis, and Orientalis, each of which is associated with a major pandemic (1, 17). KIM belongs to biovar Mediaevalis. We chose to sequence the genome of KIM10+, a KIM derivative, because it has been widely used in research and is thus more genetically and physiologically characterized than other strains. The genome of a biovar Orientalis strain, CO92, was also recently sequenced (33), providing an opportunity to examine the two genomes for differences associated with the biovar and its emergence or dominance, even though only a few hundred years separate the pandemic events by which these two biovars are defined.
KIM and many other Y. pestis strains harbor three plasmids. The KIM plasmids are known as pPCP1, pMT1, and pCD1 (9.5, 100.9, and 70.5 kb, respectively). Their sequences have all been decoded as part of this project (22, 28, 38), each revealing genes or loci necessary for, or contributing to, pathogenicity.
MATERIALS AND METHODS
Sequencing.
Genomic DNA of Y. pestis KIM10+ (13) was prepared from a culture grown in heart infusion broth at 30°C. A lysozyme-sodium dodecyl sulfate-proteinase K procedure followed by phenol and chloroform extractions was used to isolate and purify genomic DNA. A whole-genome shotgun library was prepared using nebulization to mechanically shear the genomic DNA (29). The plasmid pMT1 was present in the genomic DNA preparation, while pPCP1 and pCD1 were absent (37). Fragments with sizes of 1 to 2.5 kb were prepared by agarose gel electrophoresis, end repaired, and cloned into the M13 Janus vector (7). Random phage clones were isolated. Their DNAs were purified and used as templates for shotgun sequencing reactions using dye-terminator chemistry. The data were collected on ABI377 and 3700 automated sequencers.
Approximately 80,000 sequences were collected to give the whole genome sixfold coverage. A pBluescript plasmid library with an insert size of 5 to 6 kb was also constructed, and an additional twofold clonal coverage was obtained from this source. Some of the clones were dual end sequenced to be used as contig-linking information. Sequence data were assembled by Seqman II (DNASTAR). For the finishing steps, clonal and genomic PCR techniques, as well as primer walking, were used. The assembly and ordering of contigs were confirmed by a whole-genome physical map made by optical mapping with XhoI restriction enzyme (26, 27). This optical map was also very useful for confirming the absence of accidental rearrangements of the data, such as large inversions caused by misassembly. At regular intervals, successive assemblies were made available for BLAST searching on our website (http://www.genome.wisc.edu), allowing access to the data while the project was still in progress. The annotated sequence is available at that site as well as in the public database.
Annotation software.
The genome was annotated using the multiuser web-based software environment called MAGPIE (14, 15). This system used the program Glimmer to define open reading frames (ORFs) (41). The predicted proteins were searched against the nonredundant database using BLAST (2), and the results were displayed on a gene-by-gene basis on MAGPIE's web pages. MAGPIE's automatic annotation function suggested complete annotations for all ORFs, which were individually checked and corrected, updated, or investigated further by the annotation team. These curated annotations were captured from MAGPIE and formed the basis for the GenBank submission.
Collaborative annotation project.
The MAGPIE annotation environment enabled a team of collaborators led by R.D.P., all of whom are active researchers in Yersinia biology, to contribute their collective expertise and share the distributed effort of assessing and identifying ORFs. The search outputs and autoannotations generated by the software were displayed by MAGPIE for further analysis and accessed via the World Wide Web by our collaborators from their widespread geographical locations. New or altered annotations were entered into the web pages. Each annotator focused on an assigned genome segment and also on genes from the whole sequence in functional categories appropriate to their expertise. The whole annotation project was accessible to all of the collaborators. An outline of their findings is included below, and a more detailed analysis will be published separately. We found that the web-based approach was an efficient and effective way for the community to interact and simultaneously work on annotation, despite some limitations and imperfections in the software.
Analysis.
Comparisons of the genomes of Y. pestis strains were made by the modified MEM alignment utility (34) and verified using BLAST. The parsimony analyzer used to calculate inversion numbers was kindly provided by S. Hannenhalli. The program computes an optimal sorting by reversal for signed permutations, in an implementation of the published polynomial algorithm (19). This gives the smallest number of inversions that can explain the observed changes. Other programs used are noted elsewhere in the text.
Nucleotide sequence accession number.
The sequence discussed in this paper has been submitted to GenBank under accession number AE009952.
RESULTS AND DISCUSSION
Genome features.
