Abstract
Despite the power of sequencing and of emerging high-throughput technologies to collect data rapidly, the definitive functional characterization of unknown genes still requires biochemical and genetic analysis in case-by-case studies. This often involves the deletion of target genes and phenotypic characterization of the deletants. We describe here modifications of an existing deletion method which facilitates the deletion process and enables convenient analysis of the expression properties of the target gene by replacing it with an FRT-lacZ-aph-Plac-FRT cassette. The lacZ gene specifically reports the activity of the deleted gene and therefore allows the determination of the conditions under which it is actively expressed. The aph gene, encoding resistance to kanamycin, provides a selectable means of transducing a deleted locus between strains so that the deletion can be combined with other relevant mutations. The lac promoter helps to overcome possible polar effects on downstream genes within an operon. Because the cassette is flanked by two directly repeated FRT sites, the cassette can be excised by the Flp recombinase provided in trans. Removing the cassette leaves an in-frame deletion with a short scar which should not interfere with downstream expression. Replacements of yacF, yacG, yacH, yacK (cueO), yacL, ruvA, ruvB, yabB, and yabC made with the cassette were used to verify its properties.
Years were required to sequence the first bacterial genomes; with present technology, small genomes can be sequenced in days (although annotation and analysis is still a slow process). In addition, high-throughput techniques, such as microarrays for transcriptional analysis and recently developed chips for proteomic analysis, permit the acquisition of massive amounts of information in single, albeit complex to interpret, experiments. Despite the availability of these technologies, the fully annotated sequences of over 80 species of prokaryotes, and rapid and widely available software for protein sequence comparison and analysis, at least half of the open reading frames (ORFs) listed for each sequenced species correspond to genes of either hypothetical or unknown function. This is true even for so well studied an organism as the model bacterium Escherichia coli K-12, which has about 4,300 annotated genes. According to Serres et al. (18), about 1,250 ORFs have been assigned possible functions which await confirmation, but a further 850 still have entirely unknown functions. Since the genome was originally annotated in 1997, only about 4% of the genes have had definite functions assigned on the basis of experiment or very close homology with a gene of verified function.
A popular approach to the functional characterization of an uncharacterized gene is to delete its coding sequence from the genome and look for consequent phenotypic changes. Methods to precisely delete E. coli genes are based on promoting genetic exchange between an altered locus engineered in vitro and the target chromosomal locus. There are two classes of method currently favored. The first employs linear DNA, which can be exchanged by a double crossover to replace the chromosomal target with engineered DNA in a single step (14). The most recent variant of this technique obviates the need for cloning altogether by using primers which need include no more than 50 bp of homology flanking the target gene to amplify a selective marker. The linear product is introduced by electroporation into a host expressing the lambda Red-Gam recombination system. Gam inhibits the RecBCD nuclease, which normally degrades linear DNA, and Red is able to mediate recombination between the very short regions of homology flanking the selective marker to replace the target gene with the engineered linear fragment (7, 20).
The second class of methods uses DNA circles, usually conditionally replicating plasmids (which are immune to exonuclease digestion), as delivery systems. The plasmids, which incorporate cloned DNA flanking the target, are forced to integrate into one of the homologous regions by selection for retention of a plasmid marker under restrictive conditions. Longer regions of homology, about 400 bp each, are required to achieve good rates of recombination using host enzymes. The later return of survivors to permissive conditions allows cointegrate resolution. Selection against the plasmid favors survivors in which allele exchange has occurred, and screening is used to identify them (8, 11). Another plasmid-based method has been described which uses a suicide plasmid carrying the desired target gene mutation and a unique recognition site for the meganuclease I-SceI. When integrated into the chromosome, the plasmid is flanked by the wild-type target locus and the constructed replacement, which includes DNA homologous to the target. Provision of I-SceI in trans will result in a double-strand cut, which will stimulate recombination between the alleles, and the engineered version can be selected (16).
The first method, when successful, can be very fast, as no cloning steps are required. However, the length of the replacing sequence is limited by the need to accurately amplify it, and acceptable rates of recombination and replacement are not necessarily easy to achieve. Methods of the second type have at least one cloning step (plus a second if a selective marker is used) and therefore require more time to complete, typically several weeks. However, a large replacement can be made without difficulty. Since each step of the procedure can be monitored for success and the number of successful replacements is expected to be high, it is easy to judge whether failures have a technical or biological basis.
Here, we report improvements and useful modifications made to the method described by Link et al. (11) in which a low-copy-number temperature-sensitive plasmid vector, pKO3, was used. First, we have modified the replacement vector to facilitate the cloning procedure. Second, we have constructed a series of removable cassettes that can be used to replace a target gene and have demonstrated that they work as designed. Each cassette is flanked by two FRT sites, in direct orientation, which allows removal of the cassette by the Flp site-specific recombinase provided in trans. Our design includes a full-length promoter-free lacZ reporter gene to allow measurement of target gene expression under a variety of conditions. To avoid transcriptional polar effects, a controllable Plac promoter has been added at the distal end of the cassette to maintain transcription of downstream genes. Although the Plac promoter will not compensate for any translational polarity which may be a feature of the operon under study, translational polarity should be restored upon cassette removal. Compared to the original pKO3 method (11), the use of a selectable reporter cassette simplifies the recovery of allele replacement mutants and extends the range of possible phenotypic studies to include gene expression measurement.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains and plasmids used in this study are described in Table 1. All strains were grown in Luria-Bertani (LB) medium (Difco tryptone, 1%; Beta Lab yeast extract, 0.5%; NaCl, 1%) with appropriate selection and growth temperatures. Kanamycin (Km), ampicillin (Ap), and chloramphenicol (Cm) were at concentrations of 50, 100, and 20 μg/ml, respectively. For counterselection of sacB plasmids, sucrose was added to a final concentration of 5%. Recovery of transformants prior to selection was done either in LB medium supplemented with glucose (0.36%) or in the SOC medium provided by the manufacturer in the case of TOP10 transformation (Invitrogen).
