Abstract
In uropathogenic Escherichia coli, P pili (Pap) facilitate binding to host epithelial cells and subsequent colonization. Whereas P pili can be produced at 37°C, the expression of these fimbriae is suppressed at 23°C. Previously, insertion mutations in rimJ, a gene encoding the N-terminal acetyltransferase of ribosomal protein S5, were shown to disrupt this thermoregulatory response, allowing papBA transcription at low temperature. In this study, we created an in-frame deletion of rimJ. This deletion relieved the repressive effects not only of low temperature but also of rich (Luria-Bertani [LB]) medium and glucose on papBA transcription, indicating that RimJ modulates papBA transcription in response to multiple environmental stimuli. papI transcription was also shown to be regulated by RimJ. papBA transcription is also controlled by a phase variation mechanism. We demonstrated that the regulators necessary to establish a phase ON state—PapI, PapB, Dam, Lrp, and cyclic AMP-CAP-are still required for papBA transcription in a rimJ mutant strain. rimJ mutations increase the rate at which bacteria transition into the phase ON state, indicating that RimJ inhibits the phase OFF→ON transition. A ΔrimJ hns651 mutant is viable on LB medium but not on minimal medium. This synthetic lethality, along with transcriptional analyses, indicates that RimJ and H-NS work through separate pathways to control papBA transcription. Mutations in rimJ do not greatly influence the transcription of the fan, daa, or fim operon, suggesting that RimJ may be a pap-specific regulator. Overexpression of rimJ under conditions repressive for papBA transcription complements the ΔrimJ mutation but has little effect on transcription under activating conditions, indicating that the ability of RimJ to regulate transcription is environmentally controlled.
A variety of environmental signals, including temperature, growth medium, carbon source, osmolarity, pH, oxygen level, and various ions, are known to regulate virulence gene expression in pathogenic bacteria. The expression of several genes is often coordinately regulated by one or more environmental cues (reviewed in references 15 and 36). Presumably, the bacterium uses these stimuli to determine whether it is within a host or, more specifically, to identify a particular environmental niche within the host. This regulation allows for a more efficient utilization of the bacterium's resources and may be necessary for productive colonization of the host.
Pyelonephritis-associated pilus (Pap) expression is regulated by both phase variation and environmental regulatory mechanisms. In many strains of uropathogenic Escherichia coli, Pap expression allows the attachment of bacteria to uroepithelial cells, facilitating colonization of the upper urinary tract (41, 42). Phase variation enables individual bacteria within a given population to alternate between two states of expression: phase ON, in which they are expressing fimbriae, and phase OFF, in which they are not expressing fimbriae (33). Phase variation is controlled at the transcriptional level by the formation of specific DNA methylation patterns of two GATC sites, GATCprox and GATCdist, within the pap regulatory region (6, 9, 52). Formation of these patterns relies upon the global regulators deoxyadenosine methylase (Dam), leucine-responsive regulatory protein (Lrp), and the cyclic AMP (cAMP) receptor protein CAP, as well as the operon-specific proteins PapI and PapB (reviewed in references 30 and 52).
We previously demonstrated that four environmental cues—low temperature, rich (Luria-Bertani [LB]) medium, glucose as a carbon source, and high osmolarity—decrease papBA transcription (6, 57, 60). These environmental cues control several E. coli fimbrial operons, confirming their importance in regulating virulence gene expression (18, 20, 21, 25, 29, 37, 39, 45, 46, 60). For the papBA operon, low temperature causes all cells to transition to a phase OFF state, both phenotypically and at the level of DNA methylation (7, 57). Glucose and high osmolarity decrease the rate at which cells transition into a phase ON state (7, 60).
Two proteins are known to be important in the regulation of papBA transcription in response to environmental conditions, H-NS and RimJ. H-NS is a histone-like nucleoid structuring protein that binds to A-T-rich bent regions of DNA and regulates the expression of a number of environmentally controlled virulence genes (1, 51, 61). Under all growth conditions, papBA transcription is decreased, relative to a wild-type strain, in an hns651 mutant, indicating that H-NS plays a positive role in papBA transcription (54, 57, 60). However, the repression caused by environmental signals is either fully or partially relieved by an hns651 mutation such that transcription approximates levels measured for the mutant under activating conditions (57, 60). Under environmentally repressive conditions, H-NS inhibits the phase OFF→ON transition and can prevent methylation of the pap GATCprox and GATCdist sites at 23°C, but not 37°C (57).
RimJ was initially identified in a thermoregulatory mutant screen in which random chromosomal mini-Tn10 (mTn10) insertions isolated within rimJ allowed papBA transcription at a low temperature (23°C) (58, 59). RimJ is the N-terminal acetyltransferase that modifies the ribosomal protein S5 (16). RimJ, unlike H-NS, is exclusively a negative regulator of papBA transcription: transcriptional levels in rimJ mutants are similar to levels measured in the wild-type strain grown under transcriptionally activating conditions (reference 59 and this study). The mechanism by which RimJ represses papBA transcription and how the modification of a ribosomal protein might be involved in this process are unknown.
In this study, we provide evidence that RimJ controls papBA and papI transcription in response to multiple environmental cues and inhibits the phase OFF→ON transition. In the absence of RimJ, papBA transcription still relies upon the regulators necessary to establish a phase ON state (Lrp, cAMP-CAP, Dam, PapI, and PapB), while our analyses indicate that RimJ and H-NS work in separate pathways to control papBA transcription. RimJ appears to be a pap-specific regulator that does not control other fimbrial operons in response to environmental conditions. Additionally, our experiments indicate that the ability of RimJ to control transcription is environmentally regulated.
MATERIALS AND METHODS
Strains and media.
The strains, plasmids, and bacteriophages used in this study are shown in Table 1. Media and antibiotics were prepared as described previously (38, 47, 60).