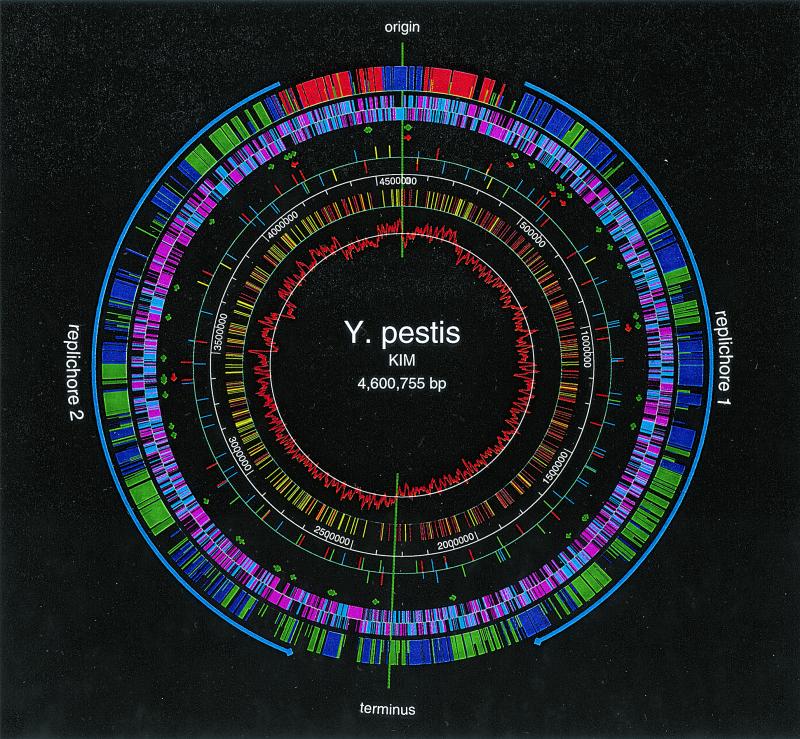
The genome of Y. pestis KIM consists of a single circular chromosome of 4,600,755 bp with an average G+C content of 47.64%. Features of the sequence are shown on the map in Fig. 1. The origin and the terminus of replication were assigned by several criteria (these have not been experimentally determined). The polarity-switching point of C-G skew was used in both cases. For the origin, the locations of DnaA boxes were used, and for the terminus, dif (the site in the center of the terminus where chromosome dimer resolution occurs) and the terC site were used. Base pair 1 of the genome was assigned between the mioC gene and the DnaA boxes within the origin of replication. As in Escherichia coli, the putative origin and terminus of replication divide the genome into two replichores, and replication presumably proceeds bidirectionally. The DNA strands that are replicated continuously in the direction from origin to terminus are leading strands. Their complementary strands are lagging strands. The DNA sequence in Fig. 1 represents the leading strand in replichore 1 and the lagging strand in replichore 2. The sequence contains many highly skewed oligomers, occurring preferentially on leading strands. The most statistically significant skewed oligomer is Chi (GCTGGTGG) (Fig. 1), which stimulates DNA repair by homologous recombination in an orientation-dependent manner (25, 30). The possibilities that Chi sites may be involved in normal DNA replication (4, 8) and in the rescue of stalled replication forks have been noted (25). Another family of octamers, with consensus RRNAGGGS (9), are highly skewed in each of the replichores, confirming our identification of both the origin and the terminus. The orientation of the map in Fig. 1 was chosen to match that of E. coli K-12 by the organization of rRNA operons and the gene content, gene orientations, and relative positions in the two replichores.
FIG. 1.
Circular genome map of KIM. The KIM genome has 4,198 ORFs, 2,254 of which are homologous to K-12 genes at the protein level (40% identity; alignments including at least 60% of both proteins). The outer circle shows the distribution of backbone ORFs: ORFs with the same replication orientations and on the same replichores (blue); ORFs with the same replication orientations but on different replichores (green); and ORFs with different replication orientations (red). The short ticks represent ORFs that have moved out of their normal K-12 context range. The second circle shows both the backbone (blue) and nonbackbone (pink) ORFs and their orientations. The arrows in the third and fourth circles show the locations and the orientations of tRNA (green) and rRNA (red) operons, respectively (not shown to scale). The fifth circle shows insertion element distribution: IS100 (red), IS1541A (blue), IS285 (green), and IS1661 (yellow). The short ticks represent partial IS elements. The sixth circle gives the scale in base pairs. The seventh circle shows highly skewed octamer Chi distribution: red and yellow indicate its distribution on the two DNA strands. The inner circle shows the C-G skew that is calculated for each sliding window of 10 kb along the genome. The maps were created by GenVision (DNASTAR).
Sequence analysis revealed 4,198 predicted ORFs with an average size of 940 bp covering 86% of the genome. Predicted ORFs smaller than 50 amino acids (aa) were only annotated if they had a convincing database match. There are 2,385 ORFs on the leading strands and 1,813 ORFs on the lagging strands. ORFs on the leading strands outnumbering ORFs on the lagging strands reflects the preference for encoding proteins in the same direction as replication.
Comparison with Y. pestis CO92.
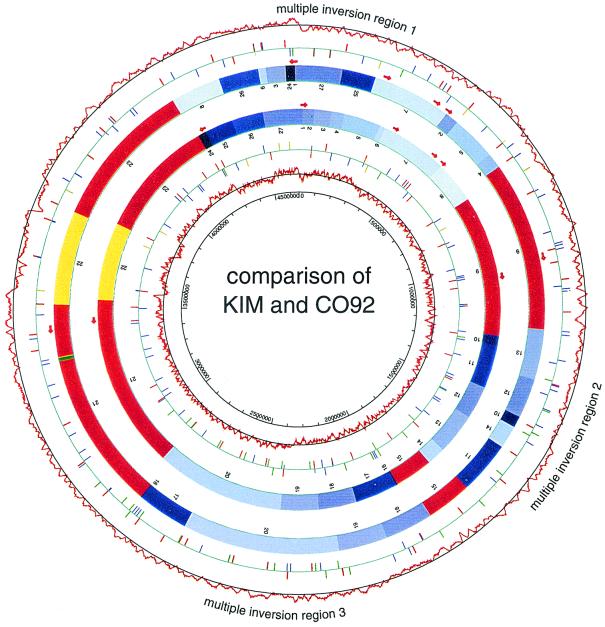
We compared the KIM genome sequence with that of Y. pestis strain CO92, recently published (33), and found that more than 95% of the sequence is shared by the two genomes. The CO92 genome is ∼50 kb larger than the KIM genome, the result of an 11-kb and many smaller insertions in CO92 relative to KIM. About 27 kb of the difference is due to insertion sequence (IS) elements, which are more numerous in CO92. CO92 also has one less rRNA operon (see below). At the protein level, 3,672 of 4,198 total ORFs match CO92 ORFs (>90% amino acid identity and >60% of each protein length in the alignment). Of the remaining 526 unmatched KIM ORFs, 318 have only 100 aa or less. Most of the unmatched ORFs encode hypothetical proteins. Although the genome sequences are very conserved, extensive rearrangements are seen. Figure 2 shows the alignment of the two genomes. For the purpose of comparison, both genomes are divided into 27 segments, each of which matches its counterpart in either direct or reverse orientation. There are three regions where multiple inversions appear to have taken place. For each region, we calculated the most parsimonious series of inversions that could account for the organizational differences between the two genomes, plotted in Fig. 3. In most steps it was possible to identify a sequence homology, mostly IS elements, that could have given rise to the proposed inversion. The most complicated region is the one that spans the replication origin, which has 12 segments and a minimum of nine inversions required to produce the observed rearrangement. Any intrareplichore inversion causes the DNA sequences involved to switch from leading strand to lagging strand or vice versa, resulting in the change of C-G skew at that particular region, which will still be detectable if the event was relatively recent and amelioration has not yet taken place (visible in Fig. 1 and 2).