TABLE 1.
E. coli strains and plasmids
| Name | Genotype or characteristics | Source or references |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | 17 |
| MG1655 | Sequenced λ− and F− derivative of K-12 | 3 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) (φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| TP8503 | Δ(lac-proB) leu thi-1 supE42 fhuA | 12 |
| XL1Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1qZΔM15 Tn10] | Stratagene |
| Plasmids | ||
| pBN | pBR322 Ω [EcoRI-HindIII: 79 bp, polylinker of pNUB193] | This study |
| pBNLC | pBN Ω [SmaI-SmaI: 4,725 bp, lacZ-lacY′-cat from pRS551-SL] | This study |
| pBNLCPS | pBNLC Ω [StuI-StuI: 242 bp, Plac promoter] | This study |
| pBR322 | Apr Tcr; cloning vector | GenBank no. J01749 |
| pBR325 | Apr Tcr Cmr; cloning vector | GenBank no. L08855 |
| pCP20 | Apr CmrrepA(Ts); pSC101-based vector expressing the Flp recombinase | 5 |
| pKD4 | Apr KmroriR plasmid containing an FRT-aph-FRT cassette | 7 |
| pKO3 | Cmr; pSC101-based vector; repA(Ts) with a sacB conferring sucrose sensitivity | 11 |
| pNUB193 | pUC19 derivative with additional restriction sites in its polylinker | New England BioLabs Inc. |
| pRS551 | Apr KmrlacZ; operon fusion vector | 19 |
| pRS551-SL | Same as pRS551 but without tryptophan terminator upstream of lacZ | This study |
| pSLO3 | AprlacZ; promoter cloning vector | 10 |
| pTOF1 | pUC18 Ω [EcoRI-HindIII: 162 bp, NotI-FRT-SmaI-FRT-NotI]; Apr | This study |
| pTOF2 and -3 | pTOF1 Ω [HincII-HincII: 1,252 bp, aph gene from pUC4K]; Apr Kmr | This study |
| pTOF20 | pTOF1 Ω [SmaI-SmaI: 4,967 bp, LacZ-cat-Plac of pBNLCS]; Apr Cmr | This study |
| pTOF24 | pK03 Ω [HincII-HincII: 1,252 bp, aph from pUC4K]; Cmr Kmr Ts Sucs | This study |
| pTOF25 | pKO3 Ω [BamHI-BamHI: 1,264 bp, aph from pUC4K]; Cmr Kmr Ts Sucs | This study |
| pTOF27 | Site-directed mutagenesis of pTOF20 adding a SnaBI site in lacY; Apr Cmr | This study |
| pTOF29 and -30 | pTOF27 Ω [HincII-HincII: 1,252 bp, aph gene from pUC4K]; Apr Kmr | This study |
| pTOF60 | Site directed mutagenesis of pTOF27 adding a SnaBI site upstream of Plac; Apr Cmr | This study |
| pTOF61 | Site-directed mutagenesis of pBR322 adding a NotI site in tet; Apr | This study |
| pTOF70 | pTOF61 Ω [NotI-NotI: 5,114 bp, FRT-lacZ-cat-Plac-FRT from pTOF60]; Apr Cmr | This study |
| pTOF72 and -73 | pTOF70 Ω [HincII-HincII: 1,252 bp, aph gene from pUC4K] Apr Kmr | This study |
| pUC4K | Apr Kmr; cloning vector | GenBank no. X06404 |
| pUC18 | Apr; cloning vector | GenBank no. L08752 |
| pUC19 | Apr; cloning vector | GenBank no. M77789 |
DNA manipulation.
For plasmid DNA purification, genomic DNA preparation, DNA cleaning, DNA gel extraction, and DNA site-directed mutagenesis, we used the Wizard Plus SV Minipreps kit (Promega), the Quantum Prep AquaPure genomic DNA isolation kit (Bio-Rad), the QIAquick PCR purification kit (Qiagen), the QIAquick gel extraction kit (Qiagen), and the QuikChange site-directed mutagenesis kit (Stratagene).
Crossover PCR (9) and analytical PCR were carried out on OmniGene and PCR Sprint cyclers (Hybaid) using Pfu DNA polymerase (Promega) and Taq DNA polymerase (Promega), respectively, according to the manufacturers' recommendations. The primers used in this study were purchased from either Sigma-Genosys or MWG-Biotech and are listed in Table 2. Restriction endonucleases were purchased from Roche, Promega, or New England BioLabs Inc.
TABLE 2.
