Cocaine (1) is a powerful stimulant and may be the most reinforcing of all drugs. Consequently, the abuse of cocaine continues to be a major societal and health problem. A myriad of medical problems, including death, often accompany cocaine use and the association of the drug with the spread of AIDS is of concern.1 Cocaine acts as an indirect dopamine agonist by blocking the dopamine transporter in the pleasure/reward center of the brain.2 This obstruction leads to an excess of dopamine in the synapses, amplifying pleasure sensation. Despite intensive effort, there is no effective pharmacotherapy for cocaine abuse.3 The inherent difficulties in antagonizing a blocker have led to the development of protein-based therapeutics designed to treat cocaine abuse. Our laboratory4 and others5 have shown that anti-cocaine antibodies can sequester cocaine, retarding its ability to enter the CNS, in an approach termed immunopharmacotherapy. A parallel strategy utilizes catalytic antibodies that are specific for the hydrolysis of the benzoyl ester of cocaine to give the nonpsychoactive products benzoate and methyl ecgonine 2 (Scheme 1).6 While the potential of this method has been demonstrated in rodent models of cocaine overdose and reinforcement, the kinetic constants of these antibodies must be improved to be a viable clinical treatment.6a,7 Alternatively, potential enzymatic therapeutics have been explored and include butyrylcholinesterase (BChE), the major cocaine-metabolizing enzyme present in the plasma of humans and other mammals,8 and the bacterial cocaine esterase (cocE).9 The efficacy of any protein-based cocaine treatment is limited by their inability to access the CNS. Thus, their success depends on peripheral contact between the protein and ingested cocaine.
An improved treatment would contact cocaine both in circulation as well as within the CNS. Filamentous bacteriophage with foreign proteins displayed on its surface are able to penetrate the CNS of mice after various routes of administration (e.g., intravenous, intraperitoneal, intramuscular, intranasal), and can be administered multiple times without visible toxic effects.10 Furthermore, bacteriophage can also diffuse into a wide variety of peripheral organs including the lung, kidney, spleen, liver, and intestine.11 The genetic flexibility of filamentous phage allows for a wide variety of proteins, antibodies, and peptides to be displayed on the protein phage coat in a methodology known as phage display.12 Filamentous bacteriophage fd (Figure 1), can be produced in high titer in bacterial culture, making production simple and economical. Indeed, we have shown the therapeutic potential of a phage-displayed cocaine-binding antibody (GNC 92H2-pVIII).13 Due to the requisite 1:1 stoichiometry of any traditional antibody pharmacotherapy, it is difficult to obtain a meaningful concentration of the therapeutic agent in vivo. However, the modest success of this study encouraged us to examine a phage-displayed catalytic protein as a cocaine therapeutic. Herein, we describe the preparation and kinetics of the first catalytic phage-displayed therapeutic with suitable rates to treat cocaine addiction.
Figure 1.
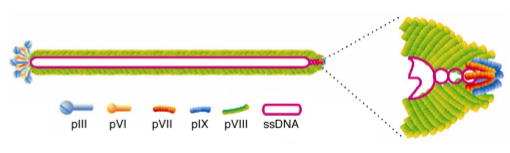
The structure of filamentous bacteriophage fd.
Cocaine esterase is a globular, 574-amino acid bacterial enzyme with a molecular weight of ~65 kDa and is the most efficient protein catalyst for the hydrolysis of cocaine characterized to date.9 The specificity rate constant of this enzyme (kcat/Km) is 103-fold higher than BChE, and 105-fold and 106-fold faster than catalytic antibodies 15A1014 and GNL3A6,6a respectively. The size and catalytic efficiency of cocE make it an ideal candidate for an improved cocaine therapy. However, an exogenous bacterial enzyme would be rapidly cleared via proteolysis and immune surveillance. Also, available protein would not be able to enter the CNS, limiting its efficacy. Bacteriophage, on the other hand, readily enter the bloodstream and cross the blood-brain barrier11 and are stable to a variety of harsh conditions, such as extremes in pH and treatment with nucleases and proteolytic enzymes. Furthermore, we and others have shown that the immune response against filamentous bacteriophage is generally slow.11,13 Thus, displaying cocE on the phage surface may overcome the inherent disadvantages of the natural enzyme and endow it with more favorable immuno/proteodynamics.
