Abstract
During quorum sensing in Vibrio fischeri, the luminescence, or lux, operon is regulated in a cell density-dependent manner by the activator LuxR in the presence of an acylated homoserine lactone autoinducer molecule [N-(3-oxohexanoyl) homoserine lactone]. LuxR, which binds to the lux operon promoter at a position centered at −42.5 relative to the transcription initiation site, is thought to function as an ambidextrous activator making multiple contacts with RNA polymerase (RNAP). The specific role of the α-subunit C-terminal domain (αCTD) of RNAP in LuxR-dependent transcriptional activation of the lux operon promoter has been investigated. The effects of 70 alanine substitution variants of the α subunit were determined in vivo by measuring the rate of transcription of the lux operon via luciferase assays in recombinant Escherichia coli. The mutant RNAPs from strains exhibiting at least twofold-increased or -decreased activity in comparison to the wild type were further examined by in vitro assays. Since full-length LuxR has not been purified, an autoinducer-independent N-terminally truncated form of LuxR, LuxRΔN, was used for in vitro studies. Single-round transcription assays were performed using reconstituted mutant RNAPs in the presence of LuxRΔN, and 14 alanine substitutions in the αCTD were identified as having negative effects on the rate of transcription from the lux operon promoter. Five of these 14 α variants were also involved in the mechanisms of both LuxR- and LuxRΔN-dependent activation in vivo. The positions of these residues lie roughly within the 265 and 287 determinants in α that have been identified through studies of the cyclic AMP receptor protein and its interactions with RNAP. This suggests a model where residues 262, 265, and 296 in α play roles in DNA recognition and residues 290 and 314 play roles in α-LuxR interactions at the lux operon promoter during quorum sensing.
Many species of bacteria regulate gene expression in a cell density-dependent manner, a phenomenon known as quorum sensing. Only when an intercellular signaling molecule, commonly termed an autoinducer, accumulates to a certain level will the expression of particular sets of genes be activated. One of the best-understood models for quorum sensing in gram-negative bacteria is the regulation of the luminescence (lux) genes in Vibrio fischeri (for reviews see references 17, 20, 42, and 45).
The lux operon, necessary for light generation, is composed of seven genes (luxICDABEG) (13). The first gene of the operon, luxI, encodes an autoinducer synthase that produces the autoinducer molecule N-(3-oxohexanoyl) homoserine lactone (10). The remaining genes in the operon encode the proteins necessary for luminescence: luxA and luxB code for the α and β subunits of luciferase, and luxC, luxD, and luxE encode components of the fatty acid reductase complex needed for synthesis of the aldehyde substrate for luciferase. However, the function of luxG is still unknown. The lux operon is activated by the product of luxR, which is located in a divergent transcriptional unit (reviewed in reference 20).
LuxR is the autoinducer-dependent transcriptional activator of the lux operon and consists of 250 amino acid residues. The N-terminal domain (NTD) is involved in autoinducer binding and modulates the activity of the C-terminal domain (CTD), which is the activation region (reviewed in reference 41). The CTD of LuxR has a helix-turn-helix motif and binds to a region of the DNA termed the lux box, which is 20 bp in length with a dyad symmetry (Fig. 1) (9, 12). The lux box is centered at the −42.5 position relative to the luxI transcription start site (11).
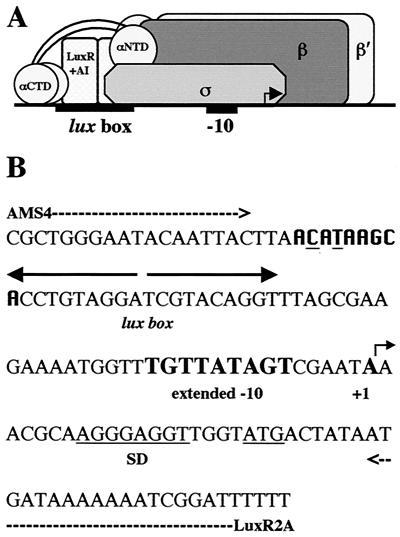
FIG. 1.
(A) Cartoon model of the proposed interactions between RNAP and LuxR at the lux operon promoter. See the text for details. (B) Nucleotide sequence of the lux operon promoter. The positions of various features of the region are highlighted as follows: dashed arrows, binding sites for primers AMS4 and LuxR2A; divergent solid arrows, lux box; boldface letters, extended −10 and transcription start site; the Shine-Delgarno (SD) sequence and initiation codon are underlined, and a putative proximal UP element half site is in boldface in a different font with the two bases that differ from the consensus underlined.