Primers used
| Name | Restriction site(s) | Sequencea |
|---|---|---|
| Primers for engineering FRT cassettes | ||
| FRTupNotI | EcoRI, NotI | GgaATTcTGTAGGCTGGAGCgGCcgCGAAGTTCC |
| FRTdownSmaI | SmaI | GAGTGCTTGCGGCAGCGcccGGGGATCTTG |
| FRTupSmaI | SmaI | TTGTGTAGcCcGGgGCTGCTTCGAAGTTCC |
| FRTdownNotI | NotI, HindIII | CCTGAaaGCTTGCGGCcGCGTGAGGGGATCTTG |
| MutLCP1 | SnaBI | CGCGTAAGGAAATCCATTAcGTACTATTTAAAAAACACAAAC |
| MutLCP2 | SnaBI | GTTTGTGTTTTTTAAATAGTACgTAATGGATTTCCTTACGCG |
| MutLCP3 | SnaBI | CGTCGCCCAATACGtAAACCGCCTCTCCC |
| MutLCP4 | SnaBI | GGGAGAGGCGGTTTaCGTAttGGGCGACG |
| MutPBR1 | NotI | CCGACCGCTTTGGCgGCCGCCCAGTCCTGC |
| MutPBR2 | NotI | GCAGGACTGGGCGGCcGCCAAAGCGGTCGG |
| NEWCAT-UP | Ecl136II | GGCGgAgcTcTTGAGTTATCGAGATTTTCAGGAGC |
| NEWCAT-DWN | SmaI, StuI | TTATTCccGgGTAGCACtAGGCcTTTAAGGGCACC |
| PLAC-1 | StuI | CAGGCATGCAgGCcTtGCGTAATC |
| PLAC-3 | StuI | GAGGAgGCctgAcgtCGCCCAATAC |
| Replacement primersb | ||
| ruvA deletion | ||
| CiRuvA | NotI | ccgttccaagcggccgcaagagcgGTGAGCAAAATCGCTCGCCCTGAC |
| CoRuvA | SalI | aaaaagtcgacAACAGCACGTCATGCGGTTCAAGG |
| NiRuvA | NotI | cgctcttgcggccgcttggaacggTCTGAGTCTGCCTATCACATGACG |
| NoRuvA | PstI | aaaaactgcagGTGATTACTCCAGCAATTTGATGC |
| ruvB deletion | ||
| CiRuvB4 | NotI | gttctgcagcggccgcgaattccgCCGCCAGAAATGCCGTAAGTC |
| CoRuvB2 | SalI | cgcacgcatgtcgacCGCAATTTGCTGTCAATGACGATAAG |
| NiRuvB3 | NotI | cggaattcgcggccgctgcagaacCATCCTTTACCTCATAACGCGGC |
| NoRuvB3 | SmaI | aaggaaaaaagcccgggcAAGCGGGTCAGGAAGCG |
| yacH deletion | ||
| CiYacH | NotI | ccgttccaagcggccgcaagagcgCGTGGAGCAAGAAGACTGGAAAGG |
| CoYacH | SalI | aaaaagtcgacATACCGAACCAGAAGTTGCATGGG |
| NiYacH | NotI | cgctcttgcggccgcttggaacggGCAAATTAGCGCCAGCACATGGGG |
| NoYacH | PstI | aaaaactgcagTACGGTATTCTTCTAGCACGACGG |
| yacL deletion | ||
| CiYacL | NotI | ccgttccaagcggccgcaagagcgGTCGTGGCGGCTTACCGCAATTTC |
| CoYacL | SalI | aaaaagtcgacCAGCTTGAGTCCGATATCGTAACC |
| NiYacL | NotI | cgctcttgcggccgcttggaacggCACTTCATGCCCCATGGACATACG |
| NoYacL | PstI | aaaaactgcagGCGAATGTCTTCCTGGCTTCTGCG |
| yacK deletion | ||
| CiYacK | NotI | ccgttccaagcggccgcaagagcgGAAGATACGGGGATGATGTTAGGG |
| CoYacK | SalI | aaaaagtcgacTCGCCGCTACGGCAGATAACTACC |
| NiYacK | NotI | cgctcttgcggccgcttggaacggCGGCAAAGCCGAAGCCACACCCAG |
| NoYacK | PstI | aaaaactgcagAGGTGTCGTAATTACTGAGGTCCC |
| yacF deletion | ||
| CiYacF | NotI | ccgttccaagcggccgcaagagcgTTTGCCATTCGTTTTATGCCGCTG |
| CoYacF | SalI | aaaaagtcgacGGCGCGTCTTATCAGGCCTAAAGG |
| NiYacF | NotI | cgctcttgcggccgcttggaacggAAACTCAATGCGCAGCCATGTACG |
| NoYacF | PstI | aaaaactgcagCACCAGATCCAGCAAGCTACTTCC |
| yacG deletion | ||
| CiYacG | NotI | ccgttccaagcggccgcaagagcgGAAGAACCAAAGCAGTGACATTTG |
| CoYacG | SalI | aaaaagtcgacCTGGAATATGAATTCCCGGACAGG |
| NiYacG | NotI | cgctcttgcggccgcttggaacggCACCACCGTTTTCCCGCAGGTTGG |
| NoYacG | PstI | aaaaactgcagGCAGCTTTGATTTACCTACATTGC |
| yabB deletion | ||
| CiYabB1 | NotI | ccgttccaagcggccgcaagagcgCTGTCGAGCATGAACCCGGTTGAG |
| CoYabB | SalI | aaaaagtcgacCGGGTTGGGTCCATACGCATGTCC |
| NiYabB1 | NotI | cgctcttgcggccgcttggaacggGTCGAGATTGACTAACGTTGCTCC |
| NoYabB | SmaI | aaaaacccgggTGTTACATGCGGCGATGAATTGCC |
| yabC deletion | ||
| CiYabC | NotI | ccgttccaagcggccgcaagagcgGGGTTACCGATGACTGAAGAGCAG |
| CoYabC | SalI | aaaaagtcgacAGCGCATTCTCTTCAAGGATCAGG |
| NiYabC | NotI | cgctcttgcggccgcttggaacggCCCATCAATGTAGATGCCATCAGG |
| NoYabC | SmaI | aaaaacccgggCACCATTGACATTTATCACCCGTG |
Restriction sites are underlined; sequences altered with respect to the template are in lowercase.
For the replacement primers, the prefixes C and N indicate the 5′ and 3′ ends, respectively, of the deleted genes, while i and o stand for inner (outward-pointing) and outer (inward-pointing) primers.
Ligations were carried out using standard procedures (17). Prior to ligation with T4 DNA ligase (Roche), vectors were dephosphorylated with calf alkaline phosphatase (Roche), and both vector and insert DNA were purified using a QIAquick PCR purification kit.
Transformation of bacteria with engineered plasmid DNA was done using a classical CaCl2 preparation unless high-efficiency transformation was needed, in which case electroporation was employed (17). Recombinant DNA was recovered in DH5α or, after site-directed mutagenesis, in XL1Blue. After the FRT cassette cloning step, recombinant DNA was recovered in TOP10 using One Shot TOP10 competent cells (Invitrogen).
Construction of lacZ-cat-Plac reporter cassette.