Expression of cocE was performed using protein III (pIII) and protein IX (pIX) of the phage coat. These ~42 kDa and ~3.7 kDa proteins, respectively, are expressed in three to five copies on opposite ends of the phage (Figure 1). These proteins were chosen because they could best accommodate a protein of the size of cocE, in contrast to major coat protein pVIII. CocE was expressed on phage by ligating the vector pCocE between two flanking SfiI restriction sites on phagemid pCGMT for cocE-pIII, 16 or pCGMT9 for cocE-pIX.17,18 E. coli cells were transformed with either phagemid and then infected with VCSM13 helper phage. After incubation and centrifugation, the pellet was resuspended in bacterial media and the culture grown at 28 ºC. Since both phage and cocE expression are temperature sensitive, 28 ºC was chosen as a compromise between optimal phage growth (37 ºC) and cocE expression (24 ºC). Under these conditions, both cocE-pIII and -pIX were reproducibly grown in high titers (~1011–1012 cfu/mL) with consistent cocaine hydrolysis activity.
The rate of hydrolysis for cocE-pIII and -pIX was measured by monitoring the increase in benzoic acid concentration over time by reversed-phase HPLC. Both cocE-pIII and -pIX displayed classic Michaelis-Menten steady-state kinetics (Table 1). Estimated values of kcat and kcat/Km are reported as ranges, assuming an average of between 0.1–5 copies of cocE per phage particle. While five copies of cocE per phage is the theoretical maximum, the lower limit of the range is a more reasonable estimate based on previous reports.19 It is encouraging to note that based on the activity of the wild-type enzyme, no less than 10% of the phage displayed an average of one copy of cocE. Furthermore, the activity of the phage does not depend on the coat protein on which cocE is expressed. Therefore, there is no interference of the enzyme due to the local conditions of the phage, such as antagonistic effects from the tethering protein or nearby pVI or pVII coat proteins. In both cases, however, cocE-pIII and -pIX are less active than the natural enzyme, primarily due to a 103-fold reduction in apparent Km. While we can exclude local phenomena on the phage surface, the reduced activity may be caused by phage itself. It is more likely that the reduction in kinetic parameters is due to misfolded enzyme, as phage expression requires higher temperature than that for cocE. Indeed, expression of native cocE at higher temperature (37 ºC) gave good yield of protein, but with little activity (data not shown). Identical to the native enzyme, cocE-phage also is able to hydrolyze cocaethylene.9 However, due to the extremely poor solubility of this substrate, we were unable to determine the kinetic parameters for this reaction.
Table 1.
Summary of Kinetic Parameters for cocE Enzymes a
| Catalyst | Kmb (μM) | kcatc (min−1) | kcat/Kmc (M−1 s−1) |
|---|---|---|---|
| cocE-pIX | 586 ± 63 | 415–8.3 | 11.8 □ 103–0.2 □ 103 |
| cocE-pIII | 412 ± 43 | 181–3.6 | 7.3 □ 103–0.1 □ 103 |
| cocEd | 0.64 ± 0.02 | 468 ± 6 | 1.2 ± 0.04 □ 107 |
See Supporting Information for procedures of kinetic experiments.
Apparent Km.
Estimated range of kcat or kcat/Km based on the possibility of 0.1–5 copies of cocE displayed per phage particle.
Values taken from ref. 9.