FIG. 2.
Comparison of KIM and CO92 at the DNA level. The outer circles show the CO92 C-G skew. The second circle shows CO92 IS elements: IS100 (red), IS1541A (blue), IS285 (green), and IS1661 (yellow); short ticks represent partial IS elements. The third circle shows CO92 rRNA operons. The fourth circle shows the CO92 genome in 27 blocks (numbered according to KIM genome order), regions that are conserved by both locations and orientations (red), a single intrareplichore inversion region (yellow), multiple-inversion regions (various blues), and genome-specific sequences (green). The inner four circles show KIM rRNA operons, the KIM genome in blocks, KIM IS elements, and KIM C-G skew. Colors are coded as for CO92.
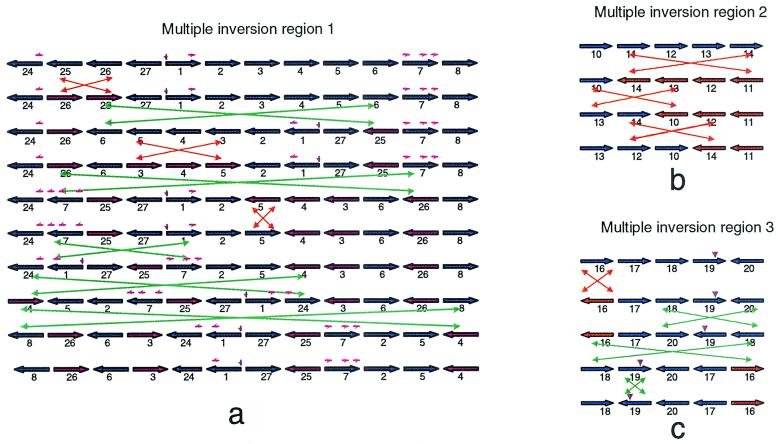
FIG. 3.
The most parsimonious series of inversions for each of the multiple-inversion regions (Fig. 2). Each horizontal arrow represents a DNA block (not shown to scale) and its orientation. Blue and red indicate opposite replication orientations of the same block. The replication origin and terminus are marked by a small brown downward arrow. The green crossed double arrows stand for interreplichore inversions, and the orange crossed double arrows stand for intrareplichore inversions. The small red arrows (not shown to scale) on some of the blocks indicate rRNA operon locations.
rRNA operons.
Rearrangement of the Y. pestis genome might also have led to the difference in the number of rRNA operons in the two strains. Of the seven rRNA operons in KIM, two are in the very highly conserved regions and the other five are in the multiple-inversion region I that is around the replication origin. As demonstrated in Fig. 3a, segment 7 has three rRNA operons, with one at the end of the segment. Both segments 24 and 1 have an rRNA operon at one end. In step 4 of the rearrangement process, 24 becomes adjacent to 7, and in step 5, it becomes adjacent to 1. Both steps bring rRNA operons together in the genome, forming large tandem repeats and providing opportunities for recombination. This could result in deletion of one of the tandem rRNA operons during the process of evolution without the loss of essential genes in between. A process of this type could explain the loss of an rRNA operon in CO92. Variant rRNA patterns (ribotypes) have been reported among Y. pestis biovar Orientalis strains isolated over the last 65 years (18), but there is no evidence of rRNA-specific rearrangements in KIM. Indeed, their conserved locations with respect to the origin of replication is remarkable in light of the level of rearrangement observed for other backbone genes (see below).
Comparison with E. coli K-12.
It has always been surprising that E. coli and Salmonella have such consistent gene order (synteny), considering their evolutionary divergence time of 110 million years. In our previous analysis of E. coli O157:H7 (34), we found by comparison with K-12 that a large proportion of both genomes is shared and colinear and that in both cases the shared regions are punctuated by islands of unique sequence, apparently acquired by horizontal transfer. In general, the shared regions include genes of central metabolism and basic conserved physiology of the Enterobacteriaceae. The islands, by contrast, frequently contained genes associated with pathogenicity or survival in the mammalian intestine, as well as many of unknown function, and some contained evidence of mobility, e.g., phage genes or IS elements. We named the shared regions “backbone” to denote the common framework in which the specialized islands are inserted. We examined KIM to determine whether this organization is detectable in Y. pestis and, if so, to what extent. In the case of the two E. coli strains, backbone regions matched at >98% nucleotide identity. The KIM and K-12 genomes do not match at this level; only 20% of randomly tested sequence reads matched K-12 at better than 60% nucleotide identity. In a random sample of KIM proteins, 45.5% matched K-12 proteins at better than 60% amino acid identity. When context is also taken into account, the extent of locally colinear segments emerges.