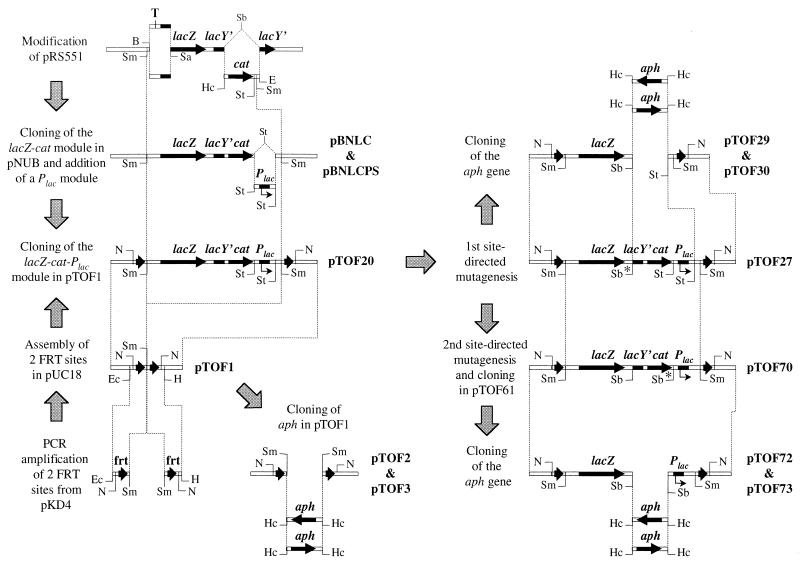
The modules of the lacZ-cat-Plac cassette were assembled in several steps (Fig. 1). First, pRS551 (19) was reconstituted with a lacZ gene lacking the upstream trp transcriptional terminator to create pRS551-SL. This was done by replacing the 2,453-bp BamHI-SacI fragment carrying the 5′ end of lacZ with the corresponding terminator-free BamHI-SacI fragment of pSLO3 (10). In a second step, cat was PCR amplified from pBR325 using the primers NEWCAT-UP and NEWCAT-DWN to engineer an upstream Ecl136II site and downstream StuI and SmaI sites, respectively. The 762-bp PCR product was cut with Ecl136II and SmaI and cloned into the unique SmaI site of pUC18. The cat gene was cut from pUC18 using Ecl136II and HincII and cloned into the SnaBI site (in lacY) of pRS551-SL. In a third step, the lacZ-cat module was cut from pRS551-SL with SmaI and cloned into the unique SmaI site of pBN to produce the plasmid pBNLC. Finally, Plac was PCR amplified from pUC19 using the primers PLAC-1 and PLAC-3 designed to flank the PCR product with StuI restriction sites. The PCR product was cut with StuI and cloned into the unique StuI site downstream of the cat gene on plasmid pBNLC. The resulting plasmid, pBNLCPS, was later used to deliver the lacZ-cat-Plac cassette.
FIG. 1.
Construction of the lacZ-cat-Plac and lacZ-aph-Plac reporter cassettes. The constructions were started with pRS551 (top left) and the amplification of FRT sites (bottom left). The large shaded arrows indicate the path of construction, which is described in detail in the text. T marks the weak transcription terminator in trp. Restriction sites are indicated with one or two letters: B, BamHI; Ec, EcoRI; E, Ecl136; Hc, HincII; H, HindIII; N, NotI; Sa, SacI; Sm, SmaI; Sb, SnaBI; and St, StuI. The short solid arrows mark FRT sites. Intact genes and their directions of transcription are shown as long solid arrows on the DNA. Untranscribed gene fragments are shown as solid bars. Vertical dotted reference lines connect points with the same DNA sequence. All named plasmids are further described in Table 1.
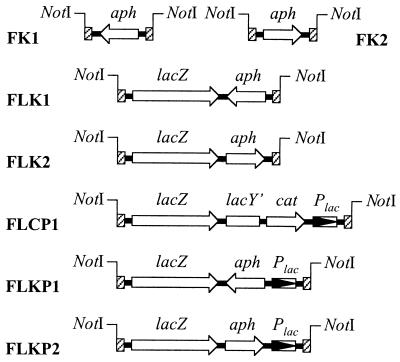
Construction of a set of FRT cassettes.
Two directly repeated FRT sites were combined in an initial construct (Fig. 1, bottom left). The sites were amplified from pKD4 using Pfu turbo (Stratagene), and two sets of primers were designed to flank the FRT product with either EcoRI-NotI and SmaI (primers FRTupNotI and FRTdownSmaI) or SmaI and NotI-HindIII (primers FRTupSmaI and FRTdownNotI). The first PCR product was cut with EcoRI and XmaI, the second was cut with XmaI and HindIII, and the products were purified and ligated to EcoRI- and HindIII-cut pUC18 in a three-molecule cloning. The recombinant plasmid pTOF1 was recovered after electroporation of DH5α, and the 162-bp insert sequence was confirmed by sequencing. The FK1 and FK2 cassettes (Fig. 2) were made by cloning the 1,252-bp HincII fragment of pUC4K, containing the aph gene (specifying kanamycin resistance), into pTOF1 at the unique SmaI site located between the two FRT sites. The recombinant plasmids pTOF2 and pTOF3, containing the aph gene in different orientations, were recovered in DH5α. The FLCP1 cassette was constructed by cloning the 4,961-bp SmaI fragment of pBNLCPS, containing the lacZ-cat-Plac module, into SmaI-cut pTOF1. The resulting recombinant plasmid, pTOF20, was mutagenized (primers MutLCP1 and MutLCP2) to generate an SnaBI site overlapping the first codon of lacY. The resulting plasmid, pTOF27, was cut with SnaBI and StuI, and the excised lacY′-cat-Plac module was replaced with the 1,252-bp HincII fragment containing the Kmr determinant of pUC4K. pTOF29 and pTOF30, bearing the cassettes FLK1 and FLK2, respectively, contain the aph gene in different orientations. pTOF27 was further mutagenized to introduce a new SnaBI site upstream of Plac using the primers MutLCP-3 and MutLCP-4. The resulting plasmid, pTOF60, contained two mutagenized Plac sequences (one originating from pUC18 and the other originating from the FLCP cassette). We separated them by transferring the 5,114-bp NotI fragment of pTOF60 containing the mutagenized FLCP cassette into the unique NotI site of pTOF61, a pBR322 plasmid with a NotI site introduced into the tetracycline resistance gene. This pBR322 derivative, pTOF70, contains a 1,548-bp lacY′-cat module flanked by two SnaBI sites, which was replaced by the 1,252-bp HincII fragment of pUC4K to generate the FLKP1 and FLKP2 cassettes of plasmids pTOF72 and pTOF73, respectively.
