SUMMARY
In the yeast Saccharomyces cerevisiae the transcription of many genes encoding enzymes of phospholipid biosynthesis are repressed in cells grown in the presence of the phospholipid precursors inositol and choline. A genome-wide approach using cDNA microarray technology was utilized to profile the changes in the expression of all genes in yeast that respond to the exogenous presence of inositol and choline. We report that the global response to inositol is completely distinct from the effect of choline. Whereas the effect of inositol on gene expression was primarily repressing, the effect of choline on gene expression was activating. Moreover, the combination inositol and choline increased the number of repressed genes compared to inositol alone and enhanced the repression levels of a subset of genes that responded to inositol. In all, 110 genes were repressed in the presence of inositol and choline. Two distinct sets of genes exhibited differential expression in response to inositol or the combination of inositol and choline in wild type cells. One set of genes contained the UASINO sequence and were bound by Ino2p and Ino4p. Many of these genes were also negatively regulated by OPI1, suggesting a common regulatory mechanism for Ino2p, Ino4p, and Opi1p. Another non-overlapping set of genes were coregulated by the unfolded protein response pathway, an ER-localized stress response pathway, but were not dependent on OPI1 and did not show further repression when choline was present together with inositol. These results suggest that inositol is the major effector of target gene expression, while choline plays a minor role.
Keywords: Microarray, Lipid Metabolism, Inositol, Choline, Unfolded Protein Response
INTRODUCTION
Phospholipids are the key structural elements of membrane-bounded organelles and play important roles in signaling and membrane trafficking pathways. Each membrane compartment is composed of a unique set of phospholipids whose biophysical properties contribute to the function of each organelle. Phospholipid metabolism is highly regulated by the cell, ensuring the biogenesis and growth of membranes by coordinating the relative rates of synthesis of individual phospholipids with numerous factors such as the availability of exogenous supplies of phospholipid precursors, growth stage, and membrane trafficking (1,2).
In many organisms, the regulation of lipid biosynthetic pathways is achieved through control of transcriptional regulatory networks. For example, lipid homeostasis in animal cells is controlled through the sterol regulatory element binding protein (SREBP) pathway (3). In the budding yeast Saccharomyces cerevisiae, a major control mechanism for the coordinated synthesis of inositol- and choline-containing phospholipids is the precise transcriptional control of genes for many of the enzymes required for phospholipid synthesis (2,4,5). The synchronized expression of these genes requires the participation of Ino2p and Ino4p, basic helix-loop-helix transcription factors, which bind as a heterodimer to a cis-acting element in the promoters of these genes called UASINO, as well as Opi1p, a negative regulator of transcription (6-15). Many of these UASINO-containing genes are maximally derepressed when the phospholipid precursors inositol and choline are absent from the growth medium in logarithmically growing cultures (16-25), suggesting a common regulatory mechanism (4).
A recent collaborative study involving our laboratory has begun to reveal how this regulation is controlled by ongoing lipid metabolism (26). When inositol is added to the medium of growing cells, a dramatic change to the pattern of phospholipid synthesis is induced. Phosphatidic acid (PA)1, a precursor for the synthesis of phosphatidylinositol, is rapidly consumed. This drop in PA levels is directly sensed by Opi1p, a component of an endoplasmic reticulum (ER)-localized lipid sensing complex (27), causing it to dissociate from the ER and translocate to the nucleus, where it participates in the repression of target genes. While Opi1p does not directly interact with the UASINO sequence (28,29), it is thought to interact with Sin3p (15), a histone deacetylase that functions as a global transcriptional repressor, and Ino2p, which potentially targets it to the promoters of UASINO-containing genes. Thus, the transcriptional regulation of UASINO-containing genes does not respond to inositol directly, but instead to a metabolic signal induced when inositol participates in lipid metabolism.
Cells grown in the absence of inositol also induce the unfolded protein response (UPR) pathway (30). The UPR pathway is an ER-localized signal transduction pathway that responds to the accumulation of unfolded proteins in the lumen of the ER as well as to secretory stress by upregulating the expression of target genes (31-35). While the mechanism for UPR induction under inositol-limiting conditions is unknown, Cox et al. (30) have suggested that the activation of the UPR might be directly involved in the mechanism by which INO1 transcription is activated. However, recent work from our laboratory has suggested that under certain conditions, UPR activation is not coupled to activation of UASINO-containing genes (35,36).
While much study of the regulatory effects of inositol and choline have focused upon the transcription of UASINO-containing genes, their effects on the expression of other genes have been largely unexplored. A recent report (37) examined the combined effects of inositol and choline on genome-wide expression in yeast, but the individual contributions of inositol and choline on gene expression were not explored. Since inositol and choline have distinct effects on different branches of phospholipid biosynthetic pathways (4) (Fig. 1), an important unanswered question is whether the presence of these phospholipid precursors individually affects the expression of the same set(s) of genes. In order to identify all of the genes whose expression is regulated by inositol and choline, we carried out a genome-wide study using cDNA microarrays to find genes whose expression is regulated, both independently and jointly, by inositol and choline. We report that inositol and choline affect the expression of different sets of genes. Furthermore, we show that OPI1 primarily regulates genes that were previously shown by Lee et al. (38) to contain a bound Ino2p and Ino4p, and these genes represent a distinct set from those genes regulated by the UPR pathway. Our results indicate that inositol, but not choline, is the major effector of Ino2p-Ino4p and UPR target gene expression. Together these results suggest that distinct pathways contribute to regulate the expression of genes in response to changes in phospholipid metabolism induced by phospholipid precursors.
Figure 1. Yeast phospholipid biosynthetic pathways.
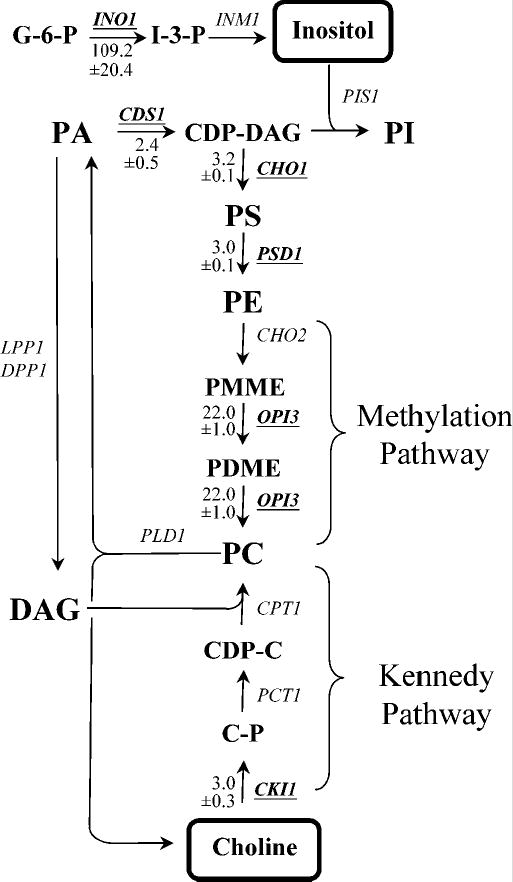
Soluble phospholipid precursors: (G-6-P, glucose-6-phosphate; I-3-P, inositol-3-phosphate; C-P, choline phosphate; CDP-C, cytidinediphosphate choline). Phospholipids: (PA, phosphatidic acid; DAG, diacylglycerol; CDP-DAG, cytidinediphosphate diacylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PMME, phosphatidylmonomethylethanolamine; PDME, phosphatidyldimethylethanolamine; PC, phosphatidylcholine). Arrows represent routes of metabolic conversion. The names of the structural genes for enzymes catalyzing specific metabolic conversions are shown adjacent to the arrows. Genes that were shown in the present study to be repressed in the presence of inositol and choline (Table II and Fig. 4) are underlined. Values indicate the fold level of repression in response to the combination of inositol and choline was well as the standard deviation in the level of expression (−/+) for indicated genes. The positions at which inositol and choline enter the metabolic pathway are boxed.
EXPERIMENTAL PROCEDURES
Strains and Media
The wild type strain BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ 0, ura3Δ0) derived from S288C (39) and an isogenic opi1Δ strain (MATα, his3Δ1, leu2Δ0, lys2Δ 0, ura3Δ0, opi1::KanMx) in which the OPI1 gene was replaced with KanMx (purchased from Research Genetics) were used for the cDNA microarray experiments. Strains were maintained on YPD plates (1% yeast extract, 2% Bactopeptone, 2% glucose, 2% agar). All experiments were performed using chemically defined synthetic complete media (16,40), containing (per liter): 20 g glucose, 5 g ammonium sulfate, 1 g potassium phosphate, 0.5 g magnesium sulfate, 0.1 g sodium chloride, 0.1 g calcium chloride, 0.5 mg boric acid, 0.04 mg cupric sulfate, 0.1 mg potassium iodide, 0.2 mg ferric chloride, 0.4 mg manganese sulfate, 0.2 mg sodium molybdate, 0.4 mg zinc sulfate, 2 μg biotin, 400 μg calcium pantothenate, 2 μg folic acid, 400 μg niacin, 200 μg p-aminobenzoic acid, 400 μg pyridoxine hydrochloride, 200 μg riboflavin, 400 μg thiamine, 20 mg adenine sulfate, 20 mg arginine, 20 mg histidine, 60 mg leucine, 230 mg lysine, 20 mg methionine, 300 mg threonine, 20 mg tryptophan, 40 mg uracil. Where indicated, media were supplemented with 75 μM myo-inositol and/or 1 mM choline.
Cell Growth
All cultures were grown in synthetic complete growth medium lacking inositol and choline or containing combinations of inositol and choline as indicated at 30 °C with shaking for at least 12 generations prior to harvesting at mid-log phase as described below. Following overnight growth in indicated media at 30 °C, strains were diluted 1:50 in the indicated media and grown for 5-6 hours at 30 °C. 250 ml of media were inoculated at an OD600 of 0.01 and allowed to grow at 30 °C until the cultures reached mid-log phase growth (OD600 = 0.5). Triplicates were prepared for each growth condition. Cells were harvested by filtration, immediately frozen on a dry ice/ethanol bath, and stored at −80 °C. Filtration was chosen as the method for harvesting cells, as the recovery of the INO1 transcript was the most reproducible using this procedure.