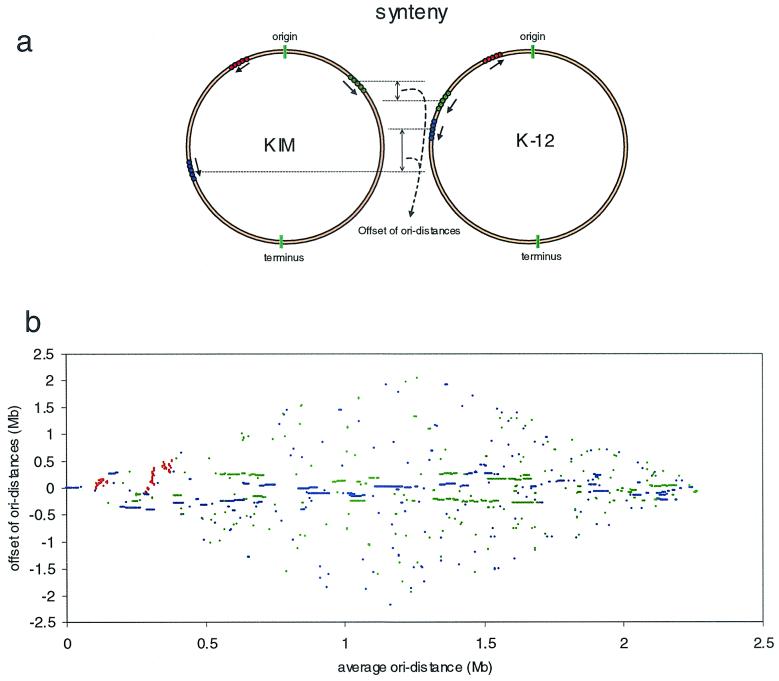
Our standard criteria for inference of orthology in the E. coli-versus-E. coli comparison were >90% amino acid identity and >60% of each protein length in the alignment. For KIM predicted proteins, we used a match of >40% amino acid identity with the K-12 protein, and alignments included >60% of both genes. This was justified when the match to K-12 was clearly the best, or the only, match, and in addition, when the adjacent encoded protein(s) also had best matches to adjacent genes in K-12. Despite their remote distance in the phylogenic tree, KIM and K-12 do in fact share many homologous genes. By these criteria, roughly 53.7% of KIM genes (2,254 of 4,198) are identified as backbone genes, with an average amino acid identity with the K-12 ortholog of 72.9% and with an average of 96.2% of the Y. pestis ORF length and 97.4% of the K-12 ORF length in the alignment. The most interesting finding is not only that so many genes are shared but that the arrangements of these backbone genes are quite similar (73% of backbone genes are in segments that are locally colinear with K-12). Figure 4a shows the synteny of orthologous genes of both genomes. We define ori distance as the distance of each gene from the origin on its replichore. An interreplichore inversion causes neighboring genes to switch from one replichore to the other, but the gene order with respect to the origin is not changed. If such an inversion happens asymmetrically around the replication origin, the ori distances of genes outside the inversion are changed. An intrareplichore inversion, however, inverts the gene order. When the offsets of the ori distances of orthologous genes from the replication origin are plotted against their average distances, a clear pattern of the arrangements of backbone genes in both genomes emerges (Fig. 4b).
FIG. 4.
Comparison of KIM and K-12 at the protein level. (a) Synteny of KIM and K-12 genomes. The backbone genes on each genome can be arranged in three different ways: genes in the same replication orientations and on the same replichores (blue), genes in the same replication orientations but on different replichores (green), and genes in different replication orientations (red). (b) Arrangement of backbone genes. x axis, the average ori distances of orthologous genes; y axis, the offset of ori distances of orthologous genes. Colors are coded as for panel a.
While the gene order of KIM appears at first glance to be totally scrambled relative to that of K-12, we found that the orientations and the ori distances of corresponding backbone genes are in fact highly conserved (±400 kb) (Fig. 4b). This form of colinearity is to be expected if the principle rearrangements in evolution occur by a series of approximately symmetrical interreplichore inversions which exchange genes from one replichore to the other without moving them very much relative to the origin (11). This type of inversion also preserves the sizes of replichores, as well as the leading strand of replication, and hence does not disrupt the strand biases characteristic of the replichore. It has happened so frequently that almost half of the backbone genes have switched to the other replichore (Fig. 4b). Only near the origin are there a few examples of intrareplichore inversions (Fig. 4b) between K-12 and KIM. The conservation of backbone structure is also observed from the relative locations of rRNA operons (five rRNA operons on replichore 1 and two on replichore 2). The finding of substantial colinearity in this sense between the backbone genomes of E. coli and Y. pestis KIM is remarkable, considering they may have been separated by as much as 500 million years of evolution from a presumed common ancestor. We estimated their time of divergence from the variability of three shared housekeeping genes, using the divergence time of 110 million years for E. coli and Salmonella (32) as a calibration, resulting in a weighted average of 375 million years with a standard error of 145 million years.
Our observations are generally in agreement with the work of Segall et al. (42), who drew attention to the conservation of gene order among members of the family Enterobacteriaceae and demonstrated that intrareplichore inversions could not be obtained experimentally by homologous recombination, though once constructed by other techniques, such inversions are stable.
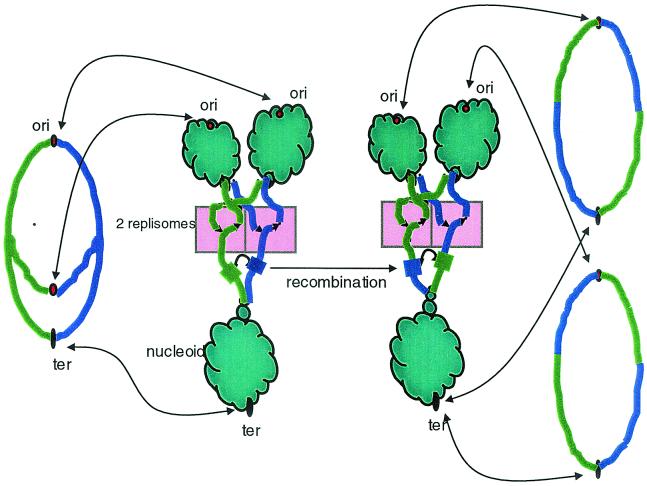
These phenomena seem more easily interpretable when we consider the possible effect nucleoid segregation during replication may have on the availability of DNA for inversional recombination (Fig. 5). If DNA exchanges are limited to rather short exposed regions near replication forks, interreplichore inversions will tend to have endpoints that are about equidistant from the origin, given that the two forks move at approximately equal rates (24). On the other hand there is little scope for intrareplichore inversion, since very little distance along a single replichore will be exposed at any given time. An exception may occur during replication initiation, since all the intrareplichore inversions between K-12 and KIM are concentrated near the origin. Larger regions of the chromosome may also be exposed during other processes, such as DNA damage repair.
