Abstract
Loss-of-function mutations in the following seven pneumococcal genes were detected and analyzed: pspA, spxB, xba, licD2, lytA, nanA, and atpC. Factors associated with these mutations included (i) frameshifts caused by reversible gain and loss of single bases within homopolymeric repeats as short as 6 bases, (ii) deletions caused by recombinational events between nontandem direct repeats as short as 8 bases, and (iii) substitutions of guanine residues caused at an increased frequency by the high levels of hydrogen peroxide (>2 mM) typically generated by this species under aerobic growth conditions. The latter accounted for a frequency as high as 2.8 × 10−6 for spontaneous mutation to resistance to optochin and was 10- to 200-fold lower in the absence of detectable levels of H2O2. Some of these mutations appear to have been selected for in vivo during pneumococcal infection, perhaps as a consequence of immune pressure or oxidative stress.
Streptococcus pneumoniae (the pneumococcus) is an aerotolerant, catalase-deficient streptococcal species that resides predominately on the surface of the human airway. This pathogen is characterized by an impressive degree of interstrain diversity, as demonstrated by the ability of different isolates to synthesize the 90 currently described types of capsular polysaccharide, its immunodominant antigen. In addition to this interstrain diversity, it displays intrastrain variation in expression of many of the factors that contribute to host-bacterial interaction, including surface proteins, teichoic acid, and the production of high levels of hydrogen peroxide (35, 46). The pneumococcus phase varies between two phenotypes, distinguished by differences in colony opacity, that allow the organism to either colonize the mucosal surface of the nasopharynx or infect the bloodstream (23, 45, 47).
Recently, information on the complete genome sequences of two unrelated strains of S. pneumoniae has become available (19, 41). Comparison of the two complete pneumococcal genomes reveals a difference in size of 122,222 bp and 193 open reading frames with an overall level of nucleotide sequence similarity of <90%. These observations suggest that, as a species, S. pneumoniae has a highly plastic and varied genome and that its diversity may be important in its capacity to adapt to its human host as well as to different host environments. However, it should be noted that variability tends to be concentrated in limited regions, with much of it in several gene clusters encompassing only 160 kb of the genome (15). Factors that may contribute to this genetic diversity include recombinational events occurring both within and between strains, the frequency and distribution of insertion sequences that make up about 5% of the genome, and the presence of large numbers of unstable duplications (41, 43). Transformation events may be especially relevant, because the pneumococcus is naturally competent, and interstrain differences suggest that transformation-mediated horizontal gene transfer may be common. The role of transformation events in the dissemination of antibiotic resistance phenotypes between pneumococci as well as from other oral streptococci to the pneumococcus, for instance, has been extensively documented (3, 7, 8).
Another observation derived from whole genomic analysis is the occurrence of large numbers of incomplete genes or gene fragments, which are unlikely to have been generated solely by transformation-mediated homologous recombinational events. In the complete genome of strain R6, at least 60 genes or >2% of the open reading frames with orthologous genes in other species are partial or truncated (19). This is finding was unexpected considering S. pneumoniae's relatively compact genome, which varies in size from 2.04 to 2.16 Mb in the completely sequenced strains (19, 41). It has been proposed that one source of incomplete genes might be frameshift mutations generated by slipped-strand mispairing within iterative DNA motifs referred to as “microsatellites,” as has been demonstrated in many other species that depend on phase variation as a mechanism of adaptation (31, 41, 42). Eighteen percent of the genes in the TIGR4 isolate were reported to contain short-sequence DNA repeats, although in few instances did these contain greater than nine tandem repeats (41). In other bacterial species, most of the previously characterized repetitive DNA sequences that vary at a detectable frequency contain considerably longer repeating units, although there are reports of short iterative sequences as short as 7 repeats that are subject to slippage (6, 16). There have been limited reports of this type of mechanism in any gram-positive species, and to our knowledge, there are no prior examples of phase variation generated by changes in the length of iterative sequences in the pneumococcus (25).
The purpose of this study was to characterize factors other than transformation events that may contribute to the genetic instability of the pneumococcus. Our results demonstrate that spontaneous mutations occur at a high frequency and that many of these events involve rearrangements of short tandem or nontandem DNA repeats or oxidative damage caused largely by endogenous hydrogen peroxide production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. pneumoniae strains used in this study are described in Table 1. S. pneumoniae was grown in tryptic soy broth with or without 1.5% agar at 37°C. Unless otherwise noted, bacteria were grown to mid-log phase (A620 = 0.3) in 5-ml liquid cultures without shaking. When specified, approximately 80 U of bovine catalase per ml (Worthington Biochemical, Freehold, N.J.) was added to liquid cultures at the time of inoculation. This amount of catalase was sufficient to reduce the concentration of H2O2 below the limit of detection. Anaerobic growth conditions were obtained with the BBL GasPak System (Becton Dickinson, Cockeysville, Md.). Catalase (5,000 U) was applied to the agar surface prior to plating and incubation in candle extinction jars. Colony morphology was determined on transparent medium under magnification and oblique, transmitted illumination as previously described (47). Unless otherwise stated, chemicals and reagents were purchased from Sigma-Aldrich Chemical Co. (St. Louis, Mo.).