FIG. 2.
FRT cassettes constructed and used in this work. The small hatched rectangles at the ends of each cassette represent FRT sites. Genes and their directions of transcription are indicated by open arrows. The constructions are described in the text and in the legend to Fig. 1.
Construction of replacement vectors.
The two replacement vectors, pTOF24 and pTOF25, derive from plasmid pKO3 (11). The three essential features of pKO3 are maintained in pTOF24 and pTOF25. The plasmids are unable to replicate at temperatures above 42°C, as they encode a temperature-sensitive replication protein (repAts). In addition, they contain a Cmr (cat) marker for selection and a counterselective marker originating from Bacillus subtilis, sacB. The sacB gene encodes a levan sucrase, which catalyzes sucrose hydrolysis and levan extension. In E. coli, the products of this reaction are toxic, and the cells are sucrose sensitive. To construct pTOF24 and pTOF25, the short polylinker, SmaI-NotI-BamHI-NotI-SalI, of pKO3 was modified by cloning the 1,252-bp HincII fragment containing the Kmr determinant of pUC4K into the unique SmaI site of the pKO3 polylinker. Although the SmaI site was lost, a PstI site, provided by the insert, was added. This new vector, pTOF24, is used for SalI-PstI cloning. pTOF25 was obtained by cloning the 1,264-bp BamHI fragment containing the Kmr determinant of pUC4K into the unique BamHI site of pKO3; pTOF25 is used for SalI-SmaI cloning. Cloning into pTOF24 or pTOF25, using SalI-PstI or SalI-SmaI, leads to a screenable loss of Kmr.
Availability of material.
All plasmids, FRT cassettes, constructs, and strains, as well as sequence or general information concerning the materials used to perform the genetic alterations described in this article, are available by contacting the authors.
RESULTS
An improved gene replacement procedure.
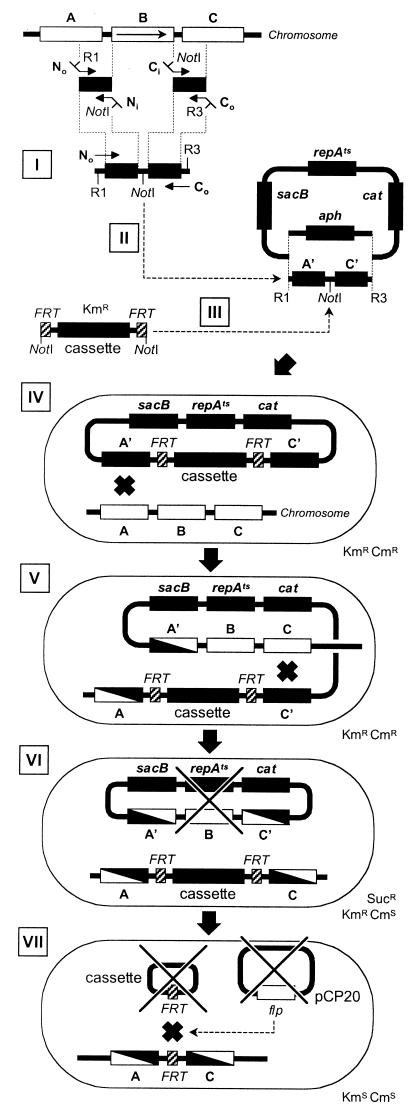
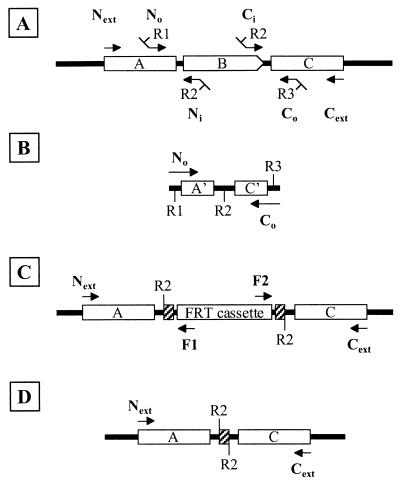
Precise deletions of E. coli ORFs were performed using a method based on that described by Link et al. (11). The step-by-step procedure and the sites of annealing of the primers used in this study are described in Fig. 3 and 4, respectively. The replacement procedure starts with the in vitro construction of a deletion cassette using a two-step crossover PCR protocol (9). In the first step, ∼400-bp segments corresponding to the regions flanking the locus or gene targeted (arms) are independently amplified using the pairs of primers No-Ni and Ci-Co (Fig. 4A). N (N-terminal) and C (C-terminal) primers anneal upstream and downstream of the target gene, respectively, while “i” (inside) and “o” (outside) indicate whether the priming site is closer to or further from the target gene. The Ni and Ci inside primers are designed to leave the ends of the targeted genes intact in order to retain the original translational signals in the final construct. In addition, Ni and Ci contain 24-nucleotide-long 5′ tails with complementary sequences. Each tail also contains a NotI restriction site for later FRT cassette insertion. In a second PCR step, the previously amplified arms are fused together using the complementary ends (provided by Ni and Ci tails). Typically, 1 μl of each arm PCR mixture is used as a template, and the fusion product is amplified with No and Co primers (Fig. 4B). In addition to their priming sequences, No and Co primers each contain an 11-nucleotide tail providing convenient restriction sites for cloning (Table 2).
FIG.3.