FIG. 5.
Accessibility of DNA for recombination near replication forks. On the left, the theta structure of the replicating chromosome is shown, with arrows indicating that the nonreplicating DNA is bound with proteins in nucleoids (in the middle). The two replichores are colored green and blue, and a region of homology is indicated by thickened segments. A recombination event between homologous regions would result in the interreplichore inversion shown on the right.
The codon usage of backbone genes and KIM-specific genes was analyzed. Some rare codons, such as AGA and AGG for arginine, are used differently in backbone and KIM-specific genes. KIM-specific genes use as much as threefold more AGA and AGG codons for arginine than backbone genes. They also use the rare codon AUA for isoleucine two times more than backbone genes. This difference is also seen in the K-12 genome between backbone and strain-specific genes. Codons are also used differently in ORFs on the leading strands and ORFs on the lagging strands. The leading-strand ORFs contain slightly more G and T than the lagging-strand ORFs; therefore, they are C-G and A-T skewed. The C-G skew for the leading strand ORFs is particularly strong in the third codon position (data not shown).
Phage, ISs, and other repeats.
Four types of IS elements were found in the KIM genome. IS1541A is the most abundant (49 complete, 6 partial, 3 interrupted by IS100). There are 35 copies of IS100, 19 copies of IS285, and 8 intact and 2 partial copies of IS1661. All of the partial IS1541A elements have an adjacent IS100, suggesting that loss of part of IS1541A was initiated by IS100 insertion. Two other complex groups consist of IS100 flanked on one side by a partial IS1541A and on the other by a partial IS1661, suggesting an insertion by IS100 into IS1541A and IS1661, followed by recombination between the two IS100 elements. These observations, and the instability of the pigmentation locus, pgm, also bounded by IS100 elements, show that this IS is very actively mobile in KIM. The IS content of KIM is slightly lower than that of CO92, but the same types of elements occur in both.
Some E. coli strains elaborate a heat-stable enterotoxin, EAST1. The coding sequence is homologous to a central region of IS285 on the opposite strand from the transposase. In KIM, 18 of the 19 IS285 copies are identical in this region, having two in-frame stop codons preventing expression, as previously observed (47). The nonidentical copy has 133 single-nucleotide differences and a 10-bp deletion but also has both stop codons.
The KIM genome contains six regions resembling phage (Table 1). The most complete is a cryptic lambdoid prophage of 41 kb, located inside a 46-kb island with ORFs similar to those for most of the lambda head and tail proteins and bounded by a 31-bp direct repeat. The integrase gene is disrupted by IS100. An ORF in the Q-lysis interval is positioned for potential transcriptional control by Q, reminiscent of the Shiga toxin phage in E. coli EDL933 (39). However, this ORF has no characteristic that suggests a function in pathogenesis. No tRNAs are encoded in the prophage. Two genes encoding phage holin and endolysin are found between the tca and tcc genes of the insecticidal toxin subunits. A single-stranded prophage observed in the CO92 genome is not present in KIM.
TABLE 1.
Coordinates and characteristics of phage and the largest island regions
| Start | End | Size | Recombinase | RNA | Contents |
|---|---|---|---|---|---|
| 185492 | 211201 | 25,710 bp | Insecticidal toxin subunits; TcaA has internal deletion; phage ORFs between toxin genes; hypotheticals | ||
| 262478 | 310975 | 48,498 bp | IS cluster at 3′ end | IcmF-like fragments and ORFs similar to OI no. 7 of EDL933; Rhs; regulator; ribokinase | |
| 581694 | 641978 | 60,285 bp | Type III secretion system (one ORF disrupted); hmu (hemin uptake); Tellurite resistance; chaperone and usher; putative adhesin | ||
| ≈834807 | ≈842229 | ≈7.4 kbp | Phage remnant; no flanking repeats; integrase; no other phage-like features | ||
| 1258841 | 1313395 | 54,555 bp | IS100 | Sugar transport; UreA; putative enzymes; hypotheticals | |
| ≈1362250 | ≈1368074 | ≈5.8 kbp | Phage remnant; no flanking repeats; integrase disrupted by a transposase; no other phage-like features | ||
| 1679400 | 1733523 | 54,124 bp | tRNA-Arg | yfu Fe uptake; fimbriae; VgrG; Pseudomonas-like hypotheticals; IS100 | |
| 2410398 | 2456467 | 46,070 bp | Integrase | Lambdoid phage: integrase with IS100 insertion; excisionase; Q; lysis and tail fiber genes; 31-bp direct repeat at boundary | |
| 2595153 | 2701942 | 106,790 bp | Integrase, IS100, IS285 | leuZ, tRNA-Asn | Composite island; yersiniabactin synthesis genes inserted near tRNA-Asn; integrase; tRNA duplication at boundary; phage remnant integrated at leuZ; two sets of transport genes; hms Fe uptake; fimbriae |
| 2952460 | 3008203 | 55,744 bp | serT | Many hypotheticals; fatty acid synthesis; cytotoxic necrotizing factor pseudogene (three pieces) | |
| 3389236 | 3400385 | 11,150 bp | ssrA | Cryptic prophage, somewhat P4-like; 85-bp direct repeat; integrase ≈800 bp from ssrA 5′ end (IS1541 in between); integrase; primase; repressor; replication genes | |
| ≈3574541 | ≈3601511 | ≈27 kbp | tRNA-Gly | Phage remnant: no flanking repeats; integrase ∼169 bp from tRNA-Gly 3′ end (glyU?); integrase; primase; regulatory gene | |
| 3691992 | 3876361 | 184,370 bp | pheV | TadA-F (biofilm formation); IS200; Fe transport; fimbriae and flagellar genes; IS1397; Rhs; adhesins; nonribosomal peptide synthase; IS100; enterotoxin; secretion proteins; aerobactin Fe uptake; quorum-sensing (HSL) genes; many hypotheticals | |
| 3923286 | 3980985 | 57,700 bp | ileX | TerXY; hemolysin; large hypothetical ORFs; transport genes; multidrug resistance | |
| 4061656 | 4088198 | 26,543 bp | Hypothetical ORFs |
We searched the genome for REP (BIME) elements and other repeat features characterized in the E. coli genome. If KIM contains such elements, they are not sufficiently similar to the E. coli sequences for recognition by a consensus search, with one exception. Two Rhs elements are present in Y. pestis. These elements are highly conserved regions of ∼10 kb containing several ORFs, including the very large “core” ORF with a repeat peptide motif (21). Though the products encoded by the element are thought to be associated with the cell surface, their functions are unknown. E. coli strains contain five to seven Rhs elements. Their G+C contents suggest that they originated outside E. coli, and similar elements have been found in several other species.