TABLE 1.
S. pneumoniae strains used in the study
| Strain | Genotype or phenotype | Source or reference |
|---|---|---|
| P10 | Type 9V | Clinical isolate |
| P24 | R6, nonautolytic | 37 |
| P43 | Type 22, optochin sensitive | Clinical isolate |
| P44 | Type 22, optochin resistant | Clinical isolate |
| P62 | P10, expresses <0.02 mM H2O2 | 35 |
| P303 | Type 6A | Clinical isolate |
| P324 | Type 6B | Clinical isolate |
| P763 | P324, opaque variant, lacks XbaI methylation | This study |
| P765 | P324, transparent variant, XbaI methylation | This study |
| P806 | Opaque revertant of P765, lacks XbaI methylation | This study |
| P833 | Type 23F clinical isolate, truncated 22-kDa PspA | 26 |
| P878 | D39spxB::TnphoA(erm), expresses <0.02 mM H2O2 | 35 |
| D39 | Type 2 | 19 |
| R6 | D39, unencapsulated | 19 |
| WG44.1 | PspA mutant transformant derived from D39 | 28 |
| TIGR4 | Type 4 genome sequence strain | 41 |
Phenotypic analysis.
Altered expression of PspA and SpxB in P833 and P62, respectively, was described previously (26, 33). Truncation of LytA in the nonautolytic strain P24 was confirmed with a polyclonal antiserum to LytA in Western immunoblots. The effects of stationary-phase autolysis due to the expression of LytA on the growth characteristics of R6 and P24 were compared as previously described (37). Methylation of XbaI sites was assessed by the presence or absence of digestion of chromosomal DNA with XbaI (New England Biolabs, Beverly, Mass.). Altered migration of the teichoic acid in WG44.1 was detected in Western immunoblots with monoclonal antibody (MAb) TEPC-15 recognizing the phosphorylcholine epitope as previously described (48). Differences in teichoic acid were confirmed by demonstrating reduced incorporation of [3H]choline added to the growth medium in WG44.1 compared to its parent strain as previously described (23). Additional confirmation of the altered teichoic acid in WG44.1 was provided by mass spectroscopy.
Genotypic analysis.
Pneumococcal chromosomal DNA for use as templates for PCR was isolated as previously described (34). DNA for direct nucleotide sequencing was generated by PCR with Taq polymerase (Promega, Madison, Wis.) with the primers specified in Table 2 as previously described for the gene encoding the ATPase c-subunit (atpC) (36). These were also used as primers for nucleotide sequencing on gel-purified PCR products. All mutations were confirmed on both strands of PCR products and in PCR products generated in independent experiments.
TABLE 2.
Sequences of the primers used in this study
| Gene | Primersa |
|---|---|
| spxB | F-CGGAATTCCTGACCAGTTCCCTTCTG |
| R-CGCGTCGACTTATTTAATTGCGCGTGATTG | |
| pspA | F-AACTGAAGAGAAAGCAAAGC |
| R-CGTCTAACTCATCAATCTTATCAC | |
| xba | F-TTGTTTTATACCCAAATATGAATAGAGACGTAG |
| R-ATCACAATGAACGAAAATACTGCCAC | |
| lytA | F-GCGCGGATCCATGGAAATTAATGTGAGTA |
| R-GGGAATTCTTATTTTACTGTAATCAAGCCATC | |
| licD2 | F-GATTATATGACCCTCCACC |
| R-GCTATGACTATACCACTCTTGC | |
| atpC | F-TCGAAAAGTGGATCAACAACTATCC |
| R-TGGGAAAGAAGAAGTAACAAACTCG |
F, forward; R, reverse.
Optochin resistance.
Resistance to ethylhydrocupreine (optochin) was determined by the addition of a concentration equal to twice the previously determined MIC for each strain to be tested. The concentration of optochin added to the growth medium was 4 μg/ml unless otherwise specified. The rate of spontaneous optochin resistance was calculated by growth of a single optochin-sensitive colony to the stationary phase and plating of 0.1 to 0.5 ml of culture under selective conditions. Numbers of optochin-resistant colonies were compared to colony counts in the absence of the antimicrobial compound. Mutation rates represent the ratio of optochin-resistant to -sensitive colonies and are expressed as ranges in at least three independent determinations in duplicate. When specified, following growth to an A620 of 0.4, 1.0 ml of cell culture was removed, and hydrogen peroxide at the concentration specified was added from a 9.8 M stock solution. The cells were incubated for an additional 60 min prior to plating as described above.
Hydrogen peroxide measurement.