Assuming the frequency of cocE incorporation relative to native phage coat protein is low (i.e., the lower estimate is accurate), the kcat of cocE-phage approaches that of the natural enzyme. In this case, cocE-pIX achieves a therapeutically relevant kcat/Km (~104 M−1 s−1);6a importantly, this value is greater than that of any known catalytic anti-cocaine antibodies and only recently obtained by a designed mutant BChE.20
While the relevance of phage-displayed cocE in vivo has not been examined, these results demonstrate a potential method for catalytic cocaine degradation in both the CNS and the periphery possessing both suitable kinetic parameters and pharmacological profile for mammalian administration. Ultimately, testing cocE-phage constructs in animal models of cocaine addiction is required prior to advancement to human models and will be the subject of future studies.
Supplementary Material
Scheme 1.
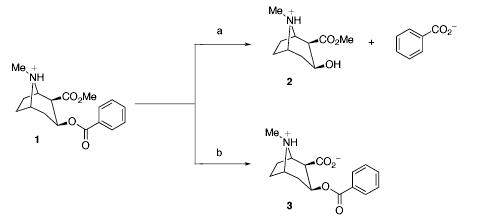
Hydrolysis Products of Cocaine Ester Cleavage a
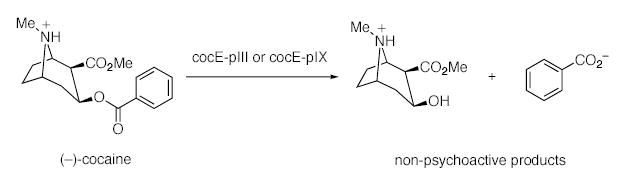
Cocaine is among the most reinforcing of all drugs of abuse, yet no effective pharmacotherapy is available. Herein we report the development and characterization of phage-displayed cocaine esterases with pharmacologically relevant kinetic parameters (kcat/Km ~ 104 M−1 s−1).
Acknowledgments
We gratefully acknowledge Dr. Neil C. Bruce of the University of Cambridge for the generous gift of the cocaine esterase vector pCocE. This work was supported by the National Institutes of Health (DA 08590 and DA 17228) and the Skaggs Institute for Chemical Biology.
Footnotes
Supporting Information Available: Experimental procedures for the expression of cocE-phage and kinetic experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a Brody SL, Slovis CM, Wrenn KD. Am J Med. 1990;88:325–331. doi: 10.1016/0002-9343(90)90484-u. [DOI] [PubMed] [Google Scholar]; b Des Jarlais DC, Frieland SR. Science. 1989;245:578. doi: 10.1126/science.2762809. [DOI] [PubMed] [Google Scholar]
- 2.a Ritz MC, Lamb RC, Goldberg SR, Kuhar M. J Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]; b Withers NW, Pulvirenti L, Koob GF, Gillin JC. J Clin Psychopharmacol. 1995;15:63–78. doi: 10.1097/00004714-199502000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Mendelson JH, Mello NK. New Engl J Med. 1996;334:965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 4.a Carrera MRA, Ashley JA, Parsons LH, Wirsching P, Koob GF, Janda KD. Nature. 1995;378:727–730. doi: 10.1038/378727a0. [DOI] [PubMed] [Google Scholar]; b Carrera MRA, Ashley JA, Zhou B, Wirsching P, Koob GF, Janda KD. Proc Natl Acad Sci USA. 2000;97:6202–6206. doi: 10.1073/pnas.97.11.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Carrera MRA, Ashley JA, Wirsching P, Koob GF, Janda KD. Proc Natl Acad Sci USA. 2001;98:1988–1992. doi: 10.1073/pnas.041610998. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Carrera, M. R. A.; Trigo, J. M.; Roberts, A. J.; Janda, K. D. Pharmacol. Biochem. Behav. 2005, in press. [DOI] [PubMed]
- 5.a Fox BS, Kantak KM, Edwards MA, Black KM, Bollinger BK, Botka AJ, French TL, Thompson TL, Schad VC, Greenstein JL, Gefter ML, Exley MA, Swain PA, Briner TJ. Nat Med. 1996;2:1129–1132. doi: 10.1038/nm1096-1129. [DOI] [PubMed] [Google Scholar]; b Kantak KM, Collins SL, Lipman EG, Bond J, Giovanoni K, Fox BS. Psychopharmacology. 2000;148:251–262. doi: 10.1007/s002130050049. [DOI] [PubMed] [Google Scholar]