Gene replacement-deletion procedure. Wild-type genes are indicated by open boxes, while engineered sequences or plasmid genes are indicated by solid boxes. B, gene targeted for deletion; A and C, genes flanking B; sacB, levan sucrase gene, toxic in the presence of sucrose (Suc); repAts, temperature-sensitive system of replication; cat and aph, genes for Cmr and Kmr, respectively. The steps of the procedure are as follows. (I) Construction of a deleted locus by crossover PCR. (II) Cloning of the crossover PCR product into pTOF24 or pTOF25 using the restriction sites (R1 and R3) provided by the outside primers (No and Co). The Kmr gene (aph) of the vector is replaced by the insert, and transformants are selected on LB-chloramphenicol medium at 30°C. (III) Cloning of an FRT cassette between the two arms of the crossover PCR insert into the unique NotI site initially provided by the internal primers Ni and Ci. Kmr is reestablished by the acquisition of the FRT cassette, and transformants are selected on LB-chloramphenicol-kanamycin plates at 30°C. (IV) First homologous recombination taking place after the introduction of the replacement plasmid in a Δlac strain. (V) Selection of integrants on LB-kanamycin-chloramphenicol plates at 42°C. (VI) Excision of the plasmid after a second homologous recombination. Allele replacement and concomitant plasmid loss (×) are selected for on LB-kanamycin-sucrose plates at 37°C. (VII) Excision of the FRT cassette after transformation with pCP20. The circular form of the cassette does not replicate and disappears, while pCP20 can be cured at 42°C. The resulting strain is Kms Cms Aps.
FIG. 4.
Primer design for allele replacement construction and deletion confirmation. Intergenic chromosomal DNA is shown as solid, and FRT DNA is hatched. (A) Chromosomal DNA with genes A, B, and C. Primers No plus Ni and Co plus Ci were used to make arms for B deletion, using chromosomal DNA as a template. R1, R2, and R3 are restriction sites. (B) The arms made in panel A are annealed at R2, reamplified, and cloned. (C) An FRT cassette is inserted at R2, and the resultant replacement plasmid is introduced into the chromosome and resolved to make a replacement. Primers Next plus F1 and Cext plus F2 are used to demonstrate that the replacement is correctly structured. (D) The chromosomal replacement after Flp-mediated cassette deletion. Arrows indicate primer position and direction.
The PCR fusion product is cloned in one of the two pKO3-modified replacement vectors, pTOF24 or pTOF25, using a PstI-SalI or an SmaI-SalI restriction strategy, respectively. Recombinant plasmids are recovered by transformation with selection on LB-chloramphenicol agar plates. In recombinants, the Kmr gene in the vector is replaced by the PCR fusion product and can be identified by Kms screening; this is much simpler than the PCR screening required when using pKO3 (11). Finally, one of the FRT cassettes is cloned into the unique NotI restriction site at the junction of the fused PCR arms, and recombinant plasmids are recovered by transformation with selection on LB-chloramphenicol-kanamycin plates (Kmr is reestablished by cloning the FRT cassette).
Next, a Δlac strain is transformed with the construct and allele replacement is started by streaking transformants on LB-chloramphenicol-kanamycin plates at 42°C. The antibiotics maintain selection for both plasmid and cassette, while the elevated temperature prevents autonomous replication of the plasmid. Thus, only integrants resulting from a homologous recombination between one of the arms of homology and the corresponding wild-type locus survive. Integrants are recognized as a small number of large colonies on a background of microcolonies. One integrant is repurified on the same medium at 42°C, and selective pressure is released by overnight culture at 30°C in LB medium, allowing the cointegrate to resolve by means of a second homologous recombination. Serial dilutions are plated on LB-sucrose-kanamycin plates at 37°C. Sucrose kills bacteria containing a functional sacB gene, either in the chromosome or on an autonomously replicating plasmid, while kanamycin selects those retaining the cassette, indicating allele replacement. The frequencies of Kmr sucrose-resistant (Sucr) colonies are quite variable from one experiment to another, but generally the colony counts on LB-kanamycin-sucrose and LB-kanamycin (or LB) are similar. This suggests that (i) the cointegrate is not stable and is quickly resolved when selective pressure is withdrawn and (ii) the plasmid is often lost during excision. When an attempt is made to replace an essential gene, the frequency of Kmr Sucr colonies falls dramatically by several orders of magnitude. In the method of Link et al. (11), no selective cassette was used for replacement and therefore the Sucr Cms bacteria isolated at the end of the replacement process might either be parent-like (if both insertion and resolution take place in the same arm) or deletants (if insertion and resolution take place in different arms). At this step, Sucr bacteria which still retain plasmid are frequently isolated. Therefore, plasmid loss must be confirmed by screening survivors for the loss of Cmr. In our hands, 3 to 100% of Kmr Sucr survivors are Cms, with an average of 50%. It is not known whether plasmid-retaining, Sucr colonies have a mutated sacB gene or whether sucrose toxicity is not lethal in some SacB+ bacteria. Screening bacteria isolated without sucrose counterselection at the excision step, however, reduces the recovery of replacement clones by a factor of about 6.
At the end of the replacement process, Kmr Sucr Cms colonies are analyzed by PCR to confirm that the cassette has replaced the wild-type locus. To do this, each junction between the cassette and the chromosome is amplified using a primer pair that primes (i) in the FRT cassette and facing outward (F1 or F2 primers) and (ii) in the flanking region facing toward the cassette (Next or Cext primers) (Fig. 4C). It should be noted that Next and Cext prime sequences that have not been cloned during the crossover PCR; otherwise, no distinction could be made between a gene replaced by an FRT cassette and the replacement plasmid containing the deletion cassette.