RNA Isolation and Microarray Analysis
Total RNA was extracted by the hot acid phenol method (41,42). For some experiments, total RNA was further purified using the RNeasy RNA purification kit (QIAGEN, Inc.), or poly(A) RNA was isolated using oligo dT cellulose as described (41,42). mRNA was reverse-transcribed from 20 μg purified total RNA using 2 μg oligo dT (18-mer) primers (New England Biolabs) or 2.5 μg mRNA using 3 μg random (9-mer) primers (Invitrogen) by incubating with 400 units of SuperScript II (Invitrogen) plus 500 μM each dATP, dCTP, dGTP, 200 μM amino-allyl dUTP (Sigma) and 300μM dTTP in a final volume of 40 μl at 42 °C for 2 hours. After cDNA synthesis, the remaining RNA was hydrolyzed by adding 10 μl each 1M NaOH and 0.5 M EDTA and incubating at 65 °C for 15 min followed by neutralization with 10 μL 1M HCl. The resulting cDNAs were purified using the QIAQuick PCR purification kit (QIAGEN, Inc.), substituting kit buffers with KPO4 buffers. Purified cDNAs were labeled with monofunctional reactive Cy3 or Cy5 dye esters (Amersham Pharmacia) in the presence of Na2CO3 pH 9.0 for 1 hour at room temperature and subsequently quenched with 1M NH2OH. Following an additional QIAQuick purification, 20 pmoles of Cy3-and Cy5-labeled cDNA probes were combined and hybridized to Corning CMT Yeast-S288c Gene Arrays (version 1.32) in 5 × SSC, 25 % formamide, 2.5 % SDS, and 100 hg/ml salmon sperm DNA at 42 °C for 14 – 16 hours. After washing, hybridized microarray slides were simultaneously scanned with 532 nm and 635 nm lasers using a GenePix 4000B array scanner (Axon Instruments, Inc.).
Statistical Analysis
Three replicates were analyzed for each experimental condition. Image analysis for each array was processed using the GenePix Pro 4.0 (Axon Instruments, Inc.) software package, which produces (R, G) fluorescence intensity pairs for each gene. After image acquisition, individual data spots on each microarray were visually inspected for size, signal-to-noise ratio, background level, and uniformity. Using these quality control criteria, about 30 % of the spots for each triplicate set of experiments were discarded due to poor spot quality, a conventional practice for microarray data analysis (43). Normalization was conducted as follows: Let M=log2(R/G), A=0.5* log2(R*G). The log ratio M is known to be dependent on overall spot intensity A (44). To remove this systematic variation, intensity-dependent normalization was conducted for each array replicate. The intensity-dependent trend was fitted using the LOWESS fit function (45) in S-PLUS, following which the log ratio values (M) were normalized by subtracting the trend values. The student t-test was performed on normalized M, for each gene for which three high quality replicates were available using the null hypothesis of no change in expression, (i.e., normalization to M = 0). A conservative significance level of p-value equal to 0.025 corresponding to a false detection rate (FDR) of q=0.075 was chosen. Hence the percentage of false positives among the significant tests is controlled to be at most 7.5%, a conservative approach that is expected to reduce false positive identifications to near zero. See Storey (46) and Storey and Tibshirani (47) for details on relating adjusted p-values to FDR and the expected number of false positive tests. The t-statistic was computed for each gene, and genes were then ranked according to their p-values. Genes were considered to have differential expression in the red channel vs. the green channel (or vice versa) if they exhibited a two-fold change while also having a p-value smaller than 0.025.
Northern Slot Blot Analysis
Approximately 250 ng of unfractionated mRNA was heated at 65 °C for 15 min in 3 volumes of denaturing buffer (66 % formamide, 7 % formaldehyde, 26 mM MOPES, 66 mM sodium acetate, 1.3 mM EDTA), placed on ice after addition of an equal volume of ice-cold 20 × SSC, and spotted on BrightStar-Plus (Ambion, Inc.) nylon membranes using a manifold slot blot apparatus as described (41). Strand-specific 32P-labeled riboprobes were synthesized from plasmids pJH310-INO1 (16), pSPACT (36), pBDG456-Ty1 (kind gift of D. Garfinkel), pBDG458-Ty2 (kind gift of D. Garfinkel), pSJ29-KAR2, pSJ30-PDI1, pSJ31-VTC3 and pSJ32-HO by in vitro transcription according to manufacturer’s instructions (Promega). pSJ29-KAR2 was constructed by PCR amplifying the KAR2 ORF and inserting the 878-bp HindIII-XbaI fragment into pGEM1 (Promega). pSJ30-PDI1 was constructed by PCR amplifying the PDI1 ORF and inserting the 861-bp HindIII-SalI fragment into pGEM1. pSJ31-VTC3 was constructed by PCR amplifying the VTC3 ORF and inserting the 703-bp PstI-XbaI fragment into pGEM1. pSJ32-HO was constructed by PCR amplifying the HO ORF and inserting the 365-bp PstI-BamHI fragment into pGEM1. Membranes were hybridized with either INO1, KAR2, PDI1, VTC3, HO, ACT1, Ty1, or Ty2 probes in formamide hybridization buffer and washed to a final stringency of 0.2 × SSC/0.1 % SDS at 60 °C as described (16,41). Quantitation was performed by analysis on a STORM 860 phosphorimager (Molecular Dynamics) and analyzed with ImageQuant software. The data were normalized by dividing the total counts per minute for the INO1, KAR2, PDI1, VTC3, HO, Ty1, or Ty2 probe by the total counts per minute for the ACT1 probe and expressed as the fraction of the amount of mRNA from cells grown in the absence of inositol and choline.
Construction of Gene Disruption Alleles
Complete disruptions of VTC1, VTC3, and VTC4 genes were constructed by PCR-mediated gene replacement as described (48) in the wild type strain BY4742. The plasmid pFA6a-His3MX6 (kind gift of M. Longtine) was used as a template to generate PCR fragments for the gene disruptions. The entire open reading frame of each gene was replaced with the HIS3 marker gene. Histidine prototrophs were screened by colony PCR to verify integration at the correct genetic locus.
Assays for Ino− and Opi− Phenotypes
To test for Ino− phenotypes, strains were grown to mid-log phase in synthetic complete media containing 75 μM inositol, washed once with water, and 10-fold serial dilutions spotted on plates containing synthetic complete medium lacking inositol. Strains were incubated for 3 days at 18 °C, 24 °C, 30 °C, and 37 °C. To test for Opi− phenotypes (49,50), strains were grown as described above and spotted on plates containing synthetic complete medium lacking inositol and incubated at 30 °C and 37 °C for 2 days. The plates were then sprayed with a suspension of a diploid tester strain (AID) homozygous for ino1 and ade2 and incubated for an additional 2 days.
RESULTS
Global analysis of inositol and choline on gene expression in yeast
Previous studies have shown that the presence of phospholipid precursors in the medium of logarithmically growing cultures of Saccharomyces cerevisiae regulates the transcriptional response of a number of genes. These genes include those encoding the structural enzymes for phospholipid biosynthesis (4) as well as genes encoding protein folding chaperones induced by the UPR pathway (51). The goal of the present study was to identify, in a comprehensive manner, the complete set of genes whose expression levels change in response to the presence or absence of the phospholipid precursors inositol and choline using cDNA microarray technology.
The cDNA microarray experiments performed in this study are shown in Table I and are summarized below. First, to determine the individual effects of inositol and choline on genome-wide expression, the relative mRNA abundance from cells grown in the presence of either inositol or choline was measured by comparing to transcript levels from cells grown in the absence of both inositol and choline (Table I, Experiments 1 and 2). Next, to characterize the combined effects of inositol and choline on genome-wide expression, the relative mRNA abundance from cells grown in the presence of both inositol and choline was compared to transcript levels from cells grown in the absence of inositol with or without choline (Table I, Experiments 3 and 4). For every experiment, fluorescently labeled cDNA probes were synthesized from RNA extracted from cells grown to mid-logarithmic phase in chemically defined synthetic media and competitively hybridized to commercial spotted cDNA microarrays containing 6,135 unique Saccharomyces cerevisiae open reading frames (ORFs). Each experiment was performed in triplicate. To assess which ORFs showed differential regulation from each experiment, rigorous statistical criteria were utilized as described in the “Experimental Procedures”. Briefly, after normalization, the ratio for each spot from the array that showed a p-value ≤ 0.025 over three replicates were considered to be differentially expressed. After statistical analysis, data collected from these experiments were compared and analyzed as described below. The complete dataset is available at our laboratory website (http://www.mbg.cornell.edu/Henry_Lab.cfm).
Table I. List of Experiments Performed in this Study.
I+ or I− indicates the presence or absence of 75 μM inositol. C+ or C− indicates the presence or absence of 1 mM choline.
| No. | Experiment Name | Experimental sample | Reference sample |
|---|---|---|---|
| 1 | Inositol dependent | Wild type I+C− | Wild type I−C− |
| 2 | Choline dependent | Wild type I−C+ | Wild type I−C− |
| 3 | Inositol + Choline dependent (i) | Wild type I+C+ | Wild type I−C− |
| 4 | Inositol + Choline dependent (ii) | Wild type I+C+ | Wild type I−C+ |
| 5 | OPI1 dependent (+ Inositol) | opi1Δ I+C− | Wild type I+C− |
| 6 | OPI1 independent (− Inositol) | opi1Δ I−C− | Wild type I−C− |
| 7 | opi1Δ control | opi1Δ I+C− | opi1Δ I−C− |
In the first experiment (Table I, Experiment 1), which measured the independent effects of inositol on genome-wide expression, a total of 32 genes showed a 2-fold or greater change in expression that met our statistical criteria (Fig. 2A). Among these genes, 29 were downregulated in the presence of inositol as compared to cells grown in its absence. These genes are listed in Table II. As expected, the transcript levels of numerous genes previously shown to be affected by inositol levels were downregulated when inositol was present, including INO1 (16), OPI3 (18,21), ITR1 (23), and PSD1 (25). INO1 showed the greatest difference in expression, consistent with previous reports that this gene is the most highly regulated of the UASINO-containing genes (2). A large number of genes involved in protein folding and secretion, including genes not previously shown to be regulated by inositol, were also downregulated when inositol was present. Surprisingly, only three genes—HO, which encodes the endonuclease involved in mating-type switching, and two uncharacterized ORFs—showed the reverse pattern and were upregulated in the presence of inositol (Table III), suggesting that the presence of inositol induces the expression of a very small number of genes.
Figure 2. Differential effects on the regulation of gene expression by inositol and choline.
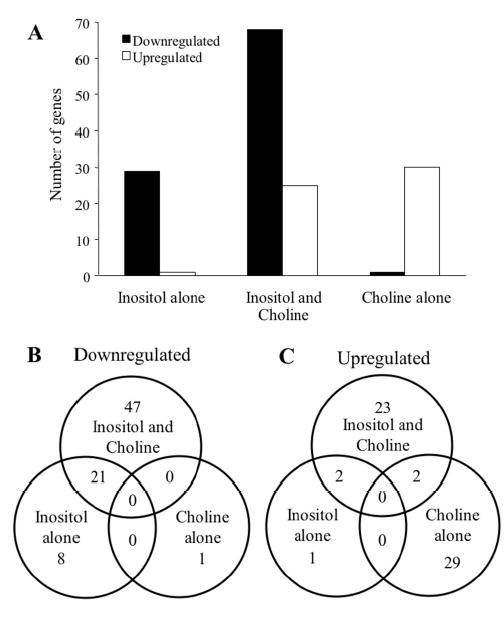
A. The total number of genes that showed a 2-fold or greater change in expression levels are shown for genes identified in experiments for inositol alone (Experiment 1), inositol and choline (Experiment 3) and choline alone (Experiment 2). B. Venn diagrams of downregulated genes identified in Experiments 1, 2, and 3 are shown. Numerals indicate the number of overlapping and non-overlapping genes. C. Venn diagram of upregulated genes identified in Experiments 1, 2, and 3 are shown.