Islands.
Y. pestis-specific regions are interspersed among the colinear backbone segments. These islands are of all sizes, and some include many ORFs of unknown function, as well as gene groups with putative functions but uncertain substrates, such as transporters. Other islands contain well-characterized segments, such as the yersiniabactin region of the high pathogenicity island (6, 16, 20). Table 1 shows the extents and contents of some of the larger island regions, including phage regions. Some islands show characteristics of classical pathogenicity islands (distinct G+C content, integrase, and insertion near a tRNA), and the yersiniabactin region is an example. Inserted near tRNA-Asn with a CP4-57-like integrase adjacent, the region has a consistently higher G+C content than flanking areas (59.6% versus a 47.6% average for the whole genome). A 10-nucleotide repeat of the end of the tRNA was found about 35 kb distant, just after the end of the section with high G+C content. In contrast, in the area in which the insecticidal toxins are encoded, none of these features were found, except for large fluctuations in G+C content. The region containing the type II secretion genes has a strikingly lower G+C content than average for the KIM genome (35 versus 47.6%), but no tRNA or recombinase is associated with this region.
Genes and functions. (i) Energy metabolism.
In general, the Y. pestis genome contains energy genes typical of a member of the family Enterobacteriaceae, with a few exceptions. Most genes are intact and have a high level of identity with their E. coli homologs. Y. pestis uses respiration (aerobic) and fermentation (anaerobic) to produce energy. Hydrogenases are widespread in bacteria and catalyze both the production and consumption of hydrogen gas. Three distinct multisubunit hydrogenases (nickel enzymes) of E. coli and the ancillary enzymes for utilization of the nickel cofactor are absent from the Y. pestis genome. There is no high-affinity nickel transporter like nikABCDE of E. coli. It is possible that low-affinity transporters may be able to import nickel.
The formate dehydrogenases H and O are present. In fdoG, as in E. coli, an opal stop codon is presumably translated as selenocysteine. This exceptional translation event requires the proteins encoded by selA, selB, and selD, as well as the selC tRNA. In the KIM genome, the entire set of sel genes is present and presumably functional, but in CO92, selB is interrupted by a +1 frameshift. In both genomes, fdoG has the opal codon, so that gene is presumably nonfunctional in CO92. Interestingly, in both Y. pestis genomes, the opal codon in fdhF is replaced by a cysteine codon, which should be a functional substitution. This may be a second example of adaptation to an environment in which micronutrients are unavailable. Other electron donors are present, but no glucose dehydrogenase gene was found. Glycerol fermentation is a defining phenotype of biovar Mediaevalis; both glpD and glpABC systems are present. Genes for biosynthesis of quinones are present. Electron acceptors, such as fumarate and nitrite reductases, are present, though not all of those found in E. coli. The nar nitrate reductase systems of E. coli are missing, and the nap homologs, though present, are inactivated by a frameshift mutation in napA, accounting for the characteristic inability of biovar Mediaevalis to reduce nitrate to nitrite. All enzymes involved in the anaerobic dissimilation of pyruvate in E. coli are present in KIM.
Curiously, a KIM locus, hpa, encodes a pathway in aromatic catabolism characterized in E. coli W but not found in K-12. The proteins encoded by this locus, also in Salmonella enterica serovar Dublin and other bacteria, degrade 4-hydroxyphenylacetic acid. In several more steps, compounds are produced that feed back into energy metabolism. Three of the enzymes in this pathway are also similar to ORFs in E. coli C.
(ii) Transport systems.
Mechanisms of iron acquisition have been studied in Y. pestis (16, 35, 36). The genome sequence revealed eight intact putative transport systems for iron and two for heme. Five of these loci are newly discovered. Two previously unknown multidomain “factory proteins,” putative nonribosomal peptide synthetases (NRPS) were found, in addition to the HMWP1 and HMWP2 proteins, whose roles in yersiniabactin synthesis have been elucidated (16). One NRPS system is encoded adjacent to a putative iron-siderophore transport system. Several redundant transport systems found in E. coli are apparently absent from KIM, for example, the high-affinity nickel transport system (NikA to -E; see above), and the AqpZ aquapore is also missing.