The model for transcriptional activation of the lux operon, under investigation here, is proposed to be similar to that in the cyclic AMP receptor protein (CRP) model at a class II-type promoter. Activation of transcription at class II CRP-dependent promoters demonstrates how an activator can make multiple contacts with RNAP during initiation of transcription (reviewed in reference 7). At class II CRP-dependent promoters such as the galP1 promoter, the DNA site for CRP is centered near position −41 and overlaps the −35 determinant for RNAP. In this case, RNAP contacts the promoter upstream as well as downstream of the CRP dimer. The upstream interaction is mediated by the α-subunit CTD (αCTD) of RNAP, and the downstream interaction is mediated by the remainder of RNAP (2, 3, 6). It is hypothesized that a LuxR multimer (presumably a homodimer) positioned to make contact with the DNA (lux box) might also be in contact with the αCTD, αNTD, and sigma subunits of RNAP (Fig. 1).
In addition to activator-RNAP contacts, interactions between the αCTD and DNA next to the activator-binding site also contribute to the stability of the complex in both class I- and class II-type promoters (7, 19). αCTD-DNA interactions with bases in the minor groove and with the DNA backbone along the minor groove can involve either upstream (UP) element-like sequences or nonspecific DNA sequences (5, 26, 30, 32-34, 48). There is also evidence that suggests some activators can alter the way that the αCTD interacts with the minor groove (1, 28, 30). Therefore, the αCTD could also be involved in DNA binding in the lux system, and LuxR could be affecting the nature of this interaction. However, the precise interactions that occur between activators, RNAP, and the target DNA cannot be predicted based solely on the position at which the activator binds the DNA. By investigating several different promoters, universal interactions can be identified, as well as unique contacts made by specific activators. This information can then be used to expand our knowledge and establish common trends in transcription activation among different promoters.
The focus of this project was to establish the roles of individual residues in the αCTD of RNAP in LuxR-dependent transcription of the lux operon. Experiments were performed using Escherichia coli and its RNAP due to the ease with which manipulations of the system can be made in comparison to V. fischeri. While the genomic sequence of V. fischeri is not available, an amino acid alignment of the αCTDs of E. coli K-12 and Vibrio cholerae shows the two to be 98% identical and 100% similar between residues 249 and 329 (only the two extreme C-terminal residues differ). Using E. coli as a heterologous host, it has previously been demonstrated that the αCTD plays a role in LuxR-dependent transcription initiation (40). Through the in vivo and in vitro analysis of mutant RNAPs (for reviews, see references 7, 19, and 23), individual residues in the αCTD that are involved in the mechanism of activation can be identified by significant decreases or increases in the rates of transcription compared to the wild-type. Further studies can then determine whether these residues are involved in protein-protein or protein-DNA interactions. Unfortunately, purification of full-length LuxR has been unsuccessful so far; however, in vitro work can be performed with a truncated form of LuxR (LuxRΔN) that is capable of the autoinducer-independent activation of the lux operon. Experiments have been designed to establish a correlation between the in vivo and in vitro studies using LuxRΔN with the intention of then using the in vivo data acquired for full-length LuxR as a reflection of its native activity.
MATERIALS AND METHODS
Strain construction.
E. coli JM109 (47) was used as the host organism for in vivo studies. A three-plasmid system was constructed by sequentially transforming three plasmids into JM109 (40). One plasmid (pAMS129) codes for the lux operon (luxICDABE) and kanamycin resistance on an RSF1010-based vector (40). The second plasmid encodes LuxRΔN (pAMS122) or LuxR (pAMS121) under the control of the isopropylthiogalactoside (IPTG)-inducible tac promoter and chloramphenicol resistance on a pACYC-based vector (40). The third plasmid codes for either wild-type or mutant α subunits of RNAP under the control of the IPTG-inducible lac promoter and ampicillin resistance on a pBR-based vector. The wild-type α was encoded on plasmid pREIIα (4) or pHTf1α (43). Variant forms of these two parent plasmids code for alanine substitutions at positions 273 to 329 (18, 25, 46) or 255 to 271 (18, 43) and 162 to 165 (31) in the αCTD, respectively. Transformants carrying all three plasmids were selected on Luria-Bertani agar medium containing 100 μg of kanamycin/ml, 100 μg of ampicillin/ml, and 20 μg of chloramphenicol/ml.
Luminescence assays.
Strains were grown overnight at 30°C in Luria-Bertani broth containing kanamycin, ampicillin, and chloramphenicol at the appropriate concentrations and were then diluted 1:1,000 in the same medium supplemented with 1 mM IPTG. Strains expressing LuxR from pAMS121 were also grown in the presence of 200 nM N-(3-oxohexanoyl) homoserine lactone (Sigma, St. Louis, Mo.). When this primary subculture had grown to an optical density at 600 nm (OD600) of 0.1, it was diluted to an OD600 of 0.025 in the same medium and grown to a final OD600 of 0.5. The luminescence output from 20 μl of culture was measured over a 4-s integration period in a TD-20/20 luminometer (Turner Designs, Sunnyvale, Calif.) with a sensitivity range over several log units. Cells from 0.5-ml aliquots of each culture at an OD600 of 0.5 were also harvested via centrifugation and frozen at −70°C for use in luciferase assays.
Luciferase assays.