Table II. Genes Downregulated by Inositol.
Description of Experiments are listed in Table I. Negative and positive values indicate downregulation and upregulation, respectively. Expression ratios in boldface indicate ≥ 2-fold difference. Overlapping expression ratios ≤ 2-fold meeting p-value cutoff of 0.025 are listed for comparison.
| Fold Difference
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Expt 1 I+ | Expt 2 C+ | Expt 3 I+C+ (i) | Expt 4 I+C+ (ii) | Expt 5 opi1Δ I+ | Expt 6 opi1Δ I− | Expt 7 control | Description |
| Ino2p-Ino4p Target Genes* | ||||||||
| INO1 | −49.7 | −109.2 | −156.1 | 96.8 | inositol-3-phosphate synthase | |||
| OPI3 | −3.6 | −22.0 | −20.5 | 5.8 | 1.6 | phosphatidyl-N-methylethanolamine N-methyltransferase | ||
| PSD1 | −2.0 | −1.4 | −3.0 | −2.3 | 2.6 | 1.5 | phosphatidylserine decarboxylase | |
| CHO1 | −1.4 | −3.2 | −2.8 | 2.2 | CDP-diacylglycerol-serine O-phosphatidyltransferase | |||
| CDS1 | −1.4 | −2.4 | 2.1 | 1.5 | phosphatidate cytidylyltransferase | |||
| CKI1 | −3.0 | 1.9 | choline kinase | |||||
| ERG20 | −2.0 | −2.0 | 1.6 | dimethylallyltranstransferase | ||||
| OLE1 | −2.1 | stearoyl-CoA 9-desaturase | ||||||
| SAH1 | −1.8 | −3.4 | −2.8 | 1.8 | 1.3 | adenosylhomocysteinase | ||
| ITR1 | −2.6 | −8.6 | −7.0 | 3.6 | 1.6 | myo-inositol transporter | ||
| HNM1 | −2.7 | −2.1 | 1.3 | 1.3 | choline transporter | |||
| INO2 | −2.4 | −2.0 | 1.8 | transcription factor | ||||
| PHO81 | −2.4 | −2.2 | 2.1 | cyclin-dependent protein kinase inhibitor | ||||
| PBI2 | −1.9 | −2.3 | −3.5 | 1.7 | endopeptidase inhibitor | |||
| SRO77 | −2.6 | −2.2 | 2.6 | 1.7 | Golgi to plasma membrane transport | |||
| YIP3 | −2.2 | −3.1 | −3.1 | 2.4 | interacts with YPT proteins | |||
| ADO1 | −1.4 | −2.7 | −2.3 | 1.7 | 1.5 | adenosine kinase | ||
| GSC2 | −1.8 | −2.2 | −2.4 | −1.6 | 1,3-beta-glucan synthase | |||
| KNH1 | −1.9 | 2.4 | KRE9 homolog | |||||
| ORM2 | −2.4 | −3.0 | response to unfolded protein | |||||
| NRG2 | −3.2 | −4.5 | 2.2 | transcriptional repressor | ||||
| RMD12 | −1.9 | −2.2 | 2.2 | mitochondrial protein required for sporulation | ||||
| TYE7 | −1.7 | −2.3 | 1.4 | −1.1 | transcription factor | |||
| YEL073C | −3.3 | −1.4 | −4.9 | −4.2 | 5.1 | 1.8 | unknown | |
| YER034W | −2.1 | unknown | ||||||
| YJR008W | −2.2 | 3.2 | unknown | |||||
| YLR132C | −2.8 | unknown | ||||||
| YNL208W | −1.7 | −1.8 | −2.4 | 1.5 | unknown | |||
| UPR Target Genes* | ||||||||
| KAR2 | −2.4 | −2.3 | −3.0 | −1.9 | chaperone | |||
| SCJ1 | −1.8 | −1.9 | −2.2 | 1.3 | co-chaperone | |||
| ERO1 | −2.6 | 1.7 | −2.3 | −3.0 | −2.8 | electron carrier | ||
| FPR2 | −2.3 | −2.5 | peptidyl-prolyl cis-trans isomerase | |||||
| MPD1 | −2.1 | −2.1 | −2.8 | protein disulfide isomerase | ||||
| PDI1 | −2.2 | −1.8 | −2.4 | protein disulfide isomerase | ||||
| MCD4 | −1.8 | −1.9 | −2.1 | −2.3 | 1.2 | GPI anchor biosynthesis | ||
| DER1 | −1.6 | −2.1 | protein degradation | |||||
| SIL1 | −2.3 | −1.8 | −2.2 | −2.8 | interacts with Kar2p | |||
| SWA2 | −1.6 | −2.0 | −2.2 | 1.3 | auxilin-like protein | |||
| ATG17 | 1.7 | −1.7 | −2.1 | kinase activator | ||||
| DOG1 | −1.5 | −1.9 | −2.1 | 1.1 | 2-deoxyglucose-6-phosphatase | |||
| ECM27 | −1.7 | −2.2 | −2.4 | ExtraCellular Mutant | ||||
| MNN4 | −1.6 | −2.0 | mannosyltransferase | |||||
| PAU2 | −2.2 | seriPAUperin family | ||||||
| YJR107W | −1.6 | −2.1 | −2.1 | lipase | ||||
| YBL101W-A | −2.4 | TyA Gag protein | ||||||
| YFR026C | −2.6 | −2.1 | −2.9 | unknown | ||||
| YGR294W | −1.4 | −2.3 | unknown | |||||
| YHL046C | −2.0 | unknown | ||||||
| YIL176C | −2.0 | 1.4 | unknown | |||||
| YMR040W | −2.5 | −6.6 | unknown | |||||
| YMR184W | −2.2 | −2.3 | −2.7 | −1.3 | unknown | |||
| Other Genes | ||||||||
| MET13 | −1.4 | −1.7 | −3.3 | −2.1 | methylenetetrahydofolate reductase | |||
| SAM2 | −1.8 | −2.6 | −2.0 | 2.6 | 1.4 | 1.3 | methionine adenosyltransferase | |
| PHO84 | −7.5 | −9.8 | 2.4 | inorganic phosphate transporter | ||||
| VTC1 | −2.2 | −2.4 | −2.2 | 1.8 | Vacuolar Transporter Chaperone | |||
| VTC3 | −3.8 | −4.2 | −3.8 | Vacuolar Transporter Chaperone | ||||
| VTC4 | −2.4 | −2.4 | −1.9 | 1.5 | Vacuolar Transporter Chaperone | |||
| YHR138C | −2.1 | endopeptidase inhibitor | ||||||
| YDR034C-C | 1.5 | −2.0 | TyA Gag protein | |||||
| YDR210W-A | 1.4 | −2.1 | TyA Gag protein | |||||
| YFL002W-B | −1.9 | −3.2 | −3.5 | TyA Gag protein | ||||
| YGR161W-A | −1.3 | −2.0 | −2.7 | TyA Gag protein | ||||
| YLR410W-A | −2.2 | −3.0 | TyA Gag protein | |||||
| YOR192C-A | −2.1 | TyA Gag protein | ||||||
| YOR343W-A | −1.8 | −1.7 | −2.3 | TyA Gag protein | ||||
| BNA2 | −3.7 | −3.5 | −6.6 | 1.7 | tryptophan 2,3-dioxygenase | |||
| ALD2 | −1.5 | 1.6 | −2.1 | aldehyde dehydrogenase | ||||
| AMS1 | −2.2 | −3.5 | −2.6 | alpha-mannosidase | ||||
| APE2 | −2.0 | leucyl aminopeptidase | ||||||
| ARP10 | −2.1 | 2.0 | −1.4 | Actin-Related Protein | ||||
| ATP14 | −1.7 | −1.9 | −2.1 | hydrogen-transporting ATP synthase | ||||
| BIO5 | −2.1 | permease | ||||||
| CAR1 | −1.5 | −1.8 | −2.0 | arginase | ||||
| FMP46 | −2.0 | −2.6 | 1.8 | Found in Mitochondrial Proteome | ||||
| FOL1 | 2.0 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | ||||||
| GAL4 | −1.6 | 2.0 | transcriptional activator | |||||
| GPG1 | −2.7 | signal transducer | ||||||
| GWT1 | −1.5 | −2.0 | −2.0 | GPI anchor biosynthesis | ||||
| HIT1 | −2.1 | HIgh Temperature growth | ||||||
| HNT3 | 2.1 | Histidine triad NucleoTide-binding | ||||||
| HSP26 | −1.4 | −2.5 | −1.9 | chaperone | ||||
| HSP30 | −2.1 | −2.9 | −1.4 | heat shock protein | ||||
| HXT12 | −2.5 | −3.7 | HeXose Transporter | |||||
| IDS2 | −1.5 | −1.9 | −2.0 | IME2-Dependent Signaling | ||||
| KTR2 | −2.0 | 1.2 | mannosyltransferase | |||||
| MED11 | −2.2 | RNA polymerase II transcription mediator | ||||||
| MER1 | −2.3 | 1.6 | pre-mRNA splicing factor | |||||
| MOG1 | −2.2 | −2.2 | RAN protein binding | |||||
| PRB1 | −1.9 | −2.3 | −3.3 | serine-type endopeptidase | ||||
| RTS3 | −1.7 | −2.2 | protein phosphatase type 2A | |||||
| SML1 | −1.4 | −2.0 | −2.0 | enzyme inhibitor | ||||
| SNA3 | −2.0 | −2.1 | Sensitivity to Na+ | |||||
| SPG5 | −1.9 | −1.9 | −2.1 | Stationary Phase Gene | ||||
| SPS19 | −2.0 | −1.2 | −1.9 | 2,4-dienoyl-CoA reductase (NADPH) | ||||
| TPO4 | −2.1 | −2.3 | −2.7 | spermine transporter | ||||
| WSS1 | −2.1 | weak suppressor of smt3 | ||||||
| YHC1 | 2.3 | mRNA binding | ||||||
| YPS1 | −1.4 | 1.5 | −2.4 | aspartic-type endopeptidase | ||||
| YPS3 | −1.1 | −2.4 | aspartic-type endopeptidase | |||||
| GON3 | −1.4 | −2.4 | unknown | |||||
| ICT1 | −2.1 | −2.2 | unknown | |||||
| YDL183C | −2.0 | unknown | ||||||
| YDR210W | −1.4 | −2.9 | −2.5 | unknown | ||||
| YDR542W | −1.5 | −2.1 | 2.3 | unknown | ||||
| YER130C | −1.8 | −2.0 | unknown | |||||
| YER138W-A | −1.4 | −2.0 | −2.1 | unknown | ||||
| YIL175W | −2.3 | unknown | ||||||
| YLL053C | −2.6 | unknown | ||||||
| YLR168C | −1.5 | −2.1 | −1.8 | 2.1 | −1.2 | unknown | ||
| YLR173W | −2.1 | −1.7 | 2.0 | unknown | ||||
| YLR194C | −1.9 | −2.6 | unknown | |||||
| YLR414C | −1.3 | −1.6 | −2.0 | unknown | ||||
| YOL002C | −2.2 | unknown | ||||||
| YOR289W | −2.1 | −3.3 | 1.8 | unknown | ||||
| YPL095C | −2.7 | unknown | ||||||
Criteria for classification of Ino2p-Ino4p Target Genes and UPR Target Genes are described in the text of the Results.