(iii) DNA replication and translation.
As noted above, the structure of the replication origin is similar to that of oriC, and all the expected replication genes are present and intact, with the exception of dnaC, of which there is no trace. DnaC guides the DnaB helicase onto the DNA-replisome complex in E. coli. Although it is dispensable in some plasmid systems, it is not clear whether it is essential for genomic replication. We note that Y. pestis grows with a generation time of 1.25 h even under the best conditions. Thus, it probably does not need to initiate multiple replication forks between cell divisions as seen in E. coli at a high growth rate. It is possible that this difference changes the requirement for DnaC or that the Y. pestis functional equivalent has no sequence similarity with DnaC of E. coli. Genes for translation are similar to those in K-12 and are contained in the backbone. There are no ORFs that match the E. coli proteins PrfH (a peptide chain release factor), RimL (an acetyltransferase for ribosomal subunit protein L), YebU (a putative nucleoid protein), and YgcA (a putative RNA methyl transferase). Only one lysine tRNA synthetase is found in KIM, most similar to LysS (E. coli has LysS and -U).
(iv) Motility and chemotaxis.
An essentially complete motility and chemotaxis system is present in the KIM genome, despite the fact that Y. pestis is nonmotile. Two sets of flagellar genes were found, one similar to the systems of Salmonella and E. coli but with a truncated FlhD, a transcriptional activator for the flagellar genes. This probably accounts for the lack of motility. The second gene set is incomplete and much less similar to any characterized system. In addition to the flagellar operons, six other putative chemotaxis-transducing proteins were found besides Tsr and Tap.
(v) Secretion systems.
Searches for similarities to known protein secretion mechanisms revealed that, as expected, KIM has an intact Sec system, components of a signal recognition particle, and components that could specify a twin arginine transfer mechanism for secretion of folded proteins with redox cofactors. Similarly to CO92, KIM has a nearly complete type II secretion system: all the ORFs except the GspM homolog are present. GspM interacts with GspE, -F, and -L to form an inner membrane structure (40). While this protein is required in some systems, it is not known whether it is essential for type II secretion in Y. pestis. Such mechanisms are often used to secrete degradative enzymes, and it will be of interest to learn whether the substrates of this system have primarily nutritional or virulence roles. No obvious type IV secretion mechanism was found.
Type III secretion systems translocate effector proteins from the bacterium directly into the mammalian host target cell. Genes of a type III system are present in the KIM chromosome, but they are more closely related to the Salmonella genes located on island SPI2 than to the Yersinia enterocolitica chromosomal locus. Not all of the Salmonella genes in this group are represented in KIM. The SsaJ homolog, a lipoprotein, is disrupted by a frame shift in KIM but is intact in CO92. The type III gene complement in KIM suggests that all of the essential components are complete for secretion but not translocation. A functional type III system is encoded by the ysc genes on plasmid pCD1 in KIM, as previously described (38). Of the 22 ORFs in the chromosomally encoded locus, only 6 are significantly similar to proteins of the plasmid system. These are orthologs of SsaRSTU/YscRSTU, which form an inner membrane complex, and also YscN, a cytoplasmic ATPase, and YscC/SepC, the outer membrane component.
We examined the genome for potential adhesins that could be important for virulence or maintenance in the flea vector. There is a tadABCDEFG locus in Y. pestis KIM, as previously described for strain CO92 (23). As this locus may be an accessory for secretion and assembly of fibers that could function in biofilm formation (3), we examined the ca. 6 kb upstream of this locus for similarities to a secretin-like protein or to pilins. Although there is homology to a secretin, this ORF is split in strain KIM and, interestingly, is absent in strain CO92 and hence is unlikely to be functional in either strain of Y. pestis.
There are two ORFs (in addition to the previously reported disrupted invA) that could encode large proteins (1,050 and 3,013 aa) with significant similarity to invasin of Yersinia pseudotuberculosis and Y. enterocolitica. Two unlinked tandem pairs of potential proteins have weak similarity to YadA of the enteropathogenic yersiniae, and at least one of each pair has a potentially cleavable signal sequence and could be surface located. Two homologs of Ail of the enteropathogenic yersiniae are present in the Y. pestis KIM chromosome. There are at least 10 predicted proteins that resemble autotransporters in strain KIM. An additional autotransporter-like protein, homologous to YapB in strain CO92, is truncated and hence may not be functional in Y. pestis KIM. Both Y. pestis strains have two large (3,295 and 2,579 aa in strain KIM) predicted surface proteins with similarities to hemagglutinins and hemolysins. Two additional ORFs encode proteins with similarity to known or putative adhesins in E. coli O157:H7. One of these is more similar to the Iha adhesin (45) (54% identical over 686 aa) than it is to its closest counterpart in strain CO92 (34% identical over 683 aa). The rscBAC locus that affects systemic dissemination of Y. enterocolitica (31) is present in Y. pestis KIM, but the predicted RscA homologue, similar to that of Haemophilus influenzae HmwA, is encoded by a broken ORF. This might have the effect of enhancing the disseminative character of Y. pestis KIM, in analogy with the effect of deletion of RscA in Y. enterocolitica. Interestingly, rscA is intact in Y. pestis CO92.
(vi) Gene regulation.
Among the gene expression regulatory systems in KIM, the absence of SoxRS was surprising. Y. pestis survives phagocytosis by macrophages in vitro and certainly encounters oxidative and nitric oxide stress, two of the host's most effective defenses. SoxRS play a central role in the ability of E. coli to survive and adapt to various adverse conditions. SoxR is the redox-sensing activator of SoxS, which is a global regulator of several response genes, including Mn-superoxide dismutase. However, soxS mutants in Salmonella enterica serovar Typhimurium were neither attenuated for virulence in mice nor displayed increased sensitivity to macrophage killing (10, 12, 46). KIM does possess oxyRS, the regulator of a second set of stress response genes which include katG, encoding a catalase or peroxidase, also present in KIM.