The levels of luciferase found within cells were quantitated using a protocol based on the method of Rosson and Nealson (35). Cells harvested as described above were resuspended in 0.5 ml of lysis buffer (10 mM KPO4 [pH 7.0], 10 mM EDTA, 1 mM dithiothreitol [DTT], 0.1% bovine serum albumin, 50 μg of lysozyme per ml) and lysed via a single freeze-thaw step. Each luciferase reaction mixture contained the following final volumes of the reagents: 10 μl of crude cell extract, 10 μl of 1:1,000-diluted and sonicated n-decyl aldehyde (decanal; Sigma), 90 μl of assay buffer (10 mM KPO4 [pH 7.0], 0.1% bovine serum albumin, 1 mM DTT), and 100 μl of 50 μM reduced flavin mononucleotide. The reduced flavin mononucleotide was added directly to the tube containing the other reagents only after the tube was placed within the chamber of the luminometer. The luminescence emitted from the reaction was measured (3-s delay; 30-s integration time) with a TD-20/20 luminometer with a manual injection port.
Purification and reconstitution of wild-type and variant RNAP holoenzymes.
Sixteen RNAPs containing the α variants (residues 258 to 266, 268, 271, 273, 275, 297, 298, and Δ235) and the wild type were purified and reconstituted according to the method of Igarashi and Ishihama (22) (set 1). Additional RNAPs of interest (set 2) were purified and reconstituted as described below.
RNAP α subunits containing a hexahistidine tag between the first and second codons were prepared using plasmid pHTT7f1NHα (44). The plasmids carrying mutant rpoA alleles were constructed by replacing the HindIII-BamHI fragment, which encodes the αCTD, with fragments from plasmids encoding the different amino acid substitutions of interest (18, 46). For this study pT7-his6 alpha 278, 286, 291, 292, 295, and 300 were constructed and pT7-his6 alpha 290, 296, 301, 302, 303, 307, and 314 and the wild-type control, pHTT7His6α, were received from Richard L. Gourse's laboratory. The nondenaturing protocol used to purify the α subunit of RNAP (containing alanine substitutions at the residues identified above) from recombinant E. coli was modeled from published protocols (18, 43). The β, β′, and σ70 subunits of RNAP were purified from inclusion bodies and reconstituted with purified α subunits into RNAP holoenzyme as described by Tang et al. (44).
In vitro transcription assays.
Purified pAMS1300 (39) DNA was linearized with HindIII according to the manufacturer's instructions and electrophoresed on a 2% low-melting-point agarose gel. The appropriate DNA fragment was extracted from the agarose matrix using the QIAquick gel extraction kit (Qiagen, Valencia, Calif.) and quantitated by comparison to a mass ladder (Bio-Rad, Hercules, Calif.). Single-round transcription assays were performed from this template as previously described (39). All reaction mixtures contained linearized pAMS1300, the luxI promoter-containing DNA template (1.3 nM), LuxRΔN (10 μM), and mutant RNAP at an activity level equivalent to that of the wild type (either 30 [set 1] or 54 [set 2] nM). The RNA-1 transcript produced from the template serves as an internal control for LuxRΔN-independent RNAP activity. Quantitation of radiolabeled transcripts was performed using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
DNA mobility shift assays.
Three DNA templates were used during DNA mobility shift assays. To obtain a template which contained the lux operon promoter region, a portion of the plasmid pJE202 (14), containing the luxI regulatory region, was amplified via PCR with primers AMS4 (5′ CGCTGGGAATACAATTAC 3′) and LuxR2A (5′ AAAAAATCCGATTTTTTTATCAT 3′) (Sigma-Genosys, The Woodlands, Tex.). The 132-bp PCR fragment that was generated was ligated into pGEM T-easy vector (Promega, Madison, Wis.), and the resulting plasmids were transformed into E. coli DH5α (21). Strains containing the ampicillin-resistant recombinant plasmids were selected for and screened for the presence of the desired construct via LacZ complementation and for the correct restriction endonuclease recognition sites. The nucleotide sequences of both strands of the cloned PCR product were verified by the Virginia Tech Sequencing Facility using T7 and SP6 primers. The final construct was designated pAHF100.
Two control DNA templates were also generated. A 228-bp region of plasmid pRLG4238 (16) containing the rrnB P1 promoter region with a strong UP element was amplified via PCR with the vector-specific primers 1620 (5′ GCGCTACGGCGTTTCACTTC 3′) and 3038 (5′ CGTATCACGAGGCCCCTTTCG 3′) (Sigma Genosys) to be used as a positive control for α binding. The 104-bp pUC multiple cloning site was amplified from pUC19 by using the universal forward and reverse primers (47) for use as a negative control in the assay.
Radiolabeled DNAs for the mobility shift experiments were generated by incorporating [32P]αCTP (20 μM per 100-μl reaction mixture) into PCR mixtures. The reaction mixtures were subjected to two phenol-chloroform-isoamyl alcohol (25:24:1) extractions and an ethanol-sodium acetate precipitation. The pellets were then washed with 70% ethanol and resuspended in water. DNA concentrations were estimated by comparison to a mass ladder (Bio-Rad).