Table III. Genes Upregulated by Inositol.
Description of Experiments are listed in Table I. Negative and positive values indicate downregulation and upregulation, respectively. Expression ratios in boldface indicate ≥ 2-fold difference. Overlapping expression ratios ≤ 2-fold meeting p-value cutoff of 0.025 are listed for comparison.
| Fold Difference
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Expt 1 I+ | Expt 2 C+ | Expt 3 I+C+ (i) | Expt 4 I+C+ (ii) | Expt 5 opi1Δ I+ | Expt 6 opi1Δ I− | Expt 7 control | Description |
| HO* | 2.6 | 3.4 | 4.1 | homothallic switching endonuclease | ||||
| PRY3* | −1.7 | 1.7 | 2.2 | Pathogen Related in Yeast | ||||
| SCW11* | 1.3 | 2.2 | 2.1 | glucan 1,3-beta-glucosidase | ||||
| WSC4* | 2.1 | −2.0 | transmembrane receptor | |||||
| YML131W* | 1.8 | 2.0 | −1.5 | 1.2 | unknown | |||
| AEP2 | 2.1 | ATPase ExPression | ||||||
| AEP3 | −2.2 | ATPase ExPression | ||||||
| AGA1 | 2.7 | −3.2 | cell adhesion molecule binding | |||||
| ARO10 | 1.3 | 2.8 | 2.6 | 1.7 | carboxy-lyase | |||
| BAP3 | 1.5 | 1.5 | 2.0 | −1.5 | amino acid transporter | |||
| BDP1 | 1.4 | −2.1 | transcription factor | |||||
| CDC13 | −2.1 | single-stranded DNA binding | ||||||
| COG4 | 1.3 | 2.0 | 1.7 | Conserved Oligomeric Golgi complex | ||||
| COS111 | −2.0 | Ciclopirox Olamine Sensitive | ||||||
| CUP1-1 | 2.4 | 1.8 | copper ion binding | |||||
| CUP1-2 | 2.3 | 1.8 | copper ion binding | |||||
| CWP1 | 1.9 | 2.0 | structural constituent of cell wall | |||||
| DAL81 | 1.5 | 2.6 | 2.3 | −1.8 | −1.6 | transcription factor | ||
| DSE1 | 1.7 | 1.2 | 3.2 | 3.1 | Daughter Specific Expression | |||
| ELG1 | 1.5 | 2.2 | −1.2 | Enhanced Level of Genomic instability | ||||
| FAR11 | 2.0 | −1.4 | Factor ARrest | |||||
| GCV2 | 1.4 | 2.2 | glycine dehydrogenase (decarboxylating) | |||||
| GLG1 | −2.0 | glycogenin glucosyltransferase | ||||||
| HIR2 | −2.2 | transcription corepressor | ||||||
| IDH1 | 1.6 | 2.1 | 2.5 | −1.5 | isocitrate dehydrogenase (NAD+) | |||
| IDH2 | 1.8 | 2.2 | −1.3 | isocitrate dehydrogenase (NAD+) | ||||
| IMD1 | 1.4 | 2.0 | 2.3 | IMP Dehydrogenase | ||||
| IMD2 | 1.4 | 1.9 | 2.4 | IMP dehydrogenase | ||||
| ISR1 | 1.5 | 2.4 | protein kinase | |||||
| MCD1 | 1.5 | 2.1 | 2.0 | −1.3 | Mitotic Chromosome Determinant | |||
| MCM6 | 2.0 | chromatin binding | ||||||
| MET10 | 1.8 | 2.6 | 2.6 | −1.8 | sulfite reductase (NADPH) | |||
| MET17 | 2.3 | O-acetylhomoserine aminocarboxypropyltransferase | ||||||
| MF(ALPHA)2 | 1.7 | 2.9 | −1.4 | pheromone | ||||
| MGM1 | 1.5 | 2.4 | −1.6 | dynamin GTPase | ||||
| MMT2 | 1.7 | 2.1 | −1.4 | mitochondrial iron accumulation | ||||
| MNT4 | −2.4 | alpha-1,3-mannosyltransferase | ||||||
| MTR10 | 2.1 | 1.9 | −1.4 | −1.5 | nuclear localization sequence binding | |||
| MUC1 | 2.1 | signal transducer | ||||||
| OPT1 | 2.3 | 4.2 | −1.6 | oligopeptide transporter | ||||
| ORC1 | 2.2 | −1.4 | −1.9 | ATPase | ||||
| PIK1 | 1.7 | 2.0 | −1.4 | −1.3 | 1.4 | 1-phosphatidylinositol 4-kinase | ||
| SPC105 | 1.3 | 2.0 | −1.8 | structural constituent of cytoskeleton | ||||
| SST2 | 1.8 | 2.4 | 2.6 | −1.8 | 1.2 | GTPase activator | ||
| SUL2 | 3.3 | sulfate transporter | ||||||
| SUN4 | 2.3 | 3.0 | glucosidase | |||||
| YCR024C | −2.2 | asparagine-tRNA ligase | ||||||
| YNR047W | 2.3 | −1.4 | −1.3 | protein kinase | ||||
| YOL155C | 1.3 | 2.5 | glucosidase | |||||
| YOR022C | −2.2 | phospholipase | ||||||
| YPL137C | 2.1 | 1.9 | unknown | |||||
| YBR168W | 1.9 | 2.5 | −1.3 | −1.6 | 1.4 | unknown | ||
| YLR460C | 3.3 | 2.0 | unknown | |||||
| YBL086C | 2.0 | unknown | ||||||
| YPR097W | 2.0 | −1.5 | −1.6 | 1.5 | unknown | |||
| YNL321W | 1.4 | 1.6 | 1.8 | −2.0 | −1.5 | unknown | ||
| YLR413W | 2.2 | 3.0 | −2.1 | unknown | ||||
| YCR023C | 2.7 | −1.7 | −1.6 | 1.5 | unknown | |||
| YIL169C | −2.0 | unknown | ||||||
Swi5p bound genes identifed by Lee et al. (38)
In the next experiment (Table I, Experiment 2), which measured the independent effects of choline on global gene expression, 32 genes showed a 2-fold or greater change in expression levels (Fig. 2A). Interestingly, the pattern of expression was reversed from the pattern described in the previous experiment. Whereas a majority of the genes showing differential expression in response to inositol were downregulated in its presence, a majority of those genes showing a change in expression when choline was present were upregulated. In all, 31 genes were upregulated in the presence of choline, while only a single gene was downregulated (Table IV). Functional classification of the genes whose expression is affected by choline did not reveal any discernable pattern. For example, none of the detected genes were related to choline or lipid metabolism, such as phosphatidylcholine metabolism or choline transport. Furthermore, none of the genes whose expression was affected by choline showed any overlap with the genes differentially expressed in response to inositol, suggesting that choline, by itself, has no significant effect on the differential expression of genes repressed or activated in the presence of inositol. Thus, the global transcriptional effect of choline appears to be completely distinct from the effect of inositol.
Table IV. Genes Regulated by Choline.
Description of Experiments are listed in Table I. Negative and positive values indicate downregulation and upregulation, respectively. Expression ratios in boldface indicate ≥ 2-fold difference. Overlapping expression ratios ≤ 2-fold meeting p-value cutoff of 0.025 are listed for comparison.
| Fold Difference
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Expt 1 I+ | Expt 2 C+ | Expt 3 I+C+ (i) | Expt 4 I+C+ (ii) | Expt 5 opi1Δ I+ | Expt 6 opi1Δ I− | Expt 7 control | Description |
| FDH2 | −2.0 | formate dehydrogenase | ||||||
| UBP9 | 3.3 | ubiquitin-specific protease | ||||||
| YMR111C | 3.0 | unknown | ||||||
| YDR282C | 2.9 | unknown | ||||||
| KAR3 | 2.8 | 1.9 | microtubule motor | |||||
| DMC1 | 2.6 | −1.4 | single-stranded DNA binding | |||||
| RNY1 | 2.5 | −1.8 | endoribonuclease | |||||
| YNL119W | 2.5 | −1.6 | unknown | |||||
| VPS16 | 2.5 | 2.1 | −1.8 | −2.0 | Vacuolar Protein Sorting | |||
| YSW1 | 2.3 | −2.1 | protein expressed specifically in spores | |||||
| SUL1 | 2.2 | sulfate transporter | ||||||
| MSC1 | 2.2 | Meiotic Sister-Chromatid recombination | ||||||
| YDR223W | 2.2 | −1.5 | unknown | |||||
| VMR1 | 2.2 | ATP-binding cassette (ABC) transporter | ||||||
| VPS30 | 2.2 | Vacuolar Protein Sorting | ||||||
| RAD4 | 2.2 | −1.6 | damaged DNA | |||||
| CHK1 | 2.1 | −1.5 | −1.7 | protein kinase | ||||
| UBP11 | 2.1 | −1.4 | 1.4 | ubiquitin-specific protease | ||||
| CTR86 | 2.1 | essential protein of unknown function | ||||||
| VIK1 | 2.1 | −1.5 | microtubule motor | |||||
| YIL082W-A | 2.1 | 2.4 | −2.1 | Hypothetical ORF | ||||
| NPR2 | 2.1 | −2.0 | Nitrogen Permease Regulator | |||||
| CAF120 | 2.1 | −2.2 | CCR4 Associated Factor 120 kDa | |||||
| PET127 | 2.0 | 2.9 | 2.6 | −1.6 | −1.6 | PETite colonies | ||
| YFL034W | 2.0 | 2.3 | −1.4 | −1.7 | unknown | |||
| PUT1 | 2.0 | −1.9 | proline dehydrogenase | |||||
| GPH1 | 2.0 | glycogen phosphorylase | ||||||
| YOR154W | 2.0 | −1.5 | −1.5 | unknown | ||||
| RRD2 | 2.0 | protein phosphatase type 2A regulator | ||||||
| SSA4 | 2.0 | 2.0 | −1.4 | chaperone activity | ||||
| DCS2 | 2.0 | DeCapping Scavenger | ||||||
| PIG1 | 2.0 | protein phosphatase regulator | ||||||
| PKH2 | 1.9 | −2.0 | protein kinase | |||||
| DIA1 | 1.9 | −2.3 | Digs Into Agar | |||||
However, it has been reported that addition of choline to medium containing inositol increases the extent of the repression of certain phospholipid biosynthetic genes compared to inositol alone. For example, it was originally reported that INO1 expression is repressed by an additional 2-3 fold in cells grown in inositol-containing medium when choline is present compared to cells grown in inositol-containing medium when choline is absent (16). Similar simultaneous effects of inositol and choline have also been observed for CHO1 (17,20), OP13 (21), CHO2 (21), CKI1 (52), and CPT1 (53). All of the above genes have been shown to contain an active UASINO element in their promoters, suggesting that the additive repressive effect of choline in the presence of inositol might be specific to this particular set of genes. For this reason the UASINO sequence has been referred to as the inositol/choline responsive element (ICRE) (19). One prediction from the aforementioned studies is that the combination of inositol and choline might have a general repressive effect on the entire set of genes affected by inositol alone. To address this possibility, the combined effects of inositol and choline on genome wide expression were determined by comparing the relative mRNA transcript levels from cells grown in the presence of inositol and choline to transcript levels from cells grown in the absence of inositol and choline (Table I, Experiment 3). These are conditions that have typically been utilized to measure the effect of phospholipid precursors on gene expression (16,19,37,52). The genes identified in Experiment 3 were subsequently compared to the set of genes regulated by inositol alone (Table I, Experiment 1). Interestingly, the number of genes showing a statistically significant change in expression level by 2-fold or greater increased from 32 when only inositol was present to 93 when choline was present in addition to inositol. The transcript levels of 68 genes were downregulated, while 25 genes were upregulated in the presence of both inositol and choline compared to the absence of inositol and choline (Fig. 2A).