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Strain, plasmid, or bacteriophage | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| MC4100 | F−araD139 Δ(lacIPOZYA-argF)U169 rpsL thi-1 | 11 |
| NH757 | B178 hns651 tyrTβ::Tn10 | 22 |
| DL479 | MC4100 λ246 lysogen (papBA-lacZYA) rimJ-2::mTn10 | 58 |
| DL812 | MC4100 λMW01 lysogen (fanABC′-lacZYA) | 53 |
| DL1504 | MC4100 λ354 lysogen (papBA-lacZYA) | 9 |
| DL1509 | DL1504 rimJ-2::mTn10 | This work |
| DL1530 | MC4100 λ366 lysogen (daa-lacZYA) | 55 |
| DL1910 | DL1504 Δlrp | 54 |
| DL1947 | DL1504 hns651 | 54 |
| DL2208 | MC4100 λ354-15 lysogen (papBA-lacZYA lysogen with ATG start codon of papB changed to CTG) | 57 |
| DL2838 | MC4100 λ491 lysogen (papI-lacZYA) | 57 |
| DL3052 | MC4100 λ354-73 lysogen (papBA-lacZYA lysogen containing papI frameshift mutation) | 57 |
| DL2873 | DL1910 rimJ-2::mTn10 | This work |
| DL3089 | DL1504 Δcrp-45 zhd-3083::Tn10 | D. A. Low |
| AAEC198A | MG1655 ΔlacZYA fimA-lacZYA | 5 |
| CWZ381 | DL812 rimJ-2::mTn10 | This work |
| CWZ382 | DL1530 rimJ-2::mTn10 | This work |
| CWZ387 | MC4100 ΔrimJ | This work |
| CWZ388 | DL1504 ΔrimJ | This work |
| CWZ395 | CWZ388 containing pCWZ101 and pMV101 | This work |
| CWZ400 | AAEC198A rimJ-2::mTn10 | This work |
| CWZ403 | CWZ388 hns651 | This work |
| CWZ405 | CWZ388 Δcrp-45 zhd-3083::Tn10 | This work |
| CWZ406 | CWZ387 hns651 | This work |
| CWZ410 | DL1504 dam-13::Tn9 | This work |
| CWZ411 | DL1509 dam-13::Tn9 | This work |
| CWZ412 | DL2838 rimJ-2::mTn10 | This work |
| CWZ418 | CWZ387 λ354-73 lysogen | This work |
| CWZ419 | CWZ387 λ354-15 lysogen | This work |
| Bacteriophages | ||
| P1L4 | Virulent phage P1 | D. A. Low |
| λ491 | papI-lacZYA fusion phage | 57 |
| λ354 | papBA-lacZYA fusion phage | 9 |
| λ354-15 | papBA-lacZYA lysogen with ATG start codon of papB changed to CTG | 57 |
| λ354-73 | papBA-lacZYA lysogen containing papI frameshift mutation | 57 |
| λ366 | daa-lacZYA fusion phage | 55 |
| λMW01 | fanABC′-lacZYA fusion phage | 10, 53 |
| Plasmids | ||
| pUHS*2 Pzl-2 | ColE1 replicon containing the Plac/ara-1 promoter | 35 |
| pMV101 | pMC9 derivative containing lacIq that is Aps Tcr | 14 |
| pMV106 | pUHS*2 Pzl-2 with replacement of Knr with Apr | 14 |
| pKO3 | pSC101 replicon containing repA (Ts) replication origin, sacB, and Cmr | 32 |
| pCWZ100 | pKO3 containing ΔrimJ deletion | This work |
| pCWZ101 | pMV106 containing rimJ under Plac/ara-1 promoter | This work |
Construction of mutant strains by P1 transduction.
The preparation of P1 lysates and P1 transductions were carried out as described previously (47, 60). rimJ-2::mTn10, dam-13::Tn9, Δcrp-45, and hns651mutant strains were created by P1 transduction of the individual mutations into the appropriate recipient strain (Table 1).
UV induction and lysogenization of UV-induced phage.
UV induction and lysogenization were performed as described previously (47, 58). UV induction was performed on DL3052 and DL2208, with the resulting phage lysates used to lysogenize CWZ387, creating CWZ418 and CWZ419, respectively (Table 1).
Construction of ΔrimJ strain.
Crossover PCR was used to create an internal, in-frame deletion within rimJ by the method of Link et al. (32). Primers rimJ(A), 5′-CGCGGATCCGGCGATACCCATTGTGGC-3′, and rimJ(B), 5′-CCCATCCACTAAACTTAAACAACTGCGATAGCCAAACAT-3′, were used to generate a 573-bp upstream fragment, and primers rimJ(C), 5′-TGTTTAAGTTTAGTGGATGGGGCATTAACTACCCCAGAC-3′, and rimJ(D), 5′-CGCGGATCCCGCGTTTACCCGGTTCGC-3, were used to generate a downstream 556-bp fragment. The two PCR products were combined in a secondary PCR using primers rimJ(A) and rimJ(D) for amplification. The BamHI-SalI-digested PCR product was cloned into BamHI-SalI-digested pKO3 to create pCWZ100 (Table 1).
pCWZ100 was transformed (12) into DL1504, and the selection for integration of the ΔrimJ deletion onto the chromosome was performed as described previously, with the exception that the incubation on sucrose was completed at 23 rather than 30°C (32). Colony PCR was used to detect clones in which the amplification of the rimJ region showed the expected decrease in size. In the resulting ΔrimJ strain, CWZ388 (Table 1), the region overlapping the deletion was sequenced to confirm the correct replacement. Steps identical to those described above were followed to construct CWZ387 (Table 1).
Construction of pCWZ101 for overexpression of RimJ.
rimJ was amplified from wild-type DL1504 chromosomal DNA using primers 5′-CGGAATTCGCGTATTAAAGACGTTAC-3′ and 5′-GCTCTAGACAAGGGCAGTAAGTTGAT-3′. The amplified fragment and pMV106 were each digested with EcoRI and BamHI and subsequently ligated to create pCWZ101 (Table 1). pCWZ101 and pMV101, containing the lacIq gene, were cotransformed (12) into CWZ388 to yield strain CWZ395 (Table 1).
Growth conditions.
Media (M9 glyc, M9 gluc, M9 NaCl, and LB) were prepared as described previously (60). For growth conditions that are activating for papBA transcription, the bacteria were cultured in 10 ml of M9 glyc at 37°C. Low temperature was tested by growing the bacteria at 23°C in M9 glyc, whereas rich medium was tested by growth of bacteria in LB broth at 37°C. Cultures grown at 37°C in M9 gluc or M9 NaCl medium were used to measure the effect of a change in carbon source and osmolarity, respectively. Glucose was substituted for glycerol in the M9 minimal medium (M9 gluc). The sodium chloride concentration was increased by 300 mM (M9 NaCl) compared to 8.5 mM sodium chloride in the M9 glyc medium to test osmolarity.
Culture inoculation and measurement of β-galactosidase activity.