Gel mobility shift assays were based on published procedures (38). Reaction mixtures (45 μl) contained 3 nM radiolabeled DNA, 50 mM NaCl, 1 mM MgCl2, 0.002 mg of calf thymus DNA/ml, acetylated bovine serum albumin (2 mg/ml), 1 mM DTT, 0.1 mM EDTA, and 10% glycerol in 40 mM HEPES (pH 7.4). The reaction mixtures were incubated for 15 min at 30°C and run on 4% polyacrylamide gels containing 10% glycerol with recirculation of the running buffer (20 mM HEPES, 3 mM NaCl, 1 mM EDTA, pH 8.0). Radioactive bands were visualized using a Storm PhosphorImager (Molecular Dynamics).
RESULTS
Effects of alanine substitutions in α on LuxRΔN- and LuxR-dependent luminescence and luciferase levels.
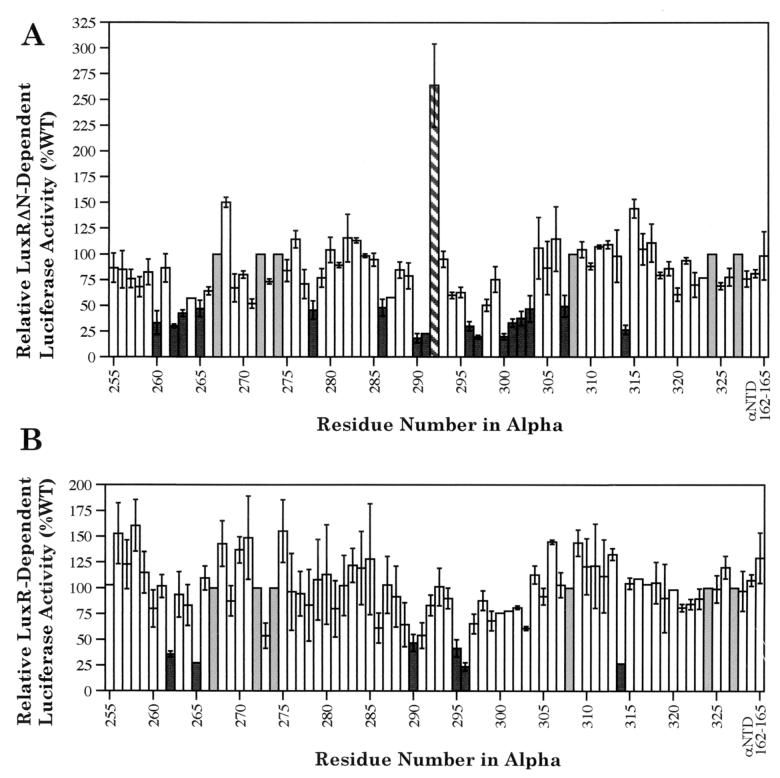
The effects of alanine substitutions in the αCTD on the rate of transcription of the lux operon were examined through in vivo luminescence and luciferase assays. Strains of recombinant E. coli containing three plasmids encoding (i) the lux operon, (ii) LuxRΔN or LuxR, and (iii) the wild type or 1 of 69 variants of α with single alanine substitutions in the CTD between residues 255 and 329 were tested. In addition, a variant of α with alanine substitutions at 162 to 165 in the αNTD was also included in the analysis. Simultaneous substitution of alanine for all four of these amino acid residues in the αNTD resulted in a defect in class II CRP-dependent transcription (31).
Luminescence assays were used as a first screen to establish whether the variant forms of α had any significant effects on activation of the lux operon (data not shown). However, other cellular metabolic functions that play an indirect role in the luminescence process, such as synthesis of precursors for the aldehyde substrate for the luciferase enzyme or production of reduced coenzymes and ATP, might also be affected by the variant RNAPs. Therefore, while the luminescence assays do serve as a useful first screen to establish which strains should be further investigated, luciferase activity levels from cell extracts were also used as a more direct way to measure the rate of transcription from the luxI promoter.
The residues identified as playing critical roles in the activation of the lux operon through luciferase assays represent a subset of those identified initially through luminescence assays. Strains expressing α variants with alanine substitutions at residues 260, 262 to 263, 265, 278, 286, 290 to 291, 296 to 297, 300 to 303, 307, and 314 had at least twofold-lower luciferase enzyme levels than the wild type in the presence of LuxRΔN. The strain expressing the α variant with a substitution at residue 292 had a >2-fold-higher luciferase level than the wild type (Fig. 2A). In the presence of LuxR, the variant forms of α that resulted in at least a twofold decrease in luciferase enzyme levels compared to the wild type had mutations in residues 262, 265, 290, 295, 296, and 314 (Fig. 2B). There were no α variants that produced a twofold increase in luciferase levels in the presence of LuxR. The finding that more residues in the αCTD are involved in LuxRΔN-dependent activation than in LuxR-dependent activation of the lux operon is consistent with previous studies which suggested some difference in the mechanism of RNAP and DNA interaction between the full-length and truncated forms of LuxR (12, 38). The in vivo luciferase assays performed also showed that the αNTD variant acted essentially as wild-type α (Fig. 2).