Comparison of the set of genes regulated by the combined effects of inositol and choline with the set of 29 genes downregulated by inositol alone revealed an overlap of 21 genes that showed at least a 2-fold or greater change in expression levels (Fig. 2B). Notably, the downregulated level of a subset of these overlapping genes was greater when cells were grown in the presence of inositol and choline, including INO1 and OPI3 (Table II), in agreement with previous reports (16,21). The presence of choline also had an additional repressive effect on other inositol-regulated genes including ITR1, YEL073C, YIP3, PSD1, and SAH1, which is reported here for the first time. However, other inositol-regulated genes did not show a significant change from their expression level when choline was present (Table II). For example, the relative change in expression of KAR2 and ERO1, genes whose expression is regulated by the UPR pathway (34,51), showed similar levels of repression in response to inositol, regardless of the presence or absence of choline in the growth media. KAR2 and ERO1 were repressed by 2.4-fold and 2.6-fold, respectively, in cells grown in the presence of inositol alone, and both were repressed by 2.3-fold when grown in the presence of inositol and choline. These results suggest that choline has an additional repressive effect on expression levels when inositol is present in the growth medium, but only on a subset of overlapping genes.
Surprisingly, only two genes—PET127 and YLR460C—which were upregulated in the presence of both inositol and choline, overlapped with the set of genes that were upregulated by choline alone (Fig. 2C), suggesting that inositol in combination with choline overwhelms the activating effect of choline alone on gene expression. To further examine the combined effects of inositol and choline on genome wide expression, the transcriptional response of cells grown in medium containing both inositol and choline was compared to cells grown in medium lacking inositol but containing choline (Table I, Experiment 4). Since choline is present in both growth conditions, this experiment measures the effect of choline on inositol-dependent gene expression, but should not include any inositol-independent effects of choline. As expected, a majority of downregulated genes identified in Experiment 1 and Experiment 3 overlapped with the set of downregulated genes identified in Experiment 4 (Table II). Furthermore, approximately one-half of the upregulated genes identified in Experiment 4 overlapped with the set of genes upregulated in Experiment 3 (Table III), suggesting that these genes are affected by inositol but not choline. Many of the upregulated genes identified in Experiments 3 and 4 also overlapped with genes upregulated in Experiment 1, but the level of differential expression in Experiment 1 was less than 2-fold in most cases (Table III). However, there was minimal overlap in Experiments 3 and 4 with genes upregulated by choline alone (Table IV). Taken together, these results suggest that inositol, but not choline, is the major effector of target gene expression.
To confirm the results from the cDNA microarray experiments for a subset of genes identified in Experiments 1, 3, and 4, INO1, VTC3, KAR2, PDI1, ACT1, HO, Ty1, and Ty2 were chosen for further characterization by quantitative Northern slot blot analysis. ACT1, which is not regulated by inositol and choline, was chosen as a control. The relative levels of mRNA from these genes was measured from wild type cells grown in the absence of inositol and choline was compared to transcript levels for cells grown in the absence of inositol and presence of choline, presence of inositol and absence of choline, and presence of inositol and choline. In agreement with the data obtained from the microarray experiments, INO1, VTC3, KAR2, PDI1, and Ty2 were downregulated in the presence of inositol. The relative levels were consistent with the microarray data (Table II, Fig. 3), and the repression levels for INO1 were also comparable to levels previously observed by slot blot analysis (16). In addition, HO was upregulated in response to inositol, in agreement with the microarray results (Fig. 3). Moreover, unlike Ty2 transcript levels, Ty1 transcript levels were not significantly affected by inositol and choline (Fig. 3, see below), which corresponds with the microarray analysis.
Figure 3. Quantitative slot blot analysis of a subset of genes regulated by inositol and choline.
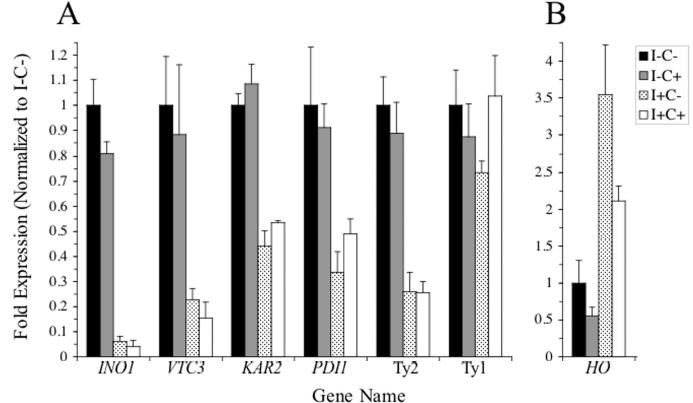
mRNA from wild type cells grown in the absence of inositol and choline (I−C−), absence of inositol and presence of choline (I−C+), presence of inositol and absence of choline (I+C−), and presence of inositol and choline (I+C+) were analyzed in triplicate by slot blot hybridization as described in the “Experimental Procedures” using the indicated probes. Fold expression for each transcript are normalized to the total counts per minute for ACT1 and expressed as the fraction of the amount of mRNA from cells grown in the absence of inositol and choline. The standard deviation for individual transcripts at each growth condition is indicated. Fold expression of each transcript is plotted in panels A and B using different scales.
The above experiments indicate that the predominant effect on genome wide expression of inositol in the growth media is repressive, and that this effect is augmented by the addition of choline. Integration of the results obtained in Experiments 1, 3, and 4 revealed that the expression levels of 110 genes were repressed by at least 2-fold by the presence of inositol. The remainder of the report will focus on the analysis of this set of inositol-repressed genes, which are shown in Figure 4. Functional classification revealed 14 genes involved in lipid biosynthesis, including INO1, OPI3, ITR1, PSD1, CHO1, CKI1, CDS1, and INO2, which have previously been shown to contain an active UASINO element and to be regulated by inositol and choline (1) (Fig. 5). Genes involved in protein folding, including KAR2, SCJ1, ERO1, FPR2, MPD1, and PDI1, which have been shown to be regulated by the UPR pathway (34), were also detected (Fig. 5). Additional functional categories were also identified (Fig. 5) and will be discussed below.
Figure 4. List of genes repressed by inositol.
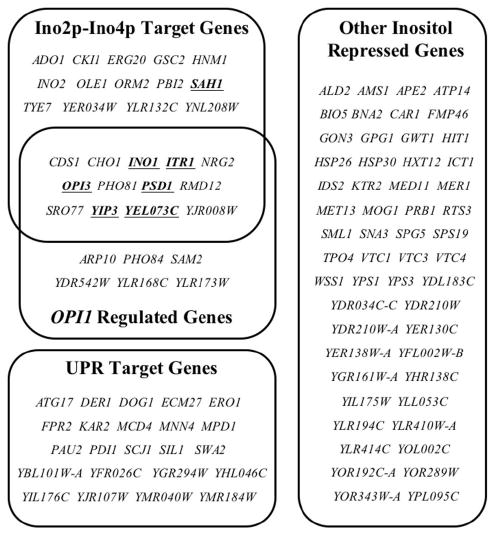
Listed are genes that showed a 2-fold or greater level of repression in cells grown in the presence of inositol identified in Experiments 1, 3, and 4. Genes that overlapped with the set of Ino2p-Ino4p target genes (38), OPI1-regulated genes (Experiment 5), and UPR target genes (34) are categorized. Criteria for classifying Ino2p-Ino4p and UPR target genes are described in the text of the Results. Genes whose repression levels were enhanced in the presence of choline (Table II) are underlined and in boldface. Genes that did not fit into these categories are classified as “Other”.
Figure 5. Functional categories of inositol-repressed genes.

Inositol-dependent genes shown in Fig. 4 that were functionally annotated in the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org) were categorized based upon their gene ontology annotation. Each section of the pie chart represents a functional category. The numerals refer to the number of members of each particular category. Bold headings indicate functional categories. Genes were assigned to subcategories according to summaries of published data listed in the SGD. (*Ino2p-Ino4p target genes identified by Lee et al. (38); +UPR target genes identified by Travers et al. (34)). Genes falling into categories with 2 or fewer members were grouped into the category “Other”. Genes with no functional annotation were grouped into the category “Unknown”.