The cassette can be removed by site-specific recombination between the FRT sites flanking the cassette by providing the cognate Flp recombinase in trans with plasmid pCP20 (5). Plasmid pCP20 is a temperature-sensitive replicon with an flp gene under the control of a λ cI857 repressor. However, as already described, the basal expression of flp at 30°C is enough to catalyze extensive FRT recombination (5). Therefore, the Flp-catalyzed FRT cassette excision is simply carried out by transforming cassette-bearing clones with pCP20 and selecting transformants at 30°C on LB-chloramphenicol (or LB-ampicillin) medium. We find that 100% of the transformants have already lost the Kmr cassette marker at this step. Next, Kms tansformants are cured of pCP20 by streaking them on LB plates at 42°C, and the loss of the plasmid is confirmed by a failure to grow on either chloramphenicol or ampicillin plates. Finally, cassette excision is checked by PCR using primers flanking the locus of interest; the PCR fragment obtained from a deleted locus will now have a reduced size compared to its corresponding wild type (Fig. 4D).
Gene replacement and cassette removal.
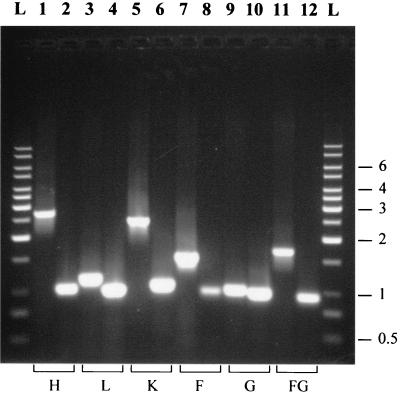
The FRT cassettes have been successfully used to construct a number of deletions. The TP8503 genes ruvA, ruvB, yacH, yacL, and yacK have been replaced by the cassette FLK2, and yacF, yacG, yabB, and yabC have been replaced by the cassette FLKP2. In all cases where it was attempted (yacH 〈 〉 FLK2, yacL 〈 〉 FLK2, yacK 〈 〉 FLK2, yacF 〈 〉 FLKP2, yacG 〈 〉 FLKP2, yabB 〈 〉 FLKP2, and yabC 〈 〉 FLKP2), replacements were easily transduced to MG1655. P1 transduction can also be used for combining a replacement with a desired second mutation. All of the cassettes could be successfully removed by the Flp recombinase. Figure 5 shows PCR amplifications of wild-type loci and their deletion derivatives after the cassettes were removed. The same gene-specific outside primers were used for both the original gene and its deletion derivative. In all cases, the deletion is associated with a decrease in PCR fragment size as expected (although it is small in the case of the yacG deletion). It should be noted that the deletion process leaves a 93-nucleotide-long in-frame scar. When the deletion is correctly designed, the scar will not contain a stop codon, and the original translational signal of the truncated protein will remain functional.
FIG. 5.
PCR amplifications of wild-type and deleted regions for five different ORFs. FRT cassette replacements were made for each of the genes yacH, -L, -K, -G, and the -FG pair. The FRT cassette was later deleted with FLp. Each lane labeled with an odd number shows amplified wild-type DNA, and the even-numbered lane to its right shows the corresponding deletion. Amplification in all cases used the appropriate Next-Cext primer pair. DNA size standards (lanes L) are on both sides of the gel, with length in kilobases indicated on the right.
Reporting the expression of the deleted gene.
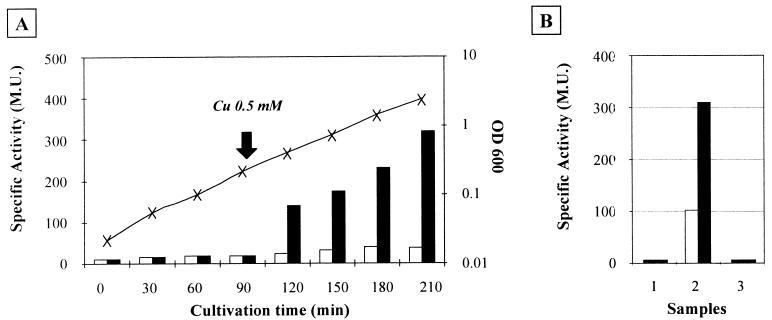
The FRT cassettes were engineered to increase the information that can be collected with a single deletion and replacement by using a cassette containing a lacZ reporter to replace the target gene. The efficacy of the reporter system was first tested with the yacK 〈 〉 FLK2 replacement. The yacK gene, now known as cueO, has recently been characterized as encoding a multicopper oxidase, and its expression has been shown to be induced by copper (15). The replacement strain TP8503 yacK 〈 〉 FLK2 was grown in LB medium to an optical density at 600 nm (OD600) of 0.2, and an aliquot was treated with 0.5 mM CuSO4. β-Galactosidase activity was assayed at regular intervals during growth (13). Figure 6A shows that yacK expression increased by a factor of 5 to 6 following the addition of copper.
FIG. 6.
Use of lacZ to report expression of deleted genes. (A) An exponentially growing culture of TP8503 yacK ↔ FLK2 was divided in two (arrow), and one half was induced by addition of 0.5 mM CuSO4. Samples of both cultures were collected for enzyme analysis. β-Galactosidase activity in the induced culture quickly reached levels about six times greater than that in the uninduced control. Solid bars, culture induced with 0.5 mM CuSO4 (Cu); open bars, control culture. “X” indicates the OD600, which was the same for both cultures. M.U., Miller units. (B) ruvA expression after SOS induction with mitomycin C. At an OD600 of 0.1, cultures were divided and inducer was added to one of the cultures. At 100 min, samples were taken for assay. Lane 1, ruv+ control; lane 2, ruvA reporter replacement; lane 3, same as lane 2 but with the reporting cassette in the opposite orientation. Open bars, β-galactosidase activity control; solid bars, induced cultures.