FIG. 2.
Effects of alanine substitutions in α on LuxRΔN-dependent (A) and LuxR-dependent (B) cellular luciferase levels in recombinant E. coli. The value for each variant form of α represents the average of two independent experiments with individual luciferase assays performed in quadruplicate. The error bars represent the range of each experiment from the mean. Luciferase activities from strains containing either of the two wild-type (WT) controls, pHTf1α or pREIIα, were set to 100% for each experiment. The solid bars highlight variants producing <50% wild-type levels of luciferase activity, the hatched bar highlights a variant producing >200% wild-type levels of luciferase activity, the open bars are used at positions where the variants exhibited luciferase levels within twofold of the wild-type, and the shaded bars are used to represent positions were alanine is already present in wild-type α.
Effects of alanine substitutions in the αCTD on LuxRΔN-dependent transcriptional activation of the lux operon in vitro.
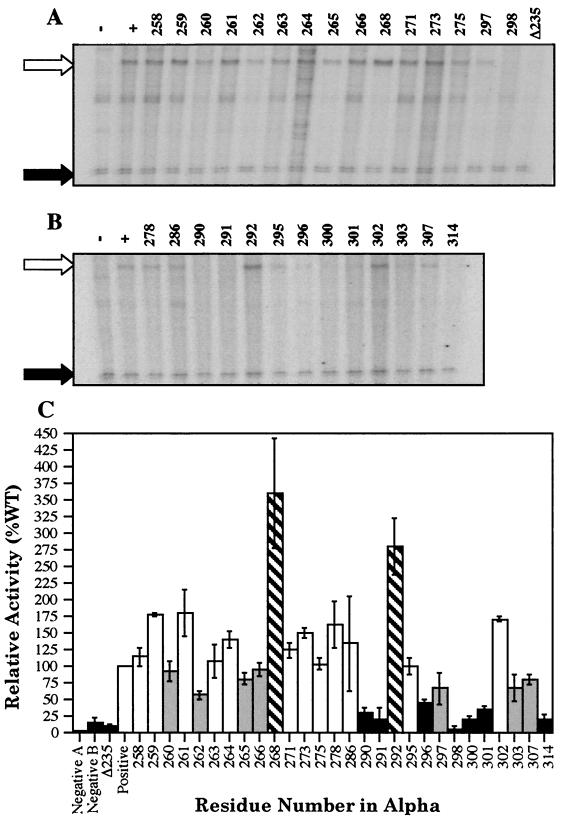
A number of α variants, including all those that limited or enhanced luciferase levels by at least twofold, were further analyzed via in vitro transcription assays. The analysis was carried out in two phases. In the first phase (set 1), the activity of a well-studied set of existing mutant RNAPs (and a wild-type control) purified in the Ishihama laboratory was examined (Fig. 3A). In the second phase (set 2), additional mutant RNAPs (and a wild-type control) of interest were reconstituted in the Stevens laboratory for use in the assays (Fig. 3B). So that data comparisons could be made between the two sets, the activities of the two wild-type RNAP controls were examined. Even though slightly different protein concentrations were used (30 [set 1] versus 54 [set 2] nM), there were approximately equal levels of activity as defined by the amount of RNA-1 transcript. The luxI/RNA-1 transcript ratio was also calculated for both, and similar values were observed (0.51 versus 0.54). Thus, the protein concentrations used for both of the reconstituted wild-type RNAPs containing wild-type α subunits were within twofold of one another, and the levels of LuxRΔN-dependent activation were roughly equal.
FIG. 3.
LuxRΔN-dependent in vitro transcription from the luxI promoter generated by wild-type or variant RNAPs. Representative results for RNAPs in set 1 (A) and set 2 (B) are shown. Transcripts produced by wild-type RNAP (set 1, 30 nM, or set 2, 54 nM) in the absence (−) and presence (+) of LuxRΔN (10 μM) are shown in lanes − and +, respectively. Lane Δ235 illustrates the results when a truncated form of RpoA missing the C-terminal 94 amino acid residues was used in the assays and serves as an additional control. The remaining lanes are labeled with the residue numbers indicating the positions of the alanine substitutions in α present in RNAP. The open arrows point to the lux mRNA products, and the solid arrows indicate the RNA-1 transcripts, which serve as an internal control for LuxRΔN-independent activity and permit normalization of the activity of the variant RNAPs under investigation. The identities of the extra transcripts produced from the DNA template in some lanes are unknown. (C) Graphical representation of the relative average value of the luxI transcript in comparison to the wild-type (WT; 100%) from two independently run experiments. The error bars represent the range of each experiment from the mean. The solid bars highlight variants (or negative controls) with <50% wild-type levels, the shaded bars highlight variants with <100% but >50% wild-type levels, the hatched bars highlight variants with >200% wild-type levels, and the open bars are used at positions where alanine substitutions in α had >100% but <200% wild-type levels.