A major subset of inositol-regulated genes interact directly with Ino2p and Ino4p
Previous studies have shown that Ino2p and Ino4p act as positive transcription factors by binding to the UASINO element present in the promoters of a number of phospholipid biosynthetic genes (1). As a rationale for clustering of genes affected by the presence of inositol and choline, we reasoned that Ino2p and Ino4p should be bound to a subset of the genes that are repressed by growth in inositol containing medium. Recently Young and colleagues located the genomic binding sites of Ino2p and Ino4p (38). Throughout the remainder of this report, the set of genes reported by Lee et al. (38) that are bound by both Ino2p and Ino4p will be referred to as Ino2p-Ino4p target genes. We made use of this comprehensive data set by comparing the Ino2p-Ino4p target genes with the set of genes identified in our analysis that are regulated by inositol and choline. A total of 27 genes, which were downregulated by inositol alone or in combination with choline, overlapped with the set of Ino2p-Ino4p target genes (Fig. 4). As expected, a number of genes identified in the current study that were previously shown to be regulated by inositol, including INO1 (16), OPI3 (21), ITR1 (23), PSD1 (25), CHO1 (17,20), CDS1 (24), CK1I (52), HNM1 (54), and INO2 (55), were also reported to be bound by Ino2p and Ino4p in the study by Lee et al. (38). Interestingly, almost two-thirds of the remaining inositol-repressed genes identified here as overlapping with the set of Ino2p-Ino4p target genes (38) had not been previously shown to be regulated by inositol and choline (Fig. 4). These newly identified inositol repressed genes include those involved in membrane trafficking (YIP3 and SRO77), methionine metabolism (SAH1), and lipid metabolism (OLE1 and ERG20). In addition to its function in lipid metabolism, ERG20, which encodes farnesyl-pyrophosphate synthetase, plays a role in protein prenylation and dolichol synthesis (56). These additional genes represent a novel set that are likely to be co-regulated with phospholipid biosynthetic genes.
Importantly, none of the genes that were upregulated in cells grown in the presence of inositol or choline (Tables III and IV) were identified by Lee et al. (38) as Ino2p-Ino4p target genes.
Opi1p negatively regulates Ino2p-Ino4p bound genes, but not the unfolded protein response
Opi1p negatively regulates the transcription of INO1 and a number of phospholipid biosynthetic genes (7) by functioning through the UASINO element present in target genes (13). To ascertain the role of Opi1p on the genome-wide regulation in response to phospholipid precursors, the transcriptional response of an opi1Δ strain was compared with its isogenic wild type in the presence of inositol (Table I, Experiment 5). Of the 24 genes upregulated by 2 fold or greater in the opi1Δ strain, 19 overlapped with the set of genes negatively regulated by inositol; a set which includes genes that are further repressed by the presence of choline in addition to inositol (Table II). Furthermore, more than two-thirds of these coregulated genes were reported by Lee et al. to contain a bound Ino2p and Ino4p (38) (Fig. 4), indicating that Opi1p primarily functions through genes regulated by Ino2p and Ino4p. Taken together, these findings corroborate the role of Opi1p as a negative transcriptional regulator of genes repressed by phospholipid precursors (7,26).
Walter and colleagues first reported that the UPR pathway is activated when cells are grown in the absence of inositol (30). When the UPR is triggered, the transcription of UPR target genes is induced to protect cells from secretory stress. Inositol starvation has also been shown to induce the expression of two UPR target genes—KAR2 (30) and PDI1 (57)—as well as a UPRE-lacZ reporter construct (35,36). A genome wide study of UPR target genes (34) identified genes including those required for folding and transport of proteins through every step of the secretory pathway. Comparison of the UPR target genes identified by Travers et al. (34) with the set of genes negatively regulated by inositol revealed 23 UPR target genes that were regulated in response to the presence of inositol (Fig. 4). These included many of the so-called canonical UPR genes involved in protein folding, including KAR2, EUG1, ERO1, PDI1, SCJ1, MPD1, and FPR2 (34), as well as numerous genes of unknown function. Since the expression levels of these genes were not influenced by the presence of choline in the growth media (Table II), these data show that the UPR pathway is activated solely as a consequence of the lack of inositol. Furthermore, the set of UPR target genes induced by inositol starvation did not overlap with the set of inositol-repressed Ino2p-Ino4p target genes (Fig. 4). Two Ino2p-Ino4p target genes (38), INO1 and OPI3, which are repressed in the presence of inositol (Fig. 4, Table II), have also been classified as UPR target genes (30,34). However, since both genes have been previously demonstrated to contain an active UASINO element (8,21), we chose to classify these genes in the set of Ino2p-Ino4p target genes and omit them from the UPR target gene list for the purpose of our analysis.
Consistent with the previous report by Travers et al. (34), none of the inositol-regulated UPR target genes overlap with the set of OPI1-regulated genes (Fig. 4), suggesting that activation of the UPR pathway represents a separate response to inositol starvation from the metabolic response that coordinates the regulation of the set of Ino2p-Ino4p target genes. As a further test of this hypothesis, the transcriptional profile of opi1Δ and wild type strains grown in the absence of inositol were compared and analyzed (Table I, Experiment 6). Under these growth conditions Ino2p-Ino4p target genes will be expressed in both strains; in opi1Δ cells since the Opi1p negative regulator is missing, and in wild type cells since inositol is absent (2,7). As expected, none of the Ino2p-Ino4p target genes were differentially expressed to a significant degree in the opi1Δ strain as compared to the wild type strain grown in the absence of inositol. However, opi1Δ cells also overproduce and excrete inositol into the growth medium (7,49,58). For this reason any inositol-dependent, but non-Ino2p-Ino4p target genes are expected to be repressed in opi1Δ cells regardless of inositol supplementation when compared to wild type cells grown in the absence of inositol. Indeed, several UPR genes—ERO1, MCD4, SIL1, ATG17, DOG1, and MNN4 (Table II)—were downregulated in the opi1Δ strain compared to the wild type strain grown in the absence of inositol. Thus, in an opi1Δ strain, Ino2p-Ino4p target genes are constitutively expressed, but the UPR is not constitutively activated.
In a final control experiment, the gene expression in the opi1Δ strain grown in the absence of inositol was compared to expression in the same strain grown in the presence of inositol (Table I, Experiment 7). As discussed above, it is expected that the addition of inositol to the medium should make little difference to genome wide expression in an opi1Δ strain since these cells excrete inositol into the growth medium. Indeed, no significant changes in gene expression were detected when opi1Δ cells were grown in the absence compared to the presence of inositol. Taken together the results of experiments 5, 6, and 7 suggest that Opi1p negatively regulates Ino2p-Ino4p target genes, but not UPR target genes.
Other categories of genes regulated by inositol
Functional classification of genes repressed in the presence of inositol included a subset of genes that are coregulated by the PHO pathway, including PHO81, PHO84, VTC1, VTC3, and VTC4 (59) (Fig. 5). The PHO pathway, which consists of at least 22 genes (59), is induced under low phosphate conditions to allow the cell to survive when phosphate is limiting. Since the cells were grown under high phosphate conditions in the experiments reported here, these results show that a subset of PHO pathway genes are induced when inositol is limiting. PHO81 encodes a cyclin dependent kinase, which initiates the PHO pathway (60), and PHO84 encodes a high affinity inorganic phosphate transporter (61). VTC1, VTC3, and VTC4 gene products are involved in polyphosphate accumulation by the vacuole (59), but also function together in a complex during late stages of vacuole fusion (62-64). Interestingly, two additional genes—PBI and YHR138C—which function in early stages of vacuole fusion (65) but not the PHO pathway, were also identified in the present study. Taken together, these results suggest a combined function for these genes under conditions of nutrient-limited growth. Given their overlapping roles in storage of polyphosphate and maintenance of vacuole morphology, VTC1, VTC3, and VTC4 genes may also play a role in inositol storage in the vacuole. If the vacuole is an important inositol storage compartment, it is possible that disruption of VTC1, VTC3, or VTC4 might lead to growth defects in the absence of inositol. However, neither vtc1Δ, vtc3Δ, nor vtc4Δ mutants showed diminished growth in the absence of inositol (Ino− phenotype) or an overproduction of inositol (Opi− phenotype) (data not shown).
Functional classification also revealed eight Ty2 element TYA GAG ORFs to be regulated by inositol and choline (Fig. 5). Ty elements are members of a family of eukaryotic elements called retrotransposons that resemble animal retroviral genomic RNA in structure (66). S. cerevisiae has five distinct retrotransposon families, designated Ty1-Ty5 (67). Previously, it was reported that the expression of Ty1-H3, a Ty1 element retrotransposon closely related to Ty2 elements (67), is regulated by inositol and choline (68) and contains a UASINO sequence in its promoter (1). However, while Northern slot blot analysis confirmed that Ty2 element transcript levels are approximately 4-fold lower in cells grown in the presence of inositol and choline compared to strains grown in the absence of inositol and choline (Fig. 3), Ty1 element transcript levels were not significantly altered in cells grown in the absence or presence of inositol and choline (Fig. 3). Retrotransposons can act as mutagens that may allow cells to adapt to stressful environments (69). Given that the UPR, a stress response pathway, is activated when cells grown in the absence of inositol, it is tempting to speculate that mobilization of Ty elements might also be related to stress produced by growing cells in media lacking inositol. However, since only a single Ty2 GAG ORF was detected as a UPR target (34), activation of the remaining seven elements might indicate a stress response in addition to the UPR pathway in cells grown in the absence of inositol.
DISCUSSION
We have presented a comprehensive genome wide expression analysis designed to identify the sets of genes whose transcript levels in dividing cells are influenced by the presence of the phospholipid precursors, inositol and choline. The independent effects of inositol and choline on genome wide expression have not been examined previously and are reported here for the first time. Our findings indicate that inositol and choline affect the expression of completely distinct sets of genes. Moreover, while the effect of inositol on overall gene expression was primarily repressing (Fig. 2A), the effect of choline on target gene expression was activating (Fig. 2A). However, when choline was present in medium also containing inositol, many of the genes affected by inositol alone exhibited intensified repression (Table II). Not only was the degree of repression of certain genes enhanced, but also a greater number of genes were found to be differentially expressed in the presence of both precursors. This is in agreement with previous reports measuring expression of individual genes including INO1 (16), CHO1 (17,20), OP13 (21), CKI1 (52). We also show for the first time that ITR1, YEL073C, YIP3, PSD1, and SAH1 match this pattern of regulation. Conversely, only a few genes that were upregulated in the presence of choline alone were also upregulated when both inositol and choline were present (Table IV). Overall our results indicate that inositol plays the primary role in repressing the expression of genes sensitive to the presence of exogenous phospholipid precursors, whereas choline plays a minor role. Choline primarily enhances the repressive effect of inositol on a subset of inositol-responsive genes. A possible metabolic mechanism for these observations will be discussed subsequently.