- 6.a Matsushita M, Hoffman TZ, Ashley JA, Zhou B, Wirsching P, Janda KD. Bioorg Med Chem Lett. 2001;11:87–90. doi: 10.1016/s0960-894x(00)00659-4. [DOI] [PubMed] [Google Scholar]; b Landry DW, Zhao K, Yang GXQ, Glickman M, Georgiadis TM. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]; c Cashman JR, Berkman CE, Underiner GE. J Pharm Exp Ther. 2000;293:952–961. [PubMed] [Google Scholar]; d Baird TJ, Deng SX, Landry DW, Winger G, Woods JH. J Pharmacol Exp Ther. 2000;295:1127–1134. [PubMed] [Google Scholar]
- 7.Meijler MM, Matsushita M, Wirsching P, Janda KD. Curr Drug Discovery Technol. 2004;1:77–89. doi: 10.2174/1570163043484851. [DOI] [PubMed] [Google Scholar]
- 8.a Nachon F, Nicolet Y, Viquie N, Masson P, Fontecilla-Camps JC, Lockridge O. Eur J Biochem. 2002;269:630–637. doi: 10.1046/j.0014-2956.2001.02692.x. [DOI] [PubMed] [Google Scholar]; b Mattes CE, Lynch TJ, Singh A, Bradely RM, Kellaris PA, Brady RO, Dretchen KL. Tox Appl Pharmacol. 1997;145:372–380. doi: 10.1006/taap.1997.8188. [DOI] [PubMed] [Google Scholar]; c Sun H, Pang YP, Lockridge O, Brimijoin S. Mol Pharmacol. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 9.a Bresler MM, Rosser SJ, Basran A, Bruce NC. Appl Environ Microbiol. 2000;66:904–908. doi: 10.1128/aem.66.3.904-908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Nat Struct Biol. 2002;9:17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]; c Turner JM, Larsen NA, Basran A, Barbas CF, III, Bruce NC, Wilson IA, Lerner RA. Biochemistry. 2002;41:12297–12307. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 10.Frenkel D, Solomon B. Proc Natl Acad Sci USA. 2002;99:5675–5679. doi: 10.1073/pnas.072027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabrowska K, Switala-Jelen K, Opolski A, Weber-Dabrowska B, Gorski A. J App Microbiol. 2005;98:7–13. doi: 10.1111/j.1365-2672.2004.02422.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith GP. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 13.Carrera MRA, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Proc Natl Acad Sci USA. 2004;101:10416–10421. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng SX, De Prada P, Winger G, Landry DW. J Immunol Methods. 2002;269:299–310. doi: 10.1016/s0022-1759(02)00237-5. [DOI] [PubMed] [Google Scholar]
- 15.Larocca D, Burg MA, Jensen-Pergakes K, Ravey EP, Gonzales AM, Baird A. Curr Pharm Biotechnol. 2002;3:45– 57. doi: 10.2174/1389201023378490. [DOI] [PubMed] [Google Scholar]
- 16.Gao C, Mao S, Lo CHL, Wirsching P, Lerner RA, Janda KD. Proc Natl Acad Sci USA. 1997;94:11777–11782. doi: 10.1073/pnas.94.22.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao C, Mao S, Kaufmann GF, Wirsching P, Lerner RA, Janda KD. Proc Natl Acad Sci USA. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao C, Mao S, Lo CHL, Wirsching P, Lerner RA, Janda KD. Proc Natl Acad Sci USA. 1997;94:11777–11782. doi: 10.1073/pnas.94.22.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baek H, Suk KH, Kim YH, Cha S. Nucleic Acids Res. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H, Pang YP, Lockridge O, Brimijoin S. Mol Pharmacol. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