Since LuxR has not been purified, only LuxRΔN was used for the in vitro analysis. RNAP containing a truncated form of α missing the C-terminal 94 amino acid residues (Δ235) was also used in the assays as an additional negative control. Using this variant of α, it had been previously established that the C-terminal region of the αCTD is essential for transcription of the lux operon (40). Several RNAPs containing α variants exhibited twofold decreases in the rate of transcription from the luxI promoter in comparison to the wild-type (residues 290, 291, 296, 298, 300, 301, and 314). Smaller decreases were also observed in residues 260, 262, 265, 266, 297, 303, and 307 with values ranging from 57.8 to 95.9%. In addition, RNAP variants containing mutations in α at residues 268 and 292 produced >2-fold-higher rates of transcription from the luxI promoter. Somehow, the substitutions at residues 268 and 292 permit the variant RNAPs to function more effectively in the presence of LuxRΔN than the wild-type RNAP. However, none of the variant RNAPs tested was shown to display any LuxRΔN-independent activation of the luxI promoter above basal levels of transcription (data not shown).
Overall, a good correlation exists between the in vivo luciferase and in vitro transcription assay results in most of the 28 variant RNAPs tested (Fig. 2 and 3). However, results using some RNAP variants (residues 263, 266, 268, 278, 286, 298, and 320) do not show a strict correlation between the in vivo and in vitro analyses. This may be explained in part by differences in plasmid maintenance and stability in the three-plasmid system used in the in vivo assays. Despite these discrepancies, five α variants with mutations in residues 262, 265, 290, 296, and 314 were identified as having >2-fold effects in vivo (in the presence of either LuxR or LuxRΔN) and producing rates of transcription in vitro less than those of the wild-type control.
Analysis of the interactions between the αCTD, the luxI promoter, and LuxRΔN.
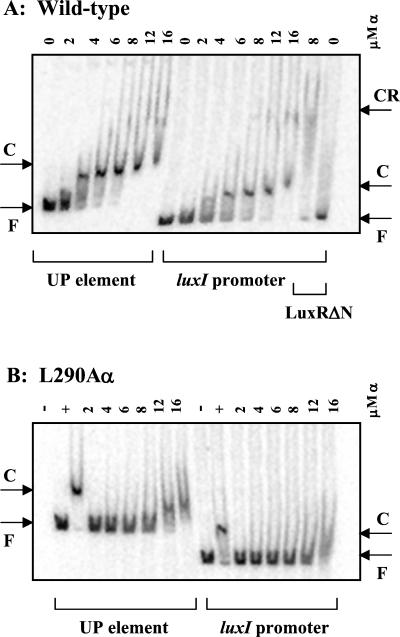
Purified wild-type α has previously been shown to have the ability to bind to the minor groove within the UP element of the rrnB P1 promoter (32, 34, 48). In this study, the ability of wild-type α to bind and recognize the UP element and the luxI operon promoter was examined. A complete shift of DNA fragments containing either the UP element or the lux DNA was observed when 8 to 12 μM wild-type α was used in the assays (Fig. 4A). Additionally, there was no shift observed with the negative control fragment containing the pUC multiple cloning site and wild-type α (data not shown). Therefore, there is a possible binding site for α somewhere in the lux DNA fragment used. By nucleotide sequence analysis, a region immediately upstream of the lux box and overlapping it by 1 bp has a potential binding site for α differing from the consensus sequence for the proximal subsite of the UP element by only two mismatches (Fig. 1B) (15, 16). It has been proposed that the A-T-rich sequences of the UP element are required to provide the optimum width of the minor groove needed for αCTD anchoring, as opposed to sequence-specific recognition (48). The lux operon promoter region has long stretches of high A+T content, and thus, it is likely that this region may contain more than one potential binding site for α. The long stretch of high A+T content upstream of the lux box may also explain why previous attempts to demonstrate α binding to the lux promoter via DNase I protection assays have not revealed any obvious footprints (38-40). Further in vitro analysis will be necessary to determine the precise binding site of α on the lux DNA.
FIG. 4.
Autoradiograms of gel mobility shift assays. The numbers above each lane refer to the concentrations of α or α variant used in the assays. The results shown are representative of two independent trials. F, free DNA; C, α-DNA complexes; CR, LuxRΔN-α-DNA complexes. (A) Interactions between purified wild-type RNAP α and a 228-bp PCR fragment containing a strong UP element from the rrnB P1 promoter amplified from pRLG4238 (UP element) or a 132-bp PCR fragment with the luxI promoter region amplified from pAHF100 (luxI promoter). The last two lanes show interactions when 10 μM LuxRΔN is added. (B) Interactions between the L290A α variant and the UP element or the luxI promoter. Control reactions containing no α (−) or 8 μM wild-type α (+) are also shown.