A distinct set of inositol-repressed genes regulated by the transcription factors Ino2p and Ino4p
Several distinct classes of genes were repressed in response to the presence of inositol or inositol plus choline in the growth media. One set included genes, previously identified by Lee et al. (38), that contain a bound Ino2p and Ino4p. Ino2p and Ino4p are members of the family of basic helix-loop-helix transcription factors, both of which are required for depression of INO1 and other coregulated genes encoding enzymes of phospholipid biosynthesis (9,11,12). Together these transcription factors form a heterodimer and bind to the repeated element UASINO found in the promoters of target genes (1,12). Using genome-wide location analysis (70), a method to identify genomic protein-DNA interaction sites using chromatin immunoprecipitation followed by DNA microarray analysis, Lee et al. (38) identified the genomic binding sites of 106 yeast transcriptional regulators, including Ino2p and Ino4p. The number of promoter regions containing both Ino2p and Ino4p using a p-value threshold of 0.01 for both factors was 189 (38). As expected, virtually all of the genes involved in lipid metabolism that were downregulated in the presence of inositol were also detected by Lee et al. (38) as Ino2p-Ino4p target genes (Fig. 5). Many of these genes had previously been shown to contain an active UASINO element in their promoters (1), confirming that both Ino2p and Ino4p play a pivotal role in the regulated expression of these genes.
A recent and more thorough genome-wide location analysis study conducted by Young and colleagues (71), which identified the genomic occupancy of all 203 known yeast transcription factors and the recognition motifs for 116 of these regulators, confirmed that Ino2p and Ino4p are bound to the UASINO sequence in the promoters of target genes throughout the genome. In addition, the more recent study by Harbison et al. (71), which refined the statistical analysis of bound genes identified by Lee et al. (38), confirmed that Ino2p and Ino4p bind to target genes as regulator pairs (12). Using a more stringent significance threshold for genes bound by Ino2p and Ino4p from the study by Harbison et al. (71), a portion of the Ino2p-Ino4p target genes reported by Lee et al. (38) that are repressed by inositol shown in Fig. 3 did not show significant binding to Ino2p. These genes include CKI1, ERG20, HNM1, PBI2, TYE7, CHO1, NRG2, PHO81, and RMD12. One explanation for the failure to detect Ino2p binding to these genes is that Ino2p levels might be reduced in cells grown in YPD medium used by Harbison et al. (71). The current study (Table II) and previous studies (55,72) showed that INO2 expression is downregulated in the presence of inositol, which is present in YPD media. Moreover, CKI1, HMN1, and CHO1 each contain a UASINO element in their promoters (1) and match the pattern of regulation of other UASINO-containing genes (17,20,52,54). Thus, it is likely that both Ino2p and Ino4p are required for their regulated expression.
In addition, our results show that Ino2p-Ino4p target genes significantly overlap with the set of OPI1-regulated genes. In the first indication that Opi1p physically interacts with any gene, a subset of the genes we identified as OPI1-regulated were shown by Harbison et al. (71) to be bound by Opi1p including, INO1, CDS1, and ITR1. Opi1p is localized at steady state to the ER in an inactive form as part of a lipid-sensing complex (27). Addition of inositol to the media of growing cells results in a rapid change in lipid metabolism, causing Opi1p to translocate to the nucleus (26). Although Opi1p does not appear to directly interact with the UASINO element (28,29), Opi1p is potentially targeted to UASINO-containing genes by a direct interaction with Ino2p, where it represses the transcription of genes by recruiting the histone deacetylase Sin3p (15). These observations suggest that Opi1p plays a central role in sensing metabolism and modulating the regulation of Ino2p-Ino4p target gene expression.
Additional potential regulatory mechanisms that mediate the differential expression of genes regulated by inositol
Aside from their role in regulating the transcription of genes that are repressed in the presence of inositol, Ino2p and Ino4p may also indirectly affect the transcription of genes that are activated in the presence of inositol by functioning through a transcriptional regulatory network. For example, both Ino2p and Ino4p bind to the SWI5 gene in YPD media containing inositol (38). SWI5 encodes a transcription factor that regulates the expression of genes expressed during the G1 phase and the M/G1 boundary of the cell cycle (73). While the differential expression of SWI5 was not detected in our study, five genes that were activated in the presence of inositol—HO, PRY3, SCW11, WSC4, and YML131W (Table III)—have been shown by Lee et al. (38) and another study (73) to be bound by Swi5p. The HO endonuclease gene, which showed the greatest differential expression in the presence of inositol, is regulated by Swi5p (73), and HO, SCW11, and PRY3 are regulated in a cell cycle dependent manner (73,74). Thus, Ino2p and Ino4p may contribute to the regulation of gene expression by controlling the expression of transcription factors that, in turn, regulate cell cycle specific genes.
The expression of other genes may be regulated in response to the enzymatic activity of genes directly regulated by Ino2p and Ino4p. For example, the product of the INO1 gene, inositol-3-phosphate synthase, catalyzes the formation of inositol-3-phosphate from glucose-6-phosphate in a reaction that is absolutely dependent upon nicotine adenine dinucleotide (NAD) (75). Interestingly, BNA2, which is required for the de novo synthesis of NAD, is highly expressed when cellular NAD levels are depleted (76) and in cells grown in the absence of inositol (Table II). Because INO1 levels are dramatically elevated in cells grown in the absence of inositol (16) (Table II), overall cellular levels of NAD may drop due to consumption by increased activity of inositol-3-phosphate synthase, activating a previously undetected feedback regulatory mechanism.
In contrast to the results presented here, a recent study by Santiago and Mamoun (37), which investigated the combined effects of inositol and choline on genome-wide expression in yeast, concluded that Ino2p and Ino4p regulate only a minor subset of genes affected by these phospholipid precursors. There are several differences between our study and that of Santiago and Mamoun in the both experimental design and findings. First, Santiago and Mamoun detected none of the UPR target genes and identified only three genes involved in lipid biosynthesis as responding to the combined effects of inositol and choline. Second, they concluded that only two genes, OPI3 and YDL241W, require Ino2p and Ino4p for inositol and choline dependent repression. However, neither INO1 nor any other gene besides OPI3, previously demonstrated to contain an active UASINO element (1), was identified in their study as being Ino2p and Ino4p dependent. We believe that their failure to identify genes such as INO1, which is clearly Ino2p- and Ino4p-dependent, is most likely due to the design of their experiment. Santiago and Mamoun compared gene expression in ino2Δ and ino4Δ cells to wild type cells, all of which were grown in the presence of both inositol and choline. In the present study, we have shown that Ino2p-Ino4p target genes are fully repressed in wild type cells grown under these conditions (Table II). Since the ino2Δ and ino4Δ strains each lack an activator required for the derepression of target genes, these mutant strains are expected to express only repressed levels of the affected genes, regardless of the growth condition. Santiago and Mamoun obtained the expected, but uninformative, result since the expression levels of Ino2p-Ino4p dependent genes in ino2Δ and ino4Δ cells did not show significant differences compared to wild type cells. Finally, these authors identified numerous genes that were not detected in our study, including genes involved in biotin synthesis and nutrient transport. For example, GIT1, which encodes a plasma membrane glycerophosphoinositol permease, was detected as a inositol and choline responsive gene in their study. However, GIT1 is known to be expressed only when both inositol and phosphate are limiting (77,78). In fact, GIT1 is fully repressed regardless of the presence or absence of inositol in high phosphate media (77,78) such as that based on standard yeast nitrogen base formula media, comparable to the media that we utilized in the present study. In addition, many other genes identified by Santiago and Mamoun overlap with genes shown by Wodicka et al. (79) to be upregulated in minimal media. Thus, essential nutrients, including vitamins and phosphate, may have been limiting or absent in the growth media utilized in the study by Santiago and Mamoun.
A distinct regulatory network for regulating UPR target genes
We have shown that a subset of genes reported by Travers et al. (34) to be regulated by the UPR pathway are also activated in the absence of inositol. The UPR is a stress response pathway that is activated under conditions in which insufficient protein folding capacity leads to an accumulation of unfolded proteins in the ER lumen, a condition referred to as ER stress (80). In yeast, UPR activation induces the transcription of target genes that increase the capacity for protein folding and secretion as well as genes for degradation of unfolded polypeptides (34). In all, 381 UPR target genes were identified in the genome wide analysis by Travers et al. (34) of cells treated with tunicamycin and dithiothreitol (DTT), conditions that are known to induce the UPR pathway. In the current study, we found 23 UPR target genes in the set of genes repressed in the presence of inositol. While we detected many canonical UPR target genes, we did not detect other targets of the UPR pathway including genes involved in ER-associated degradation (ERAD) or membrane trafficking pathways. Possibly, the UPR response in cells grown in the absence of inositol is less robust than when it is induced by tunicamycin or DTT.
Importantly, the set of UPR genes that are induced in cells grown in the absence of inositol do not overlap with the set of genes shown to contain a bound Ino2p-Ino4p by Lee et al. (38) (Fig. 4). Furthermore, the expression of the UPR target genes identified here was not affected further by the presence of choline in the growth media. These results strongly suggest that induction of UPR target genes under inositol limiting conditions is through an entirely different mechanism than genes regulated by Ino2p and Ino4p. Clearly the relationship between UPR activation with inositol starvation and Ino2p-Ino4p target gene expression is complex and worthy of continued analysis.
A mechanism for regulating the expression of inositol-responsive genes by a metabolic signal
Previous studies have suggested that the expression of INO1 and other UASINO-containing genes are regulated by a common metabolic signal that is produced by the participation of inositol and choline in lipid metabolism (4), even though these precursors enter the pathways for phospholipid biosynthesis by distinctly different routes (Fig. 1). Inositol reacts directly with CDP-diacylglycerol (CDP-DAG) to form phosphatidylinositol (PI), while choline is phosphorylated and converted to CDP-choline before it reacts with diacylglycerol (DAG) to form phosphatidylcholine (PC) via the Kennedy (CDP-choline) pathway. PC is also synthesized via the three-step phosphatidylethanolamine (PE) methylation pathway (Fig. 1). PE is derived from phosphatidylserine (PS), which is produced in a reaction that competes directly for CDP-DAG with PI biosynthesis (81). Since DAG and CDP-DAG intermediates are derived from PA (Fig. 1), PA is the common lipid precursor for the production of PI and both routes of PC biosynthesis. Moreover, PA is sensitive to the presence of inositol and choline since it is consumed in the production of inositol- and choline-containing phospholipids (4,26,81).
Several lines of evidence suggest that PA levels provide the relevant signal for the repression of Ino2p-Ino4p target genes. First, the signal controlling INO1 expression, and presumably other UASINO-containing genes, is sensitive to ongoing metabolism in all three of the interconnected pathways leading to PI and PC synthesis described above (Fig. 1). Moreover, input from these pathways appears to be additive. As shown in the current study and previous investigations of individual Ino2p-Ino4p target genes, the presence of inositol alone is enough to trigger repression (Table II). However, many Ino2p-Ino4p target genes including INO1, CHO1, OP13, CKI1, ITR1, YEL073C, YIP3, PSD1, and SAH1 (16,17,20,21,52) (Table II) show enhanced repression when choline is present with inositol, supporting the hypothesis that these two precursors affect a common signal in an additive fashion. Since PA is the common precursor at the branch point for the synthesis of PI from CDP-DAG and PC from DAG (Fig. 1), it is a logical candidate for the relevant signaling molecule.