This account presents the most unexpected and important features of the KIM genome and its features relative to CO92 and E. coli. While it is by no means a complete analysis of the genome contents, it should provide the foundation for many future experiments aimed at fully characterizing the pathogenicity and lifestyle of Y. pestis.
Acknowledgments
This work was supported by grant AI44387 from NIH/NIAID to F.R.B.
We thank the members of the UW genome sequencing team for their excellent technical support of this project. We thank B. Mishra and T. Ananthraman for contributing algorithms used in optical mapping.
Footnotes
This is paper number 3594 from the Laboratory of Genetics.
REFERENCES
- 1.Achtman, M., K. Zurth, C. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker, R. R. 2000. Yersinia pestis and bubonic plague. In S. F. M. Dworkin, E. D. Rosenberg, K.-H. Schleifer, and E. Stackelbrandt (ed.), The prokaryotes, an evolving electronic resource for the microbiological community. Springer Verlag, New York, N.Y.
- 6.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burland, V., D. L. Daniels, G. Plunkett III, and F. R. Blattner. 1993. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 21:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burland, V., G. Plunkett III, D. L. Daniels, and F. R. Blattner. 1993. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics 16:551-561. [DOI] [PubMed] [Google Scholar]
- 9.Capiaux, H., F. Cornet, J. Corre, M. I. Guijo, K. Perals, J. E. Rebollo, and J. M. Louarn. 2001. Polarization of the Escherichia coli chromosome. A view from the terminus. Biochimie 83:161-170. [DOI] [PubMed] [Google Scholar]
- 10.Dukan, S., and D. Touati. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178:6145-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:11.1-11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, F. C., A. Vazquez-Torres, and Y. Xu. 1997. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect. Immun. 65:5371-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fetherston, J. D., and R. D. Perry. 1994. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol. Microbiol. 13:697-708. [DOI] [PubMed] [Google Scholar]
- 14.Gaasterland, T., and C. W. Sensen. 1996. Fully automated genome analysis that reflects user needs and preferences. A detailed introduction to the MAGPIE system architecture. Biochimie 78:302-310. [DOI] [PubMed] [Google Scholar]
- 15.Gaasterland, T., and C. W. Sensen. 1996. MAGPIE: automated genome interpretation. Trends Genet. 12:76-78. [DOI] [PubMed] [Google Scholar]
- 16.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 17.Guiyoule, A., F. Grimont, I. Iteman, P. A. D. Grimont, M. Lefèvre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannenhalli, S., and P. Pevzner. 1995. Transforming Cabbage into Turnip (polynomial algorithm for sorting signed permutations by reversal), p. 178-189. In Proceedings of the 27th Annual ACM Symposium on the Theory of Computing. ACM Press, New York, N.Y.
- 20.Hare, J. M., and K. A. McDonough. 1999. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J. Bacteriol. 181:4896-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, C. W., C. H. Sandt, and D. A. Vlazny. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12:865-871. [DOI] [PubMed] [Google Scholar]
- 22.Hu, P., J. Elliott, P. McCready, E. Skowronski, J. Garnes, A. Kobayashi, R. R. Brubaker, and E. Garcia. 1998. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodursky, A. B., B. J. Peter, M. B. Schmid, J. DeRisi, D. Botstein, P. O. Brown, and N. R. Cozzarelli. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. USA 97:9419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett III, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J., R. Qi, C. Aston, J. Jing, T. S. Anantharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 28.Lindler, L. E., G. V. Plano, V. Burland, G. F. Mayhew, and F. R. Blattner. 1998. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 66:5731-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahillon, J., H. A. Kirkpatrick, H. L. Kijenski, C. A. Bloch, C. K. Rode, G. F. Mayhew, D. J. Rose, G. Plunkett III, V. Burland, and F. R. Blattner. 1998. Subdivision of the Escherichia coli K-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene 223:47-54. [DOI] [PubMed] [Google Scholar]
- 30.Myers, R. S., A. Kuzminov, and F. W. Stahl. 1995. The recombination hot spot chi activates RecBCD recombination by converting Escherichia coli to a recD mutant phenocopy. Proc. Natl. Acad. Sci. USA 92:6244-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, K. M., G. M. Young, and V. L. Miller. 2001. Identification of a locus involved in systemic dissemination of Yersinia enterocolitica. Infect. Immun. 69:6201-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 34.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 35.Perry, R. D., S. W. Bearden, and J. D. Fetherston. 2001. Iron and heme acquisition and storage systems of Yersinia pestis. Recent Res. Dev. Microbiol. 5:13-27. [Google Scholar]
- 36.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry, R. D., S. C. Straley, J. D. Fetherston, D. J. Rose, J. Gregor, and F. R. Blattner. 1998. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 66:4611-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2:244-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segall, A., M. J. Mahan, and J. R. Roth. 1988. Rearrangement of the bacterial chromosome: forbidden inversions. Science 241:1314-1318. [DOI] [PubMed] [Google Scholar]
- 43.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 44.Straley, S. C., and M. N. Starnbach. 2000. Yersinia: strategies that thwart immune defenses, p. 71-92. In M. W. Cunningham and R. S. Fujinami (ed.), Effects of microbes on the immune system. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 45.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor, P. D., C. J. Inchley, and M. P. Gallagher. 1998. The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect. Immun. 66:3208-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, T., and I. Taneike. 2000. The sequences of enterohemorrhagic Escherichia coli and Yersinia pestis that are homologous to the enteroaggregative E. coli heat-stable enterotoxin gene: cross-species transfer in evolution. FEBS Lett. 472:22-26. [DOI] [PubMed] [Google Scholar]