The ability of the α variants with alanine substitutions at residues 262, 265, 290, 296, and 314 to bind to the UP element and the native luxI promoter was also examined over the range of 2 to 16 μM. Variants 262A, 265A, 296A, and 314A demonstrated an inability to bind either the UP element or the lux operon promoter (data not shown), while variant 290A demonstrated roughly a fourfold reduction in its ability to bind the same sequences (Fig. 4B). This implies that these five residues are involved in DNA binding. However, the possibility that the alpha variants are not correctly folded into an active conformation is not excluded. In the case of 290A, since it retains a certain level of DNA binding, the defect in transcription activation for this variant might be due to impaired interactions with LuxR. As previously observed, LuxRΔN cannot bind to the DNA alone (Fig. 4A) (38). However, there is a weak supershift of the luxI promoter DNA fragment observed in the presence of both LuxRΔN and wild-type α (Fig. 4A). The supershift occurred at roughly the same concentration of α that was necessary to observe a shift of the DNA fragment with α alone. Therefore, in our in vitro system, it appears that α is assisting LuxRΔN in binding. The LuxRΔN-α-DNA complexes formed did not appear to be stable, as evidenced by smearing (Fig. 4A). Given the diffuse and variable nature of the supershifted band, attempts to determine the effects of the alanine substitutions in α on protein-protein interactions with LuxRΔN were not extensively pursued via this method.
DISCUSSION
The residues identified as playing critical roles in activation of the lux operon through luciferase assays represent a subset of those identified through luminescence assays. This is most likely due to additional indirect effects of the α variant on other cellular processes during the luminescence assays. Full-length LuxR and a truncated form of LuxR (LuxRΔN) were both used in the luminescence and luciferase assays so that comparisons could be made between the two. LuxRΔN is a monomer in solution and is thought to have a lower affinity for the DNA than full-length LuxR. It can only bind to the lux operon promoter in synergy with RNAP (38). Full-length LuxR is believed to bind to the DNA as a multimer, presumably a homodimer (8, 11), and recent genetic evidence confirmed that LuxR can bind to the lux box in the absence of RNAP (12).
Substitutions at six amino acid residues (residues 262, 265, 290, 295, 296, and 314) in the αCTD decreased the level of LuxR-dependent transcription twofold or more in E. coli cells (Fig. 5A). Substitutions at five of these six residues (residues 262, 265, 290, 296, and 314) also produced decreased levels of LuxRΔN-dependent transcription, as determined through a combination of in vivo and in vitro assays (Fig. 5B). In addition, substitutions at 13 other residues (residues 260, 263, 266, 278, 286, 291, 297, 298, 300 to 303, and 307) produced decreased levels of LuxRΔN-dependent transcription in at least one of the assay systems, and substitutions at two other residues (268 and 292) resulted in increased levels of transcription (Fig. 5B). The extra residues that appear to play a role only in LuxRΔN-dependent transcription are located in close physical proximity to the residues shown to be important in LuxR-dependent transcription (Fig. 5B). Differences in oligomeric states and DNA binding affinities may explain the requirement for more contact sites by RNAP with LuxRΔN. The lower binding affinity of LuxRΔN for the DNA would mean that substitutions that compromise binding even slightly might have a better chance of shifting the equilibrium to the unbound situation.
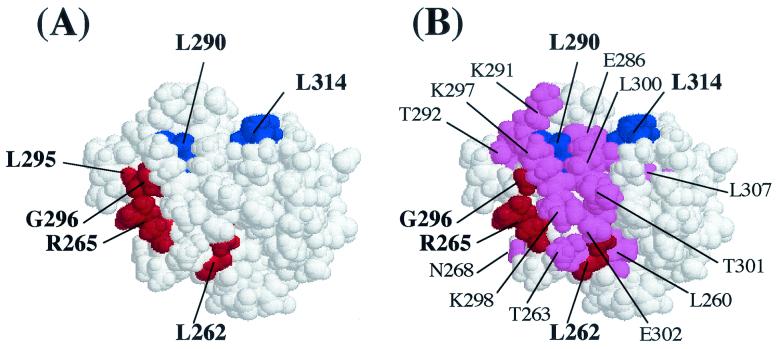
FIG. 5.
Space-filling model of the αCTD based on the atomic coordinates of Jeon et al. (24) showing the positions of some of the residues identified as important for LuxR-dependent (A) and LuxRΔN-dependent (B) activation of the lux operon. (A) Amino acid residues producing >2-fold effects on LuxR-dependent luciferase levels in vivo when changed to alanine are highlighted. The residues in red (262, 265, 295, and 296) are hypothesized to be involved in DNA recognition, and the residues in blue (290 and 314) are hypothesized to be involved in protein-protein interactions. (B) Amino acid residues producing >2-fold effect on either LuxRΔN-dependent luciferase levels in vivo or transcription rates in vitro are highlighted. The residues in red (262, 265 and 296) and blue (290 and 314) were found to be important in both LuxR- and LuxRΔN-dependent activation, while the residues in violet are unique to the LuxRΔN-dependent mechanism.