Second, mutations causing complete or partial blocks at any step in the de novo synthesis of PC from PA through the PE methylation pathway (Fig. 1) lead to the derepression of INO1 and other UASINO-containing genes even in the presence of inositol (4). In these mutants, derepression of INO1 occurs even when inositol is present, and they have an Opi− phenotype when inositol is absent. Since each block in the pathway leads to the buildup of precursors upstream of the specific metabolic lesion, including PA, derepression of Ino2p-Ino4p target genes in these mutant strains is correlated to a rise in PA (4). Moreover, supplementation with choline restores PC biosynthesis, eliminates the Opi− phenotype, and restores INO1 regulation in response to inositol in cho2 and opi3 mutants (4). Since choline enters via the Kennedy pathway and uses DAG as a precursor (Fig. 1), these results indicate that ongoing metabolism in the two routes to PC biosynthesis have additive effects on INO1 expression and that they influence the same signal as inositol. The PA signal hypothesis gained strong support from the finding that sec14ts mutants that are also blocked in the Kennedy (CDP-choline) pathway overexpress INO1 even in the presence of inositol (82). This was shown to be due to increased PC turnover by a phospholipase D (Pld1p)-dependent mechanism (83), which leads directly to increased PA levels (Fig. 1).
Finally, a recent collaborative study involving our laboratory has revealed the molecular mechanism whereby PA levels regulate the expression of INO1 (26). Opi1p, which is a member of an ER-localized lipid sensing complex (27), was shown to interact directly with PA (26), suggesting that Opi1p serves as the relevant PA sensor in the ER. The introduction of inositol was shown to result in a rapid rise in PI biosynthesis and a fall in PA levels. Concurrent with the drop in PA levels, Opi1p rapidly translocated from the ER to the nucleus, which coincided with INO1 repression (26). The results presented in the current study predict that this mechanism can be extended to all Ino2p-Ino4p bound genes that are coregulated by Opi1p.
A remaining unanswered question is why choline alone has little or no effect on expression of Ino2p-Ino4p target genes in wild type cells in the absence of inositol. One testable prediction is that the effect of choline on PA levels is less than that of inositol, whereas the combination of inositol and choline produces an enhanced effect on PA levels. Moreover, the relative effects of these two precursors on PA levels should be reversed in mutants such as cho2 and opi3 with respect to their effects on both gene expression and PA levels. We are currently testing these models.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM-19629 to SAH and NSF grant DMS 02-04252 to MTW. We are indebted to Drs. David Garfinkel and Mark Longtine for the kind gifts of plasmids. We wish to thank all members of the Henry laboratory for stimulating discussions during the course of this work.
Footnotes
The abbreviations used are: PA, phosphatidic acid; ER, endoplasmic reticulum; UPR, unfolded protein response; ORF, open reading frame; NAD, nicotine adenine dinucleotide; DTT, dithiothreitol; CDP-DAG, CDP-diacylglycerol; PI, phosphatidylinositol; DAG, diacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine
References
- 1.Greenberg ML, Lopes JM. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carman GM, Henry SA. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 3.Rawson RB. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 4.Henry SA, Patton-Vogt JL. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 5.Carman GM, Kersting MC. Biochem Cell Biol. 2004;82:62–70. doi: 10.1139/o03-064. [DOI] [PubMed] [Google Scholar]
- 6.Hoshizaki DK, Hill JE, Henry SA. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- 7.White MJ, Hirsch JP, Henry SA. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- 8.Lopes JM, Henry SA. Nucleic Acids Res. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikoloff DM, McGraw P, Henry SA. Nucleic Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosaka K, Nikawa J, Kodaki T, Yamashita S. J Biochem (Tokyo) 1994;115:131–136. doi: 10.1093/oxfordjournals.jbchem.a124287. [DOI] [PubMed] [Google Scholar]
- 11.Nikoloff DM, Henry SA. J Biol Chem. 1994;269:7402–7411. [PubMed] [Google Scholar]
- 12.Ambroziak J, Henry SA. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 13.Bachhawat N, Ouyang Q, Henry SA. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 14.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. Mol Microbiol. 2001;41:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch JP, Henry SA. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailis AM, Poole MA, Carman GM, Henry SA. Mol Cell Biol. 1987;7:167–176. doi: 10.1128/mcb.7.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodaki T, Hosaka K, Nikawa J, Yamashita S. J Biochem (Tokyo) 1991;109:276–287. [PubMed] [Google Scholar]
- 19.Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E. Embo J. 1992;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailis AM, Lopes JM, Kohlwein SD, Henry SA. Nucleic Acids Res. 1992;20:1411–1418. doi: 10.1093/nar/20.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaynor PM, Gill T, Toutenhoofd S, Summers EF, McGraw P, Homann MJ, Henry SA, Carman GM. Biochim Biophys Acta. 1991;1090:326–332. doi: 10.1016/0167-4781(91)90197-t. [DOI] [PubMed] [Google Scholar]
- 22.Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 23.Lai K, McGraw P. J Biol Chem. 1994;269:2245–2251. [PubMed] [Google Scholar]
- 24.Shen H, Dowhan W. J Biol Chem. 1997;272:11215–11220. doi: 10.1074/jbc.272.17.11215. [DOI] [PubMed] [Google Scholar]
- 25.Griac P. J Bacteriol. 1997;179:5843–5848. doi: 10.1128/jb.179.18.5843-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 27.Loewen CJ, Roy A, Levine TP. Embo J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graves JA, Henry SA. Genetics. 2000;154:1485–1495. doi: 10.1093/genetics/154.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner C, Blank M, Strohmann B, Schuller HJ. Yeast. 1999;15:843–854. doi: 10.1002/(SICI)1097-0061(199907)15:10A<843::AID-YEA424>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 30.Cox JS, Chapman RE, Walter P. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox JS, Shamu CE, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 32.Mori K, Ma W, Gething MJ, Sambrook J. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 33.Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. Embo J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 35.Chang HJ, Jones EW, Henry SA. Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang HJ, Jesch SA, Gaspar ML, Henry SA. Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santiago TC, Mamoun CB. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- 38.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 39.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Dowd SR, Bier ME, Patton-Vogt JL. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- 41.Ausubel, F. M. (2001) in Current protocols in molecular biology, J. Wiley, New York
- 42.Kohrer K, Domdey H. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Hessner MJ, Wu Y, Pati N, Ghosh S. Bioinformatics. 2003;19:1341–1347. doi: 10.1093/bioinformatics/btg154. [DOI] [PubMed] [Google Scholar]
- 44.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudoit S, Yang YH, Callow MJ, Speed TP. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- 46.Storey JD. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2002;64:479–498. [Google Scholar]
- 47.Storey JD, Tibshirani R. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Greenberg ML, Reiner B, Henry SA. Genetics. 1982;100:19–33. doi: 10.1093/genetics/100.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swede MJ, Hudak KA, Lopes JM, Henry SA. Methods Enzymol. 1992;209:21–34. doi: 10.1016/0076-6879(92)09005-n. [DOI] [PubMed] [Google Scholar]
- 51.Chapman R, Sidrauski C, Walter P. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- 52.Hosaka K, Murakami T, Kodaki T, Nikawa J, Yamashita S. J Bacteriol. 1990;172:2005–2012. doi: 10.1128/jb.172.4.2005-2012.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMaster CR, Bell RM. J Biol Chem. 1994;269:14776–14783. [PubMed] [Google Scholar]
- 54.Nikawa J, Hosaka K, Tsukagoshi Y, Yamashita S. J Biol Chem. 1990;265:15996–16003. [PubMed] [Google Scholar]
- 55.Ashburner BP, Lopes JM. Mol Cell Biol. 1995;15:1709–1715. doi: 10.1128/mcb.15.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song L, Poulter CD. Proc Natl Acad Sci U S A. 1994;91:3044–3048. doi: 10.1073/pnas.91.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. Proc Natl Acad Sci U S A. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenberg ML, Goldwasser P, Henry SA. Mol Gen Genet. 1982;186:157–163. doi: 10.1007/BF00331845. [DOI] [PubMed] [Google Scholar]
- 59.Ogawa N, DeRisi J, Brown PO. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider KR, Smith RL, O’Shea EK. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 61.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller O, Neumann H, Bayer MJ, Mayer A. J Cell Sci. 2003;116:1107–1115. doi: 10.1242/jcs.00328. [DOI] [PubMed] [Google Scholar]
- 63.Muller O, Bayer MJ, Peters C, Andersen JS, Mann M, Mayer A. Embo J. 2002;21:259–269. doi: 10.1093/emboj/21.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bayer MJ, Reese C, Buhler S, Peters C, Mayer A. J Cell Biol. 2003;162:211–222. doi: 10.1083/jcb.200212004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Z, Sato K, Wickner W. Cell. 1998;93:1125–1134. doi: 10.1016/s0092-8674(00)81457-9. [DOI] [PubMed] [Google Scholar]
- 66.Boeke, J. D., and Sandmeyer, S. B. (1991) in The molecular and cellular biology of the yeast Saccharomyces (Broach, J. R., Jones, E. W., and Pringle, J. R., eds) Vol. 1, pp. 193–261, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 67.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 68.Lamping E, Luckl J, Paltauf F, Henry SA, Kohlwein SD. Genetics. 1994;137:55–65. doi: 10.1093/genetics/137.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curcio MJ, Garfinkel DJ. Trends Genet. 1999;15:43–45. doi: 10.1016/s0168-9525(98)01643-6. [DOI] [PubMed] [Google Scholar]
- 70.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 71.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwank S, Hoffmann B, Sch-uller HJ. Curr Genet. 1997;31:462–468. doi: 10.1007/s002940050231. [DOI] [PubMed] [Google Scholar]
- 73.Dohrmann PR, Butler G, Tamai K, Dorland S, Greene JR, Thiele DJ, Stillman DJ. Genes Dev. 1992;6:93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 74.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Donahue TF, Henry SA. J Biol Chem. 1981;256:7077–7085. [PubMed] [Google Scholar]
- 76.Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. Mol Cell Biol. 2003;23:7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Almaguer C, Mantella D, Perez E, Patton-Vogt J. Eukaryot Cell. 2003;2:729–736. doi: 10.1128/EC.2.4.729-736.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almaguer C, Cheng W, Nolder C, Patton-Vogt J. J Biol Chem. 2004;279:31937–31942. doi: 10.1074/jbc.M403648200. [DOI] [PubMed] [Google Scholar]
- 79.Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 80.Papa FR, Zhang C, Shokat K, Walter P. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- 81.Kelley MJ, Bailis AM, Henry SA, Carman GM. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- 82.Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- 83.Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