Twenty microliters of a solution consisting of 3 mg of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) per ml and 0.2 mg of horseradish peroxidase per ml in 0.1 M sodium phosphate buffer (pH 7.0) was added to 180-μl aliquots of each supernatant to be tested (13). The reaction was allowed to proceed for 20 min at room temperature, and then the A560 was measured. Values were calculated by comparison to those of a standard curve generated with known concentrations of H2O2.
Nucleotide sequence accession number.
The nucleotide sequence accession numbers of the genes examined in this study include AY072940 (pspA in P833), AF467892-3 (spxB in P62 and P10, respectively), AF469000-1 (xba in P765 and P806, respectively), AF467751 (licD2 in WG44.1), AF467249 (lytA in P24), and AF467738-50 for mutations conferring optochin resistance in the atpC gene.
RESULTS
Frameshifts within short-sequence DNA repeats.
In the course of our studies, we have observed translational frameshift mutations occurring because of spontaneous variation in the length or number of homopolymeric repeats within three pneumococcal genes (Table 3).
TABLE 3.
Spontaneous frameshift mutations involving homopolymeric repeats
| Strain | Gene | Mutationa | Positionb | Effect on gene expression |
|---|---|---|---|---|
| P833 | pspA | A(8) to A(7) | 538 | Truncation of PspA (amino acid 189 of 655) |
| P62 | spxB | A(6) to A(7) | 460 | Truncation of SpxB (amino acid 160 of 591) |
| P765 | xba | T(7) to T(8) | 959 | Restored expression of XbaI methylase |
| P806 | xba | T(8) to T(7) | 959 | Truncation of XbaI methylase (amino acid 335 of 361) |
Refers to the numbers of bases in homopolymeric repeats at the position noted.
Refers to the first base in the homopolymeric repeat relative to the first base in the initiation codon.
In Western blots, P833, isolated from the middle ear of a child with otitis media, was noted to express a 22-kDa form of pneumococcal surface protein A (PspA), a protein normally migrating between 80 and 100 kDa (26). Comparison of the nucleotide sequence from P833 to the most homologous pspA sequence in current databases revealed a frameshift mutation consisting of loss of a single residue within eight intragenic tandem adenines. Loss of this base resulted in premature truncation of the protein at amino acid 189. Since this was a minimally passaged clinical isolate, it appeared likely that this event may have occurred in vivo, possibly as a result of selective immune pressure to eliminate expression of the more conserved C-terminal portion of this highly immunogenic surface protein (17, 27).
P62 was isolated as a spontaneous mutant of a type 9V clinical isolate displaying altered colony morphology during in vitro passage. Analysis of whole-cell lysates of this mutant by two-dimensional gel electrophoresis revealed loss of expression of a single protein, pyruvate oxidase (SpxB) (33). Nucleotide sequence analysis of spxB in P62 revealed a gain of a single adenine residue in a series of six intragenic tandem adenines when compared to its parent strain, P10. The gain of this adenine resulted in premature termination after expression of only 160 of 591 amino acids in the full-length SpxB. The truncated SpxB was no longer functional, because P62 did not express detectable levels of hydrogen peroxide, a by-product of the oxidative metabolism of pyruvate (35, 40). This mutant may have been selected for during in vitro growth, since spxB mutants grow more efficiently because of the absence of an inhibitory effect of endogenously generated H2O2 on colonies. It was not possible to identify spontaneous revertants of P62 expressing a full-length SpxB by screening colonies, most likely because such revertants would have a competitive growth disadvantage in vitro.
Like many pneumococcal isolates, strain P324 phase varies between opaque and transparent colony forms. Susceptibility to digestion of genomic DNA by the restriction endonuclease XbaI is associated with colony opacity in this strain (data not shown). Comparison of the nucleotide sequence of the XbaI methylase gene, xba, in phase variants of P324 revealed an additional thymine residue within a repeat of seven thymine residues in the transparent variant, resulting in a frameshift and expression of the full-length gene product. Multiple, independently derived transparent phase variants all contained the same single-base-pair insertion (data not shown). A spontaneous transparent-to-opaque revertant, P806, was associated with a loss of a thymine base at this site, resulting in a truncated gene product. Only variants of this strain with the opaque phenotype were susceptible to DNA cleavage by XbaI, because of an out-of-frame number of homopolymeric repeats causing expression of a truncated XbaI methylase (data not shown). Insertional mutagenesis of xba in this strain, however, did not affect colony morphology, suggesting that differential methylation of XbaI sites correlates with, but is not directly responsible for, phase variation in colony opacity.
Together, our findings with P833, P62, and P324 confirmed that homopolymeric repeats as short as 6 bases are unstable genetic features in the pneumococcus, because of reversible loss or gain of bases in these elements.
Mutations caused by short nontandem repetitive sequences.