The residues involved in LuxR- and LuxRΔN-dependent transcription activation are remarkably similar to those obtained in analysis of CRP-dependent transcription at class II-type promoters. Two critical determinants corresponding to sites of alpha-DNA and alpha-activator interaction have been proposed in the CRP model (7). In this model, the 265 determinant is required for αCTD-DNA interactions and consists of residues R265, N268, N294, G296, K298, S299, and E302 (7, 18, 29, 43, 49). The 287 determinant (T285, E286, V287, E288, L289, G315, R317, and L318) is not required for αCTD-DNA interaction and plays no role in UP element-dependent transcription; hence, it is proposed that it is involved in protein-protein interactions with the CRP activator (7, 18, 36). This same region has been shown to be involved in transcription activation by FNR (residues L289 and L317 [27]) and P2 Ogr (residues E286, V287, L289, L290, and L300 [46]). The 265 determinant has also been proposed to play a role in DNA recognition from Fis-dependent promoters, while the 273 determinant mediates αCTD-Fis interactions (1, 28). Interestingly, while the surfaces involved in αCTD-activator interactions do appear to be specific, recent evidence suggests, at least in the cases of CRP and Fis, that the surfaces involved in the interaction in both proteins can be the same at class I- and class II-type promoters (1, 28, 36, 37).
Based on our analysis and information derived from other well-studied activators, it appears that there are two αCTD determinants involved in LuxR- and LuxRΔN-dependent activation. One determinant is likely to be involved in αCTD-DNA interactions and has at its core amino acid residues R265 and G296 (Fig. 5). These are surface-exposed αCTD residues and have been found to be two of the most critical for direct DNA interaction (18). L262 also plays an important role in DNA binding in the lux operon promoter; however, this residue is not surface exposed and may be involved indirectly due to resultant conformational changes of the αCTD. This amino acid could be necessary in accurate protein folding to allow other residues to contact the DNA (29). The fact that only a subset of all of the residues known to be involved in DNA binding by α are essential in the presence of LuxR may be an indication that LuxR is influencing the mode of αCTD-DNA recognition. A second α determinant requiring residues L290 and L314 for its function has also been identified in this study (Fig. 5) and is likely involved in αCTD-LuxR-specific interactions. Residues 290 and 314 are both leucine, an aliphatic molecule with a hydrophobic side chain, in the wild-type protein. Consequently, it does not seem likely that these residues are directly contacting the DNA. At least α variant L290A retained some ability to bind to the DNA as measured through gel mobility shift assays. No DNA binding activity was observed for the α variant L314A. This finding could be an artifact due to the limitations of the assay system used or might suggest a role for residue 314 in allowing either proper α conformation or α-α dimerization to occur. Suppressor analysis using positive control mutants in LuxR will be employed in future studies that will attempt to more completely define the nature of the specific protein-protein contacts that are required between the αCTD and the quorum-sensing transcription factor LuxR.
Acknowledgments
We thank Tamas Gaal, Richard Gourse, and Richard Ebright for generously providing strains, protocols, training, and advice.
This work was supported by grants from the CREST fund of the Japan Science Corporation to A.I., the Virginia Tech Undergraduate Biological Sciences Initiative to R. J.B., the Thomas F. and Kate Miller Jeffress Memorial Trust to A.M.S., and the National Science Foundation (CAREER Award MCB-9875479) to A.M.S.
REFERENCES
- 1.Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Harvanen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 316:501-516. [DOI] [PubMed] [Google Scholar]
- 2.Attey, A., T. Belyaeva, N. Savery, J. Hoggett, N. Fujita, A. Ishihama, and S. Busby. 1994. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the Escherichia coli galactose operon P1 promoter. Nucleic Acids Res. 22:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyaeva, T., J. Bown, N. Fujita, A. Ishihama, and S. Busby. 1996. Location of the C-terminal domain of the RNA polymerase alpha subunit in different open complexes at the Escherichia coli galactose operon regulatory region. Nucleic Acids Res. 24:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatter, E. E., W. Ross, H. Tang, R. L. Gourse, and R. H. Ebright. 1994. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute an independently folded domain capable of dimerization and DNA binding. Cell 78:889-896. [DOI] [PubMed] [Google Scholar]
- 5.Bokal, A. J., W. Ross, and R. L. Gourse. 1995. The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J. Mol. Biol. 245:197-207. [DOI] [PubMed] [Google Scholar]
- 6.Burns, H. D., A. Ishihama, and S. D. Minchin. 1999. Open complex formation during transcription initiation at the Escherichia coli galP1 promoter: the role of the RNA polymerase α subunit at promoters lacking an UP element. Nucleic Acids Res