For the assays determining the effects of environmental stimuli on fimbrial transcription, each bacterial culture was inoculated as described previously (60). To assess the effect of rimJ overexpression in CWZ395, two phase ON (Lac+) colonies were excised from M9 glyc agar at 37°C and resuspended in 2 ml of M9 salts. Flasks containing 10 ml of the prewarmed medium (M9 glyc or LB) with the appropriate concentration of isopropyl-β-d-thiogalactopyranoside (IPTG) were inoculated with 140 μl of the colony suspension. These inoculation methods ensured that all the bacterial cultures had grown for approximately 9 to 11 generations under the new conditions prior to the measurement of β-galactosidase activity. The bacterial cultures were grown to exponential phase (optical density at 600 nm, 0.25 to 0.9), and β-galactosidase activities were measured as described previously (38). All the values for the β-galactosidase activities represent averages from two or more independent cultures grown under identical conditions.
Calculation of switch frequencies.
Phase transition rates were calculated as described previously (7, 60). Weighted averages were calculated from at least two independent analyses and are given as the number of events per cell per generation.
RESULTS
An in-frame deletion of rimJ causes a loss of papBA thermoregulation.
An in-frame deletion of rimJ was created to analyze the effect of a total loss of the RimJ protein on papBA transcription. The two originally characterized rimJ::mTn10 mutations, previously designated tcp, for thermoregulatory control of pap, are insertions within the 3′ end of rimJ, and minicell analysis demonstrated that for both insertions a fusion protein was expressed (59). Thus, the effect of the rimJ::mTn10 mutations on papBA thermoregulation could be due to either a total loss of RimJ activity in these mutants or an alteration in the levels or specificity of RimJ activity. In addition, because all of the previously mapped insertions in rimJ from our laboratory and others were in the C terminus, the possibility was raised that rimJ might be an essential gene (59, 63).
A deletion of rimJ was created by crossover PCR, cloned into the allelic exchange vector pKO3, and recombined onto the chromosome of DL1504, producing the ΔrimJ strain CWZ388 (Table 1). In the ΔrimJ mutation, the DNA sequences for the first 5 and the last 11 amino acids of RimJ are retained while the internal 179 amino acids of RimJ are replaced by an insertion that encodes 8 amino acids. The insertion is in frame, preventing any polar effects on two downstream genes of unknown function that appear to be in the same operon as rimJ.
The ΔrimJ deletion strain CWZ388 showed a phase variation phenotype at both 37 and 23°C on M9 glyc, demonstrating that the ΔrimJ mutation disrupts thermoregulation, similar to the rimJ::mTn10 insertions characterized previously (58) (data not shown). Our results also demonstrate that rimJ is not essential. Acetylation of S5 is not required for cell growth, as the ΔrimJ strain displays a growth rate similar to that of the wild-type strain DL1504 under all of the environmental conditions tested in this study (data not shown).
RimJ represses papBA transcription in response to multiple environmental cues.
To determine if RimJ controlled papBA transcription in response to environmental cues other than temperature, β-galactosidase activity was measured in the wild-type strain DL1504 and the rimJ mutant strains, CWZ388 (ΔrimJ) and DL1509 (rimJ-2::mTn10), under differing environmental conditions. A phase ON (Lac+) colony was used to inoculate each culture, ensuring that transcriptionally active cells were used to initiate the culture. Within a Lac+ colony, 20 to 50% of cells are in a phase ON state (data not shown).
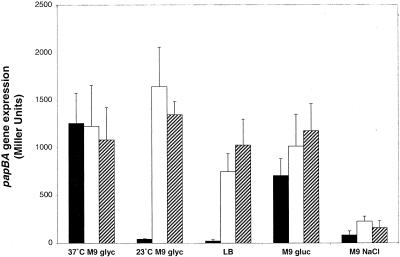
In the wild-type strain, papBA transcription is decreased by low temperature, LB medium, glucose as a carbon source, and high osmolarity compared to the activating conditions of M9 glyc at 37°C (Fig. 1) (60). The ΔrimJ mutation and the rimJ-2::mTn10 mutations relieve the repression due to low temperature and LB medium such that papBA transcriptional levels under these normally repressive conditions are similar to levels observed at 37°C in M9 glyc (Fig. 1). While we previously reported a greater reduction due to glucose (60), more recent experiments indicate that papBA transcription is decreased approximately 1.8-fold, similar to the 3.4-fold reduction measured by Båga et al. (2). Both rimJ mutations increased transcription in glucose to levels greater than that seen in M9 glyc at 37°C (Fig. 1). These results extend the function of RimJ beyond that of a thermoregulator, as RimJ responds to multiple environmental cues to control papBA transcription.
FIG. 1.
Effects of environmental stimuli on pap fimbrial transcription in wild-type and rimJ mutant strains. The bars indicate β-galactosidase activities measured in the wild-type strain DL1504 (solid bars), in strain CWZ388 containing the ΔrimJ mutation (open bars), and in strain DL1509 containing the rimJ-2::mTn10 mutation (hatched bars). β-Galactosidase activity was measured as described in Materials and Methods. Error is expressed as 1 standard deviation from the mean.
We note that RimJ is not a major regulator in response to osmolarity. While the levels of papBA transcription are slightly elevated in the ΔrimJ and the rimJ-2::mTn10 mutant strains grown in M9 NaCl compared to the wild-type strain, high osmolarity still has a repressive effect on papBA transcription in the mutant strains (Fig. 1).
RimJ controls papI transcription.
The PapI regulatory protein is necessary to establish the phase ON state and activate papBA transcription (8). papI is transcribed on a monocistronic operon, divergent from papBA (26, 57). To determine if RimJ also controls papI transcription, the rimJ-2::mTn10 mutation was transduced into DL2838, which contains a papI-lacZYA fusion, creating strain CWZ412 (Table 1). Overall levels of papI transcription were decreased in the wild-type strain DL2838 at 23 (11 ± 4 Miller units [MU] [38]) compared to 37°C (203 ± 35 MU), whereas in CWZ412, papI transcription levels were similar at 37 (157 ± 9 MU) and 23°C (120 ± 14 MU). Similar to papBA transcription, LB medium decreased papI transcription in the wild-type strain (3 ± 0 MU). The rimJ-2::mTn10 mutation increased papI transcription (28 ± 5 MU) but did not restore it to the levels seen in M9 glyc. Glucose did not greatly alter papI transcription in the wild-type (218 ± 10 MU) or the rimJ-2::mTn10 mutant (113 ± 19 MU) strain.
Maximal papBA transcription in a rimJ mutant strain requires PapI, PapB, Lrp, Dam, and cAMP-CAP.
In order to establish a phase ON state for papBA transcription, PapI, Lrp, cAMP-CAP, and Dam are required, while PapB plays primarily an indirect role in phase variation, that of activating papI transcription (reviewed in reference 30). To determine if these same regulators are still required for papBA transcription in the absence of RimJ, a rimJ mutation was tested for its effect on transcription in the absence of each individual regulator. Regardless of whether RimJ was present or absent, no phase variation was seen in strains lacking PapI, Lrp, or Dam, and transcription measured at 37 or 23°C was low (Table 2), indicating that these regulators are still required to initiate transcription in the absence of RimJ. In CWZ419 lacking PapB and RimJ, a phase variation phenotype was observed at 37°C in which Lac+ colonies displayed a pale-blue phenotype and only Lac− colonies were observed at 23°C. The phase ON cells observed in the papB ΔrimJ mutant might result from increased papI transcription due to the rimJ mutation. However, the level of papI transcription in the absence of PapB activation must not be equivalent to that in the ΔrimJ strain CWZ388, as overall papBA transcription levels are minimal (Table 2).
TABLE 2.
Effects of a rimJ mutation on papBA transcription in various mutant strains
| Strain | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|
| 37°C M9 glyc | 23°C M9 glyc | ||
| DL3052b | papI frameshift mutation | 9 ± 0 | 6 ± 0 |
| CWZ418 | papI frameshift ΔrimJ | 23 ± 1 | 11 ± 1 |
| DL2208b | papB CTG start codon mutation | 4 ± 0 | 2 ± 0 |
| CWZ419 | papB CTG start codon ΔrimJ | 35 ± 4 | 5 ± 0 |
| DL1910b | Δlrp | 4 ± 2 | 3 ± 2 |
| DL2873 | Δlrp rimJ-2::mTn10 | 3 ± 1 | 3 ± 0 |
| CWZ410 | dam-13::Tn9 | 73 ± 2 | 11 ± 0 |
| CWZ411 | dam-13::Tn9 rimJ-2::mTn10 | 68 ± 5 | 12 ± 1 |
A Δcrp-45 ΔrimJ strain was viable on LB medium but was unable to grow on M9 glyc or M9 gluc agar, indicating that the absence of both proteins was deleterious for growth on minimal medium. Measurement of transcription in the wild-type strain DL1504 in LB medium yielded 8 ± 0 MU due to the repression of LB medium on papBA transcription, while transcription was elevated to 71 ± 5 MU in the ΔrimJ strain CWZ388. The double-mutant strain CWZ403 (ΔrimJ Δcrp-45) displayed a level of papBA transcription identical to that of the Δcrp-45 strain DL3089 (5 ± 0 MU), demonstrating that papBA transcription remains cAMP-CAP dependent in the absence of RimJ. Taken together, these data indicate that all of the regulators necessary to attain a phase ON state are still required in the absence of RimJ.
RimJ inhibits the transitioning of cells to the phase ON state.
Phase transition rates were calculated to determine if the loss of repression due to the rimJ mutations could be attributed to alterations in switch frequencies. While LB medium results in repression of papBA transcription, all colonies display a uniform colony phenotype on LB medium, and phase transition rates could not be calculated.
For the wild-type and rimJ mutant strains, the phase ON→OFF rates on M9 gluc and M9 NaCl are similar to the rates calculated on M9 glyc at 37°C, indicating that the carbon source, high osmolarity, and the rimJ mutations do not greatly influence the rate at which cells transition to the phase OFF state (Table 3) (60). At a low temperature for the wild-type strain, all colonies display a phase OFF colony phenotype, correlating with a phase OFF DNA methylation state (57). The rate at which cells transition from the phase ON to a phase OFF state at 23°C in the rimJ mutants is similar to the rate on M9 glyc at 37°C. These results indicate that the absence of RimJ removes the temperature repression of phase variation, but once it is removed, cells transition to a phase OFF state at a rate similar to that under the other conditions tested.
TABLE 3.
Effects of ΔrimJ and rimJ-2::mTn10 mutations on phase transition rates for papBA operon under different environmental conditions
| Phase | Environmental conditionsa | Phase transition rateb
|
||
|---|---|---|---|---|
| Wild typec | ΔrimJ | rimJ-2::mTn10 | ||
| ON→OFF | 37°C M9 glyc | 3.37 × 10−2 | 2.60 × 10−2 | 2.20 × 10−2 |
| 23°C M9 glyc | NA | 3.97 × 10−2 | 3.68 × 10−2 | |
| M9 gluc | 4.40 × 10−2 | 1.95 × 10−2 | 4.04 × 10−2 | |
| M9 NaCl | 4.23 × 10−2 | 3.99 × 10−2 | 3.89 × 10−2 | |
| OFF→ON | 37°C M9 glyc | 3.50 × 10−4 | 1.43 × 10−3 | 8.40 × 10−4 |
| 23°C M9 glyc | NA | 2.55 × 10−5 | 6.14 × 10−5 | |
| M9 gluc | ND | 8.26 × 10−5 | 1.11 × 10−4 | |
| M9 NaCl | 1.59 × 10−4 | 7.28 × 10−5 | 4.97 × 10−5 | |
The same growth medium was used for the isolation of the initial colony (Lac+ or Lac−) and for the subsequent quantitation of phase transition rates from the initial colony.
Phase transition rates were measured in the wild-type strain DL1504, ΔrimJ mutant strain CWZ388, and rimJ-2::mTn10 mutant strain DL1509. The weighted averages were calculated from at least two independent analyses as described by Blyn et al (7). The frequencies are given as the number of events per cell per generation. The phase transition rates for DL1504 were previously published (60).
NA, not applicable; wild-type strain DL1504 does not undergo phase variation at low temperature. ND, not determined; a weighted average could not be calculated for DL1504, as no Lac+ colonies were observed in a screening of approximately 37,000 colonies from four independent analyses. An earlier study using a similar, but not identical, papBA-lacZYA transcriptional fusion yielded a phase transition frequency of 4.51 × 10−6/cell/generation (7).
In contrast, the phase OFF→ON transition rates for the papBA operon are influenced by the rimJ mutations and environmental conditions (Table 3). At 37°C on M9 glyc, the phase OFF→ON transition rates are increased in the ΔrimJ and rimJ-2::mTn10 mutant strains compared to the wild-type strain, demonstrating that the rimJ mutations increase the phase OFF→ON rate in the absence of an environmental change. The ΔrimJ and rimJ-2::mTn10 mutations allow cells to transition to the phase ON state at a low temperature and increase the phase OFF→ON transition rates on glucose compared to the wild-type strain (Table 3). While the phase OFF→ON rates are significantly increased over the wild-type rates under these conditions, they are reduced compared to the rates observed at 37°C on M9 glyc, indicating that the stimuli of low temperature and carbon source still retain a partial repressive effect on the phase OFF→ON transition rate in the absence of RimJ. This partial repression may be mediated by H-NS, which also inhibits the phase OFF→ON transition rate (60). High osmolarity also inhibits the rate at which cells transition to a phase ON state in the wild-type strain DL1504 (Table 2). Unlike the other conditions tested, the phase OFF→ON transition rates are further decreased in the rimJ mutant strains when they are grown on M9 NaCl (Table 3), in agreement with the transcriptional analyses, in which the rimJ mutations do not relieve the repression due to high osmolarity.
RimJ and H-NS control papBA transcription through separate pathways.
While the rimJ and hns651 mutant strains differ in many ways, both H-NS and RimJ have been shown to control papBA transcription in response to multiple environmental signals and share the common function of inhibiting the phase OFF→ON transition rate (57, 60), raising the question of whether RimJ and H-NS work through the same or separate regulatory pathways to control papBA transcription.
A ΔrimJ hns651 double mutant strain, CWZ403, was constructed that was viable on LB medium but was unable to grow on M9 glyc or M9 gluc agar, indicating that the absence of both proteins was deleterious for growth on minimal medium. This phenotype is independent of papBA gene expression, as the same lethality was seen in the ΔrimJ hns651 mutant strain CWZ387, which does not contain the papBA-lacZYA transcriptional fusion (data not shown). On M9 glyc, the hns651 mutant strain DL1947 has a significantly decreased growth rate and displays a mucoid phenotype, whereas the ΔrimJ mutant strain CWZ388 is indistinguishable from the wild-type strain DL1504 in growth rate and colony morphology. On LB medium, the ΔrimJ hns651 double-mutant strain grows more slowly than the wild-type or ΔrimJ strain, similar to the hns651 mutant strain DL1947. The lethality of the double-mutant strain on minimal medium indicates that RimJ and H-NS work through parallel pathways, since an additional change in phenotype in the ΔrimJ hns651 mutant strain, relative to the single mutants, would not be expected if both regulators were in the same pathway.
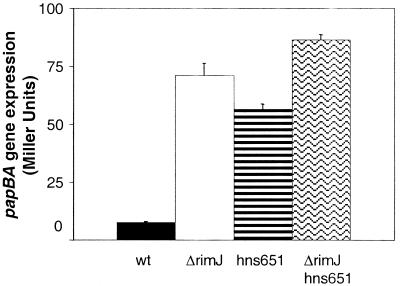
Due to the loss of viability of the double-mutant strain on M9 glyc, papBA transcription was measured after growth in LB medium at 37°C. In both of the single-mutant strains, CWZ388 (ΔrimJ) and DL1947 (hns651), the repression due to LB medium was relieved by the individual mutations compared to the wild-type strain DL1504 (Fig. 2). Overall transcription in the ΔrimJ strain was not as high as previously measured in cultures initiated from a Lac+ colony (Fig. 1). Because the strains in this experiment were initially streaked on LB medium, it could not be determined if cultures were started with a phase ON (Lac+) or phase OFF (Lac−) colony, possibly accounting for the lower level of papBA transcription. Plating of LB medium-grown colonies for CWZ388 onto M9 glyc showed an average of 5% of cells in the phase ON state, correlating with the low transcription measured in these cultures. In the hns651 strain, only a very low percentage of phase ON cells (2%) were observed even under transcriptionally activating conditions (54). In the ΔrimJ hns651 double mutant, papBA transcription was elevated to levels slightly higher than those with either mutation alone (Fig. 2). Using a nested analysis of variation, the differences in β-galactosidase activity due to the genotype of the strain were found to be statistically significant, supporting the conclusion that RimJ and H-NS work through different pathways.
FIG. 2.
Effects of the ΔrimJ and hns651 mutations on papBA transcription. The bars indicate β-galactosidase activities measured in the wild-type (wt) strain DL1504, in the ΔrimJ mutant strain CWZ388, in the hns651 mutant strain DL1947, and in the ΔrimJ hns651 double-mutant strain CWZ403. β-Galactosidase activity was measured as described in Materials and Methods. Error is expressed as 1 standard deviation from the mean.
RimJ does not control transcription of the fan, daa, or fim operon in response to environmental conditions.
The fan, daa, and fim fimbrial operons share common regulators and regulatory mechanisms with pap, leading us to hypothesize that RimJ might control their transcription. Transcription of all three fimbriae is regulated by Lrp and H-NS (5, 10, 25, 60). Transcription of the daa operon that encodes F1845 fimbriae (3) is controlled by a methylation-dependent phase variation mechanism similar to pap (55), whereas transcription of the fim operon encoding type I fimbriae (4, 13, 28) relies upon an invertible promoter phase variation mechanism (18, 43, 44). Transcription of the fan operon encoding K99 fimbriae (24) is not known to be subject to phase variation.
Previously, we showed that fan and daa transcription is repressed by the same environmental cues as that of pap—low temperature, LB medium, glucose as a carbon source, and high osmolarity (Table 4) (60). In this study, we demonstrate that fim transcription is also reduced by growth at a low temperature and in LB medium, in agreement with other studies showing that temperature and medium influence the rate at which cells transition to a phase OFF state (25) (Table 3).
TABLE 4.
Effects of the rimJ-2::mTn10 mutation on fimbrial transcription of the fan, daa, and fim operons
| Operon fusion | Relevant genotype | β-Galactosidase activitya
|
||
|---|---|---|---|---|
| 37°C M9 glyc | 23°C M9 glyc | LB | ||
| fanABC′-lacZYA | Wild type | 8,163 ± 1,526 | 95 ± 6 | 1,538 ± 188 |
| rimJ-2::mTn10 | 4724 ± 440 | 92 ± 39 | 1,977 ± 706 | |
| daa-lacZYA | Wild type | 72 ± 14 | 45 ± 4 | 7 ± 1 |
| rimJ-2::mTn10 | 36 ± 13 | 38 ± 6 | 10 ± 6 | |
| fimA-lacZYAb | Wild type | 2,271 ± 273 | 716 ± 195 | 69 ± 10 |
| rimJ-2::mTn10 | 2,036 ± 239 | 847 ± 194 | 85 ± 7 | |
β-Galactosidase activity is expressed as Miller units (38) and was measured as described in Materials and Methods. Error is expressed as ±1 standard deviation from the mean.
Strains DL812 (wild type) and CWZ381 (rimJ-2::mTn10) were used to analyze fan transcription, DL1530 (wild type) and CWZ382 (rimJ-2::mTn10) were used for daa transcription, and AAEC198A (wild type) and CWZ400 (rimJ-2::mTn10) were used for fim transcription.
For all three operons, the rimJ-2::mTn10 mutation did not relieve repression due to low temperature or growth in LB medium. The level of fan transcription in the rimJ-2::mTn10 mutant strain CWZ381 was similar to that of the wild-type strain DL812 under these conditions (Table 4). Similarly, the transcription of the daa operon in CWZ382 was not altered by introduction of the rimJ-2::mTn10 mutation, nor was the transcription of fim in CWZ400 (Table 4). Taken together, these data suggest that RimJ may be a pap-specific regulator in response to environmental conditions.
Overexpression of rimJ complements the ΔrimJ mutation at 23°C or in LB medium but does not repress papBA transcription at 37°C in M9 glyc.
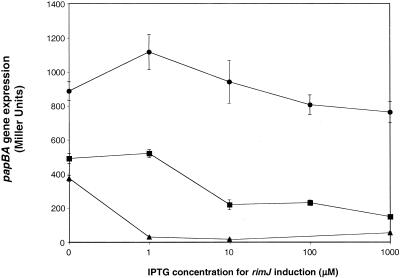
To analyze the effect of rimJ overexpression on papBA transcription, rimJ was cloned under the control of the Plac/ara-1 promoter in pCWZ101 (Table 1). At 23°C without the addition of IPTG, papBA transcription was reduced 2.4-fold compared to the level seen at 37°, indicating that some transcription of rimJ occurs in the absence of IPTG induction and that this low level partially complements the ΔrimJ mutation on the chromosome of CWZ395 (Fig. 3). At IPTG levels of 10 to 1,000 μM, papBA transcription was reduced to levels similar to those of the wild-type strain DL1504 at low temperature, demonstrating that pCWZ101 is able to fully complement the ΔrimJ mutation on the chromosome in M9 glyc at 23°C. At 37°C, papBA transcriptional levels were similar to the initial measurement made at 37°C in the absence of IPTG (Fig. 3). Thus, the overexpression of rimJ does not repress papBA transcription at 37°C in M9 glyc. Similar to the results seen at low temperature in M9 glyc, papBA transcription in LB medium at 37°C was reduced 1.8-fold in the absence of IPTG induction (Fig. 3). At increasing concentrations of IPTG (1 to 1,000 μM), the overexpression of rimJ in LB medium repressed papBA transcription. The complementation in this case was not as complete as that observed at 23°C in M9 glyc, as papBA transcription did not decrease to the levels measured for the wild-type strain in LB medium (Fig. 1).
FIG. 3.
Effect of increasing levels of rimJ on papBA transcription. The strain CWZ395 containing pCWZ101 (rimJ under the control of the Plac/ara-1 promoter) and pMV101 (lacIq) was used in this experiment. IPTG was added at concentrations ranging from 0 to 1,000 μM as indicated to induce expression of rimJ. The data points indicate β-galactosidase activities measured after growth in M9 glyc at 37°C (circles), LB at 37°C (squares), and M9 glyc at 23°C (triangles). β-Galactosidase activity was measured as described in Materials and Methods. Error is expressed as ±1 standard deviation from the mean.
DISCUSSION
In this study, we provide evidence that the function of RimJ extends beyond that of a thermoregulator. RimJ is involved in cellular response to other environmental cues, including growth (LB) medium and glucose as a carbon source. We can envision two different, and not necessarily mutually exclusive, models for the role of RimJ in decreasing papBA transcription in response to these signals. In one model, RimJ alters phase variation frequencies by decreasing the rate at which cells transition to a phase ON state and/or increasing the rate at which cells transition to a phase OFF state. Alternatively, RimJ may act to inhibit papBA transcription by a mechanism independent of phase variation.
In accordance with the first model, our results demonstrate that RimJ inhibits the transition of cells into the phase ON state. This effect on the transition rate may be the only mechanism required to account for the effect of RimJ on papBA transcription when glucose is provided as the sole carbon source. Because glucose does not alter the ON→ OFF rate (Table 3) (60), we postulate that the only effect of glucose is to prevent cells that are phase OFF from transitioning into the phase ON state due to limiting cAMP-CAP. Phase ON cells used to initiate the culture maintain a transcriptionally active state after transfer into M9 gluc, but in the absence of a mechanism to recruit new cells into the phase ON state, papBA transcription gradually decreases (Fig. 1). Growth of cultures for longer periods in M9 gluc show further reduction in overall papBA transcription and the percentage of cells in the phase ON state, consistent with this model (data not shown). The increase in papBA transcription in the rimJ mutants, relative to the wild-type strain, results from an increased frequency of cells switching to the phase ON state. It is a paradox how phase OFF cells in a rimJ mutant strain attain a phase ON state in glucose, since cAMP-CAP should be limiting. We note that the phase transition rates measured in glucose for the rimJ mutants are greater than those measured in the wild type in M9 gluc but are significantly decreased compared to the wild type and rimJ mutants in glycerol.
In contrast to glucose, low temperature and LB medium cause a more dramatic reduction in papBA transcription, suggesting that RimJ may play an additional role unrelated to inhibiting the phase OFF→ON transition. At low temperature, all of the cells transition to a phase OFF state in the wild-type strain. If temperature regulation were dependent only upon phase variation, it would suggest that at low temperature RimJ both increases the phase ON→OFF rate and decreases the phase OFF→ON rate. Yet in the rimJ mutant strains, the ON→OFF rates at 23°C on M9 glyc are basically unchanged relative to the wild-type strain at 37°C, arguing that RimJ does not function by simply altering this transition rate. Previous temperature downshift experiments show that papBA transcription is rapidly repressed within 1 generation of growth at 23°C while approximately 20% of the cells are still in the phase ON state based on analysis of the DNA methylation states (57). Thus, RimJ may have an additional role in the rapid repression of papBA transcription prior to transition to the phase OFF methylation state. Additional experiments are being pursued to understand the interrelationship between environmental regulation and phase variation, particularly in response to LB medium.
It is not known whether the acetyltransferase activity of RimJ is necessary for the regulation of papBA transcription, although the evidence presented here is suggestive. The two sequenced rimJ::mTn10 insertions are inserted between motifs A and B (59), motifs conserved in the N-acetyltransferase superfamily and that encompass the acetyl-coenzyme A binding site (19, 34, 40, 49). Minicell analysis demonstrated that fusion proteins of RimJ with the mTn10 elements are expressed (59), suggesting that it may be the disruption of the acetyl-coenzyme A site and not loss of the entire protein that leads to the loss of papBA repression. This conclusion is further supported by the observation that the rimJ-2::mTn10 and ΔrimJ mutations have similar effects on papBA transcription and phase variation. A search using only the N-terminal portion of RimJ did not detect homology to any known conserved domain: no other known function can, at present, be attributed to RimJ.
RimJ may be acting indirectly by altering the quantity of a regulatory protein or directly by modifying a protein involved in papBA transcription and influencing its activity. While it has been shown that RimJ is highly specific for its ribosomal substrate, S5, RimJ may have additional nonribosomal substrates (27, 63). Given that H-NS controls transcription of the papBA operon and that studies have indicated that H-NS is posttranslationally modified (17, 51), one possible model argues that RimJ acetylates H-NS, modulating its activity under varying environmental conditions. Our results are not consistent with this conclusion but rather indicate that RimJ and H-NS function in separate pathways. In addition, RimJ does not alter H-NS levels, as these levels remain unchanged at 23 and 37°C (57). While alternative substrates for RimJ must be considered, it is possible that the acetylation of S5 determines whether full-length papBA transcription is completed only under the activating, but not the repressive, conditions. In addition to their well-known structural roles, the ribosomal proteins S4, S10, and L4 also play roles as transcriptional antiterminators (23, 31, 48, 50, 56, 62). S5 may play a similar dual role. Studies in our laboratory are aimed at determining RimJ substrate specificity and the importance of the acetylase activity for the repression of papBA transcription.
With these ideas in mind, it is intriguing to consider how RimJ responds to environmental conditions to repress papBA transcription. Transcription of rimJ may itself be modulated by environmental signals, but this simple mechanism is undercut by the rimJ overexpression results. An inducible promoter is used in this experiment, making rimJ transcription unresponsive to environmental conditions. Nevertheless, increasing rimJ mRNA levels under activating conditions is insufficient to repress papBA transcription, indicating that functionally active RimJ protein, capable of decreasing papBA transcription, is not being produced in M9 glyc at 37°C. Thus, alternative hypotheses must be proposed for how the function of RimJ is sensitive to environmental conditions. It is possible that rimJ mRNA stability, RimJ protein stability, or RimJ translation is environmentally controlled, thus ensuring production of RimJ only under repressive conditions. Alternatively, it may be that RimJ protein is equally expressed under all growth conditions but that it is only active under repressing environmental conditions. Lastly, it may be that the substrate of RimJ is itself regulated by environmental conditions. Our preliminary experiments rule out the environmental modulation of at least one important RimJ substrate: S5 is present under all of the conditions tested, and S5 expression levels are not altered by the ΔrimJ mutation (data not shown).
From our overexpression data, we note that the function of RimJ can be modulated by multiple environmental cues and that the presence of a single repressive cue will determine the activity of RimJ. When grown in M9 glyc, the ability of RimJ to repress papBA transcription is temperature dependent. However, when the medium is changed to LB, RimJ is able to repress papBA transcription even at a higher growth temperature. Thus, while one stimulus is activating (temperature), the other stimulus is repressive (growth medium), and the repressive stimulus dictates the activity of RimJ. This represents an efficient mechanism for regulation that may be important in vivo, where it may be necessary to control virulence gene expression based on multiple environmental cues.
Taken together, our investigations of RimJ demonstrate its importance for regulating the expression of papBA expression in response to multiple environmental cues. Environmental cues play an integral role in regulating virulence gene expression that may impact a pathogen's ability to colonize a host and its survival in external environments. Consequently, RimJ may play a significant role in the adaptation of uropathogenic E. coli to changing environments.
Acknowledgments
We thank Marjan van der Woude, David Low, George Church, and Ian Blomfield for generous gifts of bacterial strains and plasmids. In addition, we thank Marjan van der Woude and Rob Dorit for critical reading of the manuscript. We also thank the Smith College students who have provided technical assistance on this project, including Olivia Carrick, Jennifer Hoot, Stephanie Jakus, Michelle Ploutz, and Angela Rasmussen.
This work was supported by National Institutes of Health grant GM62792 to Christine White-Ziegler, by the Albert F. Blakeslee Trust, and by Smith College.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Båga, M., M. Göransson, S. Normark, and B. E. Uhlin. 1985. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 4:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch, C. A., B. A. Stocker, and P. E. Orndorff. 1992. A key role for type 1 pili in enterobacterial communicability. Mol. Microbiol. 6:697-701. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. McClain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential Dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. H. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, B. A., L. B. Blyn, B. S. Skinner, and D. A. Low. 1991. Evidence for a methylation-blocking factor (mbf) locus involved in pap pilus expression and phase variation in Escherichia coli. J. Bacteriol. 173:1789-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, B. A., X. Nou, L. S. Kaltenbach, and D. A. Low. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577-588. [DOI] [PubMed] [Google Scholar]
- 10.Braaten, B. A., J. V. Platko, M. W. van der Woude, B. H. Simons, F. K. DeGraaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 12.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connell, H., L. K. Poulsen, and P. Klemm. 2000. Expression of type 1 and P fimbriae in situ and localisation of a uropathogenic Escherichia coli strain in the murine bladder and kidney. Int. J. Med Microbiol 290:587-597. [DOI] [PubMed] [Google Scholar]
- 14.Correnti, J., V. Munster, T. Chan, and M. van der Woude. 2002. Dam-dependent phase variation of Ag43 in E. coli is altered in a seqA mutant. Mol. Microbiol. 44:521-532. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. A., and J. F. Miller. 1998. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr. Opin. Microbiol. 1:17-26. [DOI] [PubMed] [Google Scholar]
- 16.Cumberlidge, A. G., and K. Isono. 1979. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J. Mol. Biol. 131:169-189. [DOI] [PubMed] [Google Scholar]
- 17.Donato, G. M., and T. H. Kawula. 1999. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. J. Bacteriol. 181:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorman, C. J., and N. N. Bhriain. 1992. Thermal regulation of fimA, the Escherichia coli gene coding for the type 1 fimbrial subunit protein. FEMS Microbiol. Lett. 78:125-130. [DOI] [PubMed] [Google Scholar]
- 19.Dutnall, R. N., S. T. Tafrov, R. Sternglanz, and V. Ramakrishnan. 1998. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94:427-438. [DOI] [PubMed] [Google Scholar]
- 20.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 21.Edwards, R. A., and D. M. Schifferli. 1997. Differential regulation of fasA and fasH expression of Escherichia coli 987P fimbriae by environmental cues. Mol. Microbiol. 25:797-809. [DOI] [PubMed] [Google Scholar]
- 22.Falconi, M., V. McGovern, C. Gualerzi, D. Hillyard, and N. P. Higgins. 1991. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 3:615-625. [PubMed] [Google Scholar]
- 23.Friedman, D. I., A. T. Schauer, M. R. Baumann, L. S. Baron, and S. L. Adhya. 1981. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc. Natl. Acad. Sci. USA 78:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaastra, W., and F. K. deGraaf. 1982. Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiol. Rev. 46:129-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Göransson, M., K. Forsman, and B. E. Uhlin. 1989. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 3:123-130. [DOI] [PubMed] [Google Scholar]
- 27.Isono, K., and S. Isono. 1980. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol. Gen. Genet. 177:645-651. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordi, B. J., B. Dagberg, L. A. de Haan, A. M. Hamers, B. A. van der Zeijst, W. Gaastra, and B. E. Uhlin. 1992. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 11:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krabbe, M., N. Weyand, and D. Low. 2000. Environmental control of pilus gene expression, p. 305-322. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 31.Li, X., L. Lindahl, and J. M. Zengel. 1996. Ribosomal protein L4 from Escherichia coli utilizes nonidentical determinants for its structural and regulatory functions. RNA 2:24-37. [PMC free article] [PubMed] [Google Scholar]
- 32.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low, D. A., E. N. Robinson, Jr., Z. A. McGee, and S. Falkow. 1987. The frequency of expression of pyelonephritis-associated pili is under regulatory control. Mol. Microbiol. 1:335-346. [DOI] [PubMed] [Google Scholar]
- 34.Lu, L., K. A. Berkey, and R. A. Casero, Jr. 1996. RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1 acetyltransferase. J. Biol. Chem. 271:18920-18924. [DOI] [PubMed] [Google Scholar]
- 35.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O and TetR/O and AraCI1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1996. Environmental regulation of virulence gene expression in Escherichia, Salmonella, and Shigella spp., p. 2803-2816. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 37.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratories, Cold Spring Harbor, N.Y.
- 39.Nagy, L. K., T. Mackenzie, D. J. Pickard, and G. Dougan. 1986. Effects of immune colostrum on the expression of a K88 plasmid encoded determinant: role of plasmid stability and influence of phenotypic expression of K88 fimbriae. J. Gen. Microbiol. 132:2497-2503. [DOI] [PubMed] [Google Scholar]
- 40.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 41.Normark, S., D. Lark, R. Hull, M. Norgren, M. Båga, P. O'Hanley, G. Schoolnik, and S. Falkow. 1983. Genetics of digalactoside-binding adhesion from a uropathogenic Escherichia coli. Infect. Immun. 41:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Hanley, P., D. A. Low, I. Romero, D. Lark, K. Vosti, S. Falkow, and G. Schoolnik. 1985. Gal-Gal binding and hemolysin phenotypes and genotypes associated with uropathogenic Escherichia coli. N. Engl. J. Med. 313:414-420. [DOI] [PubMed] [Google Scholar]
- 43.Olsen, P. B., and P. Klemm. 1994. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 116:95-100. [DOI] [PubMed] [Google Scholar]
- 44.Olsen, P. B., M. A. Schembri, D. L. Gally, and P. Klemm. 1998. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 162:17-23. [DOI] [PubMed] [Google Scholar]
- 45.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 46.Schmoll, T., M. Ott, B. Oudega, and J. Hacker. 1990. Use of a wild-type gene fusion to determine the influence of environmental conditions on expression of the S fimbrial adhesin in an Escherichia coli pathogen. J. Bacteriol. 172:5103-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratories, Cold Spring Harbor, N.Y.
- 48.Squires, C. L., and D. Zaporojets. 2000. Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54:775-798. [DOI] [PubMed] [Google Scholar]
- 49.Tercero, J. C., L. E. Riles, and R. B. Wickner. 1992. Localized mutagenesis and evidence for post-transcriptional regulation of MAK3. A putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. J. Biol. Chem. 267:20270-20276. [PubMed] [Google Scholar]
- 50.Torres, M., C. Condon, J. M. Balada, C. Squires, and C. L. Squires. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 20:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ussery, D. W., J. C. D. Hinton, B. J. A. M. Jordi, P. E. Granum, A. Seirafi, R. J. Stephen, A. E. Tupper, G. Berridge, J. M. Sidebotham, and C. F. Higgins. 1994. The chromatin-associated protein H-NS. Biochimie 76:968-980. [DOI] [PubMed] [Google Scholar]
- 52.van der Woude, M., B. Braaten, and D. Low. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4:5-9. [DOI] [PubMed] [Google Scholar]
- 53.van der Woude, M. W. 1990. Regulation of the production of fimbriae by enterotoxigenic Escherichia coli. Ph.D. thesis. Free University of Amsterdam, Amsterdam, The Netherlands.
- 54.van der Woude, M. W., L. S. Kaltenbach, and D. A. Low. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol. 17:303-312. [DOI] [PubMed] [Google Scholar]
- 55.van der Woude, M. W., and D. A. Low. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control phase variation and expression of the sfa and the daa pili operons in Escherichia coli. Mol. Microbiol 11:605-618. [DOI] [PubMed] [Google Scholar]
- 56.Warren, F., and A. Das. 1984. Formation of termination-resistant transcription complex at phage lambda nut locus: effects of altered translation and a ribosomal mutation. Proc. Natl. Acad. Sci. USA 81:3612-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 58.White-Ziegler, C. A., L. B. Blyn, B. A. Braaten, and D. A. Low. 1990. Identification of a genetic locus in Escherichia coli involved in the thermoregulation of the pap operon. J. Bacteriol. 172:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White-Ziegler, C. A., and D. A. Low. 1992. Thermoregulation of the pap operon: evidence for the involvement of RimJ, the N-terminal acetylase of ribosomal protein S5. J. Bacteriol. 174:7003-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White-Ziegler, C. A., A. Villapakkam, K. Ronaszeki, and S. D. Young. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175-185. [DOI] [PubMed] [Google Scholar]
- 62.Worbs, M., R. Huber, and M. C. Wahl. 2000. Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon. EMBO J. 19:807-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshikawa, A., S. Isono, A. Sheback, and K. Isono. 1987. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209:481-488. [DOI] [PubMed] [Google Scholar]