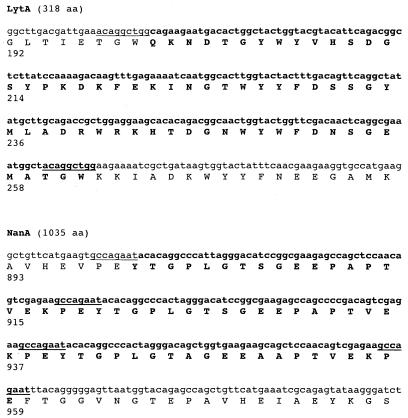
Another example of a spontaneous mutation observed through screening for altered colony morphology is P24, a minimally autolytic mutant of strain R6. P24 expresses a major murein hydrolase or autolysin (LytA) of 29 kDa rather than the expected 38.5 kDa as detected by Western analysis (data not shown) (11). The genetic basis of the altered size of LytA in P24 was a 189-bp deletion in the 3′ portion of lytA affecting the region of LytA involved in anchoring to phosphorylcholine on the cell wall teichoic acid (Fig. 1) (39). This deletion occurred between the first two of four nontandem copies of a nanomer (5′-ACAGGCTGG-3′) found within the lytA coding region. This recombinational in-frame deletion of 63 amino acids in the deduced 318-residue protein sequence of the full-length LytA results in a loss of murein hydrolase function, as determined by a lack of autolysis in mutant colonies.
FIG. 1.
Nucleotide sequence of the 3′ region of lytA and nanA. The deduced amino acid sequence and position are shown below the sequence. For lytA, a 189-bp region found in the nucleotide sequence of R6, but not the nonautolytic mutant P24, and the corresponding amino acid sequence are indicated in boldface. The positions of nontandem direct nanomer repeats flanking the deletion in the R6 lytA are shown by underlining. For nanA, a 180-bp region found in the nucleotide sequence of R6, but not TIGR4, and the corresponding amino acid sequence are indicated in boldface. The positions of nontandem direct octomer repeats, two of which flank the deletion in the R6 nanA, are shown by underlining. The predicted length of the intact protein is indicated in parentheses.
A similar deletion is present in the nanA gene encoding one of the pneumococcal neuraminidases in the TIGR4 strain used for genomic analysis (2). When compared to other pneumococcal nanA entries in the sequence databases, the 3′ coding portion of the nanA of this strain contains a 180-bp in-frame deletion occurring within two of four nontandem direct repeats of an 8-bp sequence (5′-GCCAGAAT-3′).
Therefore, the analysis of genes for two surface-anchored proteins, lytA and nanA, suggested that nontandem repetitive sequences as short as 8 bases were also examples of unstable genetic features.
Analysis of other spontaneous mutations.
The gene encoding LicD2, a putative CDP-choline transferase, contains eight tandem adenines at the 5′ end of the open reading frame and appeared to be a candidate for frameshift mutations similar to those described above. Mutants lacking a functional copy of this gene are characterized by an altered teichoic acid containing decreased amounts of phosphorylcholine (48). Forty unrelated strains were screened by Western analysis with a MAb against phosphorylcholine to look for teichoic acid units with aberrant migration. Only a single strain, WG44.1, showed a pattern suggestive of a licD2 mutant phenotype. Comparison of the nucleotide sequence of WG44.1 and its parent strain showed that, rather than a frameshift within the homopolymeric repeat of adenines, there was a single G→A base pair change causing a substitution of tryptophan for tyrosine (amino acid 35 of 267).
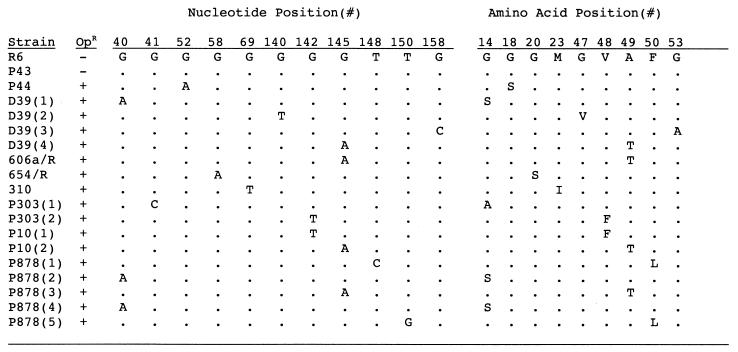
Based on the large number of different genes in which mutations were identified in this study, we sought to determine a baseline frequency of spontaneous mutation in S. pneumoniae grown in vitro. This was achieved by selecting for loss of sensitivity to the antimicrobial agent optochin, to which the pneumococcus is normally sensitive because of an H+-ATPase activity conferred by a F0/F1 class ion transport ATPase (36). The rate of loss of optochin susceptibility ranged from 0.24 to 1.8 × 10−6 in strain D39 (Table 4). Similar high rates of spontaneous mutation were found in other clinical isolates (range, 0.12 × 10−6 to 2.8 × 10−6). Nucleotide sequencing of the a- and c-subunits of the H+-ATPase genes in 12 spontaneous optochin-resistant mutants from seven unrelated isolates showed nine unique mutations. Some of these substitutions have previously been described (4, 10, 36). All nine mutations were due to a single-base-pair change involving a guanine base (G→A > G→T > G→C) resulting in single-amino-acid substitutions within the c-subunit (atpC) (Fig. 2). This finding, together with the mutation noted in licD2, suggested that guanine residues may be particularly subject to mutagenic changes in the pneumococcus.
TABLE 4.
Factors affecting the rate of spontaneous mutation conferring resistance to optochin
| Strain | Condition | Concn of H2O2 (mM)a | No. optochin resistant/optochin sensitive (10−8)b |
|---|---|---|---|
| D39 | Standard growth | 2.3 | 24-180 |
| P878(D39spxB) | Standard growth | <0.025 | <0.3-1.2 |
| D39 | + H2O2 (5 mM) | NDc | 100-190 |
| D39 | + Catalase | <0.025 | <0.3 |
| D39 | Anaerobic growth | ND | <0.3 |
| P303 | Standard growth | 2.2 | 12-80 |
| P303 | + Catalase | <0.025 | <0.3 |
| P10 | Standard growth | 2.7 | 57-280 |
| P10 | + Catalase | <0.025 | <0.3 |
The concentration of hydrogen peroxide (H2O2) in the culture supernatant was determined following growth to the stationary phase.
Values represent the range of the mean rate of spontaneous resistance in duplicate samples in at least three independent experiments.
ND, not determined.
FIG. 2.
Characteristics of mutations in atpC. The 198-bp sequence of this gene was determined in clinical isolates or P878 (D39spxB mutant), with spontaneous mutations conferring resistance to 4 μg of optochin per ml (OpR). Only those bases and corresponding amino acids that differ from the optochin-sensitive (OpS) control strain R6 are shown. The positions of the affected base and corresponding predicted amino acid changes are relative to the first nucleotide or amino acid in the gene or protein, respectively. Where indicated, multiple optochin-resistant mutants of the same strain obtained from independent experiments were analyzed. P43 and P44 were both cultured from the same infected pleural fluid.
In the case of P43/44, both optochin-susceptible and -resistant type 22 pneumococci were isolated from the same infected specimen of pleural fluid. Since it is highly unlikely that there was a simultaneous infection at the same site with two strains of the same type, the G→A mutation in P44 is likely to have occurred in vivo, generating a mixed population of optochin-sensitive (P43) and -resistant (P44) phenotypes.
Effect of hydrogen peroxide on spontaneous mutation.
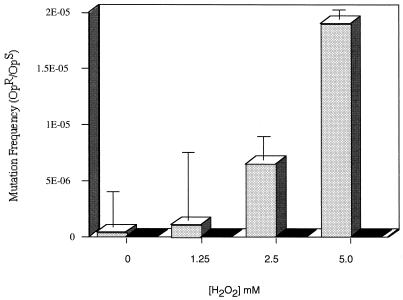
Guanine bases are particularly susceptible to oxidative damage by conversion to 8-oxoguanine (GO), with subsequent base excision and substitution catalyzed by the enzymes making up the GO system (30, 44). The pneumococcus lives in an oxygen-rich environment on the airway surface. In addition, the pneumococcus generates concentrations of hydrogen peroxide that may exceed 2 mM when grown aerobically in liquid culture without the addition of exogenous catalase (Table 4). The possibility that endogenously generated H2O2 contributes to the high frequency of mutations involving guanine residues was examined. In the presence of an exogenous source of catalase or anaerobic growth, there was no detectable H2O2 generated, and there was a >10-fold decrease in the rate of spontaneous optochin resistance in D39 and other clinical isolates (Table 4). The inactivation of spxB in D39 (P878) eliminated detectable H2O2 in culture supernatants and was associated with a similar decrease in mutation frequency during aerobic growth. Mutations involving guanine bases and causing loss of optochin sensitivity still occurred in P878, but at a much lower frequency (<0.3 × 10−8). However, unlike in the clinical isolates, not all mutations in the atpC gene of P878 involved changes in guanine. The mutations in atpC noted in the absence of high levels of endogenous hydrogen peroxide could be due to other types of oxidative stress, the minimal amounts of H2O2 still produced in the absence of pyruvate oxidase, or random background rates of substitutions. To confirm the effect of hydrogen peroxide on the mutation rate, exogenous H2O2 (0 to 5 mM) was added to D39, resulting in a dose-related increase in the rate of spontaneous optochin resistance reaching a rate of 1.9 × 10−5 in the presence of 5 mM added H2O2 (Fig. 3). Higher concentrations of exogenously added H2O2 could not be tested because of cell death. Furthermore, it was not possible to confirm the effect of exogenous hydrogen peroxide in the absence of endogenous production by using the spxB mutant, since loss of this gene results in increased sensitivity to killing by H2O2 (data not shown).
FIG. 3.
Effect of exogenous hydrogen peroxide on the spontaneous mutation rate. The frequency of spontaneous resistance to 4 μg of optochin per ml expressed as a mutation rate (optochin resistant/optochin sensitive [OpR/OpS]) in strain D39 in the absence (stippled bars) or presence (solid bars) of catalase was calculated following growth to the mid-log phase and treatment for 60 min with H2O2 added to the concentration indicated. Each value is the mean ± standard deviation from a representative experiment performed in triplicate for each condition.
DISCUSSION
In this report, we demonstrate additional sources of genetic instability in the pneumococcus that do not appear to involve homologous recombination through genetic transformation. Similar events may explain, at least in part, the large number of gene fragments found in complete genome sequences (19). One source of genetic instability was the gain or loss of a single base within short-sequence DNA repeats, presumably occurring through slipped-strand mispairing (24). A notable feature of this type of mutation, demonstrated in three separate genes—pspA, spxB, and xba—is that it may occur in particularly short iterative sequences with as few as 6 repeats. These are shorter than those reported to be subject to slippage in other species, and this suggests that postreplicative DNA proofreading may be relatively inefficient in the pneumococcus (42). In each example described here, slippage occurred within homopolymeric tracts of adenines or thymines and caused a frameshift type of mutation. Since the pneumococcus has a 60.3% A+T content, repetitive sequences with six or more adenine or thymine residues are common and widely distributed throughout the genome (19, 41). This observation suggests that the many pneumococcal genes containing similar short repeats have the potential to mutate in this manner as previously proposed (41). It remains unclear whether phase variation as detected by differences in colony opacity, which affects many pneumococcal characteristics related to host-pathogen interaction, may be due to this potentially reversible, high-frequency type of mutation (46).
Our study was unable to address the precise frequency with which these mutational events occur in short-sequence DNA repeats. The fact that similar mutations could be identified in three separate genes, however, suggests that these events are not rare. The mutations described in this report may have been selected for because of a growth advantage in the loss-of-function phenotype or were associated with other forms of variation. The rate of these mutations, therefore, is difficult to accurately determine, since it may depend on growth conditions or other factors that are difficult to control experimentally. An additional consideration is that slipped-strand mispairing in DNA microsatellites may not always be a stochastic occurrence, as has been proposed (31). In Escherichia coli, hydrogen peroxide has been shown to increase the frequency of expansions and deletions within microsatellites (21, 22). Likewise, the high levels of H2O2 produced as a by-product of oxidative metabolism could be a factor promoting the instability of short-sequence DNA repeats in the pneumococcus. In the case of P62, which contains a frameshift in spxB, the diminished hydrogen peroxide generated by this mutant may have decreased the reversion frequency to below the level of detection possible by screening. It has also been proposed that the hydrogen peroxide generated by the host may contribute to slippage in short-sequence DNA repeats (22). In this regard, the oxidative stress that accompanies inflammation may have promoted the deletion of one of eight tandem adenine bases in pspA of P833 isolated from a case of otitis media (26).
A second type of mutation noted during growth of the pneumococcus in vitro also involves repetitive DNA sequences, although in this instance, these were not tandem repeats. These in-frame deletion mutations appeared to involve recombination between identical copies of short sequences. In both examples shown, these were in genes encoding pneumococcal surface proteins. In gram-positive bacteria, genes encoding surface proteins commonly contain highly iterative domains and often vary in length because of homologous recombination within these elements (18). However, in the case of lytA in P24 and nanA in TIGR4 shown here, these recombinational events appear to have occurred between short (8 to 9 bases) nontandem repeats. The recent report of spontaneous duplications in the pneumococcal genome induced under certain growth conditions suggests that similar events, like those involving tandem repeats, may be reversible (43).
It was possible to show an association between the presence of physiologic levels of hydrogen peroxide and the overall frequency of mutations in the pneumococcus. This report differs from earlier studies in that the pneumococcus routinely exists in culture in >1,000-fold-higher concentrations of hydrogen peroxide, compared to organisms such as E. coli, where its mutagenic effect has been examined in detail (14, 20). Rather than slippage in short-sequence DNA repeats, oxidative damage due to H2O2 and leading to spontaneous resistance to optochin was shown to involve transition more than transversion mutations exclusively affecting guanine bases. These accounted for a rate of mutation 10- to 200-fold greater than that measured under conditions in which there was no detectable H2O2. However, this does not imply that hydrogen peroxide does not contribute to the gain and loss of bases in repetitive sequences, since this was not a prominent feature of the gene used in this study to assess the frequency of spontaneous mutations.
The findings in this report confirm that hydrogen peroxide produced in especially high concentrations by the pneumococcus compared to those produced by other bacterial species contributes to its genetic instability (35). The types of mutations observed at an increased rate in the presence of endogenously generated hydrogen peroxide were typical of the type and frequency of substitutions previously associated with H2O2-mediated oxidative stress (in decreasing order of occurrence, G→A > G→T > G→C) (44). Guanine residues, particularly those in motifs containing more than one guanine, have previously been shown to be hot spots for mutations caused by oxidative stress, both through direct transfer of electrons to guanine from radicals and through intrastrand transfer of electrons that originate at other bases (32). In this regard, six out of the nine guanine residues that were mutated in atpC of wild-type isolates were in motifs containing tandem guanine residues.
Under growth conditions that are tolerated without difficulty by other bacterial species, the pneumococcal DNA repair mechanisms that should correct these mutations appear to be overwhelmed. This observation provides additional evidence that DNA repair mechanisms may be relatively insufficient for aerobic growth, leading to a high rate of mutation. The pneumococcus has a well-studied mismatch repair system (Hex), which corrects many of the mismatches that result from DNA recombination, although its role, if any, in the repair of oxidative damage to DNA has not been explored (12). The pneumococcus also contains homologs to all three enzymes described in the E. coli GO repair system (29). However, one of these enzymes, mutY, an N-glycosylase endonuclease that functions in the removal of adenine bases incorporated opposite 8-oxo-G, lacks a highly conserved cysteine region that coordinates a [4Fe-4S]2+ cluster loop (38). It has been proposed that the lack of this feature is detrimental to repair of A/G mismatches and that such a handicap may be at least partly responsible for the predominance of A-T bases in the pneumococcal chromosome (38). This hypothesis is further supported by our data, which suggest that oxidative stress experienced by the pneumococcus on the surface of the airway would produce a relatively high frequency of A/G mismatches. An example of a mutation likely to have occurred during natural infection and similar to those promoted by endogenously generated hydrogen peroxide during growth in vitro was presented. The mutation identified in P44 suggests that the observations in this report based on growth in vitro may also be relevant for events occurring during replication within the host. The frequency of mutations targeting guanines as a result of oxidative stress in natural colonization or infection is unknown. However, a recent report describes four instances in which treatment of peumonococcal pneumonia with the antibiotic levofloxacin failed due to mutations in both parC and gyrA (5). The majority of these mutations involved substitution of guanine to adenine, indicative of the mutations caused by oxidative stress described in this report.
Genomic analysis confirms that the pneumococcus lacks many of the key systems, such as catalase and OxyR, employed by other bacteria to deal with oxidative stress, including the effects of hydrogen peroxide, and thereby prevent DNA damage. In this regard, competence for genetic transformation may be an important characteristic for the pneumococcus as a mechanism for repairing frequent mutations caused in large part by its own aerobic metabolism (1). In fact, competence is downregulated under anaerobic conditions when H2O2 would not be generated and the rate of spontaneous mutation is reduced (9). Investigations involving the pneumococcus need to take into consideration these high rates of spontaneous mutation and the effect of different growth conditions on the rate of these events. Of particular concern are studies such as those comparing genetically defined strains in models of pneumococcal pathogenesis, in which there may be a selective pressure promoting the growth of unexpected and undesired mutants arising because of the genetic instability of S. pneumoniae.
Acknowledgments
We thank R. Lopez for antisera to LytA and M. Nahm for analysis of teichoic acids by mass spectroscopy.
This work was supported by grants (AI38436 and AI44231) from the U.S. Public Heath Service to J.N.W.
REFERENCES
- 1.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. Ogunniyi, I. Le Thomas, J. Garel, J. Paton, and M. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018-1028. [DOI] [PubMed] [Google Scholar]
- 2.Camara, M., B. G. Boulnois, P. W. Andrew, and T. J. Mitchell. 1994. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect. Immun. 62:3688-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffey, T., C. Dowson, M. Daniels, J. Zhou, C. Martin, B. Spratt, and J. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 4.Cogne, N., J. Claverys, F. Denis, and C. Martin. 2000. A novel mutation in the alpha-helix 1 of the C subunit of the F1/F0 ATPase responsible for optochin resistance of a Streptococcus pneumoniae clinical isolate. Diagn. Microbiol. Infect. Dis. 38:119-121. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, R., R. Cavalcanti, J. Brunton, D. Bast, J. de Azavedo, P. Kibsey, C. Fleming, and D. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 6.De Bolle, X., C. Bayliss, D. Field, T. Van De Ven, N. Saunders, D. Hood, and E. Moxon. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35:211-222. [DOI] [PubMed] [Google Scholar]
- 7.Dowson, C., T. Coffey, and B. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 8.Dowson, C., A. Hutchison, J. Brannigan, R. George, D. Hansman, J. Linares, A. Tomasz, J. Smith, and B. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echenique, J. R., and M.-C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenoll, A., R. Munoz, E. Garcia, and A. de la Campa. 1994. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the F0 complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol. Microbiol. 12:587-598. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, J. L., J. M. Sanchez-Puelles, P. Garcia, R. Lopez, C. Ronda, and E. Garcia. 1986. Molecular characterization of an autolysin-defective mutant of Streptococcus pneumoniae. Biochem. Biophys. Res. Commun. 137:614-619. [DOI] [PubMed] [Google Scholar]
- 12.Gasc, A., A. Sicard, and J. Claverys. 1989. Repair of single- and multiple-substitution mismatches during recombination in Streptococcus pneumoniae. Genetics 121:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Flecha, B., and B. Demple. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J. Biol. Chem. 270:13681-13687. [DOI] [PubMed] [Google Scholar]
- 15.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 17.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1987. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol. Gen. Genet. 207:196-203. [DOI] [PubMed] [Google Scholar]
- 19.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay, J., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, A., R. Chen, and L. Loeb. 1998. Induction of microsatellite instability by oxidative DNA damage. Proc. Natl. Acad. Sci. USA 95:12468-12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, A., and L. Loeb. 2000. Microsatellite instability induced by hydrogen peroxide in Escherichia coli. Mutat. Res. 447:187-198. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J., and J. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 24.Levinson, G., and G. A. Gutman. 1987. Slipped-stranded mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 25.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, B. J. Shelvin, E. A. Graviss, and J. M. Musser. 2001. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect. Immun. 69:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCool, T. Cate, G. Moy, and J. Weiser. 2002. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 195:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel, L. S., J. Yother, M. Vijayakumar, L. McGarry, W. R. Guild, and D. E. Briles. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J. Exp. Med. 165:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejean, V., C. Salles, L. Bullions, M. Bessman, and J. Claverys. 1994. Characterization of the mutX gene of Streptococcus pneumoniae as a homologue of Escherichia coli mutT, and tentative definition of a catalytic domain of the dGTP pyrophosphohydrolases. Mol. Microbiol. 11:323-330. [DOI] [PubMed] [Google Scholar]
- 30.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moxon, E., R. Lenski, and P. Rainey. 1998. Adaptive evolution of highly mutable loci in pathogenic bacteria. Perspect. Biol. Med. 42:154-155. [DOI] [PubMed] [Google Scholar]
- 32.Núñez, M., D. Hall, and J. Barton. 1999. Long-range oxidative damage to DNA: effects of distance and sequence. Chem. Biol. 6:85-97. [DOI] [PubMed] [Google Scholar]
- 33.Overweg, K., C. D. Pericone, G. G. C. Verhoef, J. N. Weiser, H. D. Meiring, A. P. J. M. De Jong, R. De Groot, and P. W. M. Hermans. 2000. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect. Immun. 68:4604-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearce, B. J., Y. B. Yin, and H. R. Masure. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9:1037-1050. [DOI] [PubMed] [Google Scholar]
- 35.Pericone, C. D., K. Overweg, P. W. M. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pikis, A., J. Campos, W. Rodriguez, and J. Keith. 2001. Optochin resistance in Streptococcus pneumoniae: mechanism, significance, and clinical implications. J. Infect. Dis. 184:582-590. [DOI] [PubMed] [Google Scholar]
- 37.Saluja, S. K., and J. N. Weiser. 1995. The genetic basis of colony opacity in Streptococcus pneumoniae: evidence for the effect of box elements on the frequency of phenotypic variation. Mol. Microbiol. 16:215-227. [DOI] [PubMed] [Google Scholar]
- 38.Samrakandi, M. M., and F. Pasta. 2000. Hypermutation in Streptococcus pneumoniae depends on an atypical mutY homologue. J. Bacteriol. 182:3353-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Puelles, J. M., J. M. Sanz, J. L. Garcia, and E. Garcia. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89:69-75. [DOI] [PubMed] [Google Scholar]
- 40.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803-813. [DOI] [PubMed] [Google Scholar]
- 41.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 42.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waite, R., J. Struthers, and C. Dowson. 2001. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high frequency phase variation. Mol. Microbiol. 42:1223-1232. [DOI] [PubMed] [Google Scholar]
- 44.Wang, D., D. Kreutzer, and J. Essigmann. 1998. Mutagenicity and repair of oxidative damage: insights from studies using defined lesions. Mutat. Res. 400:99-115. [DOI] [PubMed] [Google Scholar]
- 45.Weiser, J. N., D. Bae, H. Epino, S. B. Gordon, M. Kapoor, L. A. Zenewicz, and M. Shchepetov. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect. Immun. 69:5430-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiser, J. N. 2000. Phase-variation of Streptococcus pneumoniae, p. 225-231. In V. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 47.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, J.-R., I. Idanpaan-Heikkila, W. Fischer, and E. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31:1477-1488. [DOI] [PubMed] [Google Scholar]