The FLK2 reporter was further tested in a ruvA replacement in strain TP8503. For this purpose, two different strains were constructed, one with lacZ placed so as to be transcribed by the ruvA promoter and the other with lacZ in reverse orientation (TP8503 ruvA 〈 〉 FLK2direct and TP8503 ruvA 〈 〉 FLK2opposite). The expression of the ruvAB operon is normally repressed by LexA unless the SOS response is activated (2). Both TP8503 ruvA 〈 〉 FLK2 strains were grown to an OD600 of 0.1, and an aliquot was treated with 2 μg of mitomycin C/ml in order to induce an SOS response. β-Galactosidase activity was assayed 100 min after induction. Figure 6B shows that the reporter system of the cassette detects a threefold induction of ruvA expression from the cassette positioned so as to be transcribed from PruvA. With the FLK2 cassette in opposite orientation, no β-galactosidase activity was detected with or without mitomycin treatment, thus demonstrating the absence of background expression from the lacZ reporter.
The polar effect of the cassette can be reversed by removal in vivo.
Because the FRT cassettes are expected to have a polar effect, gene replacement will be lethal if downstream genes in the same operon are essential for survival. To overcome this problem we constructed a set of cassettes with a distal Plac-controllable promoter oriented outward (Fig. 2). Using FLKP2, we were able to delete the first two genes of the cell division gene cluster, yabB and yabC, in strain TP8503. Strain TP8503 has the entire lac region deleted, and the absence of lacI results in constitutive expression of Plac from the cassette, thus providing downstream transcription without added IPTG (isopropyl-β-d-thiogalactopyranoside) in the TP8503 yabB ↔ FLKP2 and TP8503 yabC 〈 〉 FLKP2 replacements. However, when these deletions were transduced to MG1655, progeny were not obtained unless the bacteria were cultivated in the presence of 1 mM IPTG. IPTG was no longer required for growth after the FLKP2 cassette was removed by Flp recombinase. This demonstrates (i) that the polar effect of the cassette can be avoided by providing an active Plac and (ii) that the 93-bp scar left after removal of the cassette does not prevent the expression of the downstream genes in an operon. The genes yabB and yabC, of unknown function, have been variously described as dispensable or essential (1, 4, 6). Our results confirm those of Dassain et al. (6) and show that neither ORF is needed provided that the cell division genes immediately downstream can be transcribed.
DISCUSSION
In this paper, we have described a revised gene deletion procedure based on the method reported by Link et al. (11). Several improvements have been made. First, the replacement vectors, pTOF24 and pTOF25, contain a Kmr stuffer fragment which facilitates the screening of recombinants after the crossover PCR cloning step. Second, we have developed and tested the set of FRT cassettes presented in this paper. The use of the FRT-lacZ-aph-Plac-FRT cassette appears to provide advantages for the recovery of deletants and their analysis: (i) the reporter system allows the collection of expression data without further fusion construction and (ii) the Plac promoter overcomes possible polar effects of the cassette.
The main disadvantage of the method is the length of time required (several weeks); the linear replacement methods, in contrast, can be very much faster. Our efforts are now focused on the development of a hybrid method consisting of two sequential linear replacements. In the first step, the gene targeted for deletion would be replaced with a short FRT cassette easily amplifiable by PCR, as already described (7, 20). The second step would be a second in vivo linear replacement of the short cassette by a longer one containing lacZ and Plac, using the fact that both cassettes have extensive sequence identity at their extremities.
Of the reporter cassettes, only FLK2 and FLKP2 have been used so far (Fig. 2). The other FRT cassettes were obtained during the construction process and are listed simply as additional materials that might be of use in particular cases. We do not know if the fact that lacZ and the aph transcription are convergent (in FLK1 and FLKP1) is likely to reduce lacZ expression. This would need to be tested by any user of these constructs. It should be noted, however, that although no terminator has been reported in the original sequence downstream of aph, this sequence contains several inverted repeats that might act to reduce transcription.
Finally, we are also investigating the possible use of the method in bacteria not related to E. coli for which genetic tools are lacking. In such bacteria, the pKO3-based vectors may well not replicate but could be used as suicide vectors. In outline, a replacement plasmid constructed in E. coli would be expected to undergo integration in the chosen host, and integrants would be selected using both the plasmid and the cassette resistance markers. Resolved cointegrants in which plasmid had been lost would be selected on sucrose as for E. coli. The plasmid should be lost quickly after resolution, as it cannot replicate.
Acknowledgments
This work was supported by a Project Grant from the British Biological Sciences Research Council (BBSRC) to M.M.
We thank N. McLennan for his contributions to the earliest stages of this project.
REFERENCES
- 1.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. [DOI] [PubMed] [Google Scholar]
- 2.Benson, F. E., G. T. Illing, G. J. Sharples, and R. G. Lloyd. 1988. Nucleotide sequencing of the ruv region of Escherichia coli K-12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res. 16:1541-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Carrion, M., M. J. Gomez, R. Merchante-Schubert, S. Dongarra, and J. A. Ayala. 1999. mraW, an essential gene at the dcw cluster of Escherichia coli, codes for a cytoplasmic protein with methyltransferase activity. Biochimie 81:879-888. [DOI] [PubMed] [Google Scholar]
- 5.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 6.Dassain, M., A. Leroy, L. Colosetti, S. Carole, and J. P. Bouche. 1999. A new essential gene of the "minimal genome' affecting cell division. Biochimie 81:889-895. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 10.Liang, S. T., P. P. Dennis, and H. Bremer. 1998. Expression of lacZ from the promoter of the Escherichia coli spc operon cloned into vectors carrying the W205 trp-lac fusion. J. Bacteriol. 180:6090-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masters, M., T. Paterson, A. G. Popplewell, T. Owen-Hughes, J. H. Pringle, and K. J. Begg. 1989. The effect of DnaA protein levels and the rate of initiation at oriC on transcription originating in the ftsQ and ftsA genes: in vivo experiments. Mol. Gen. Genet. 216:475-483. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. M. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J. Biol. Chem. 275:31024-31029. [DOI] [PubMed] [Google Scholar]
- 16.Posfai, G., V. Kolisnychenko, Z. Bereczki, and F. R. Blattner. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2. [Online.] http://genomebiology.com/2001/2/9/research/0035./ [DOI] [PMC free article] [PubMed]
- 19.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 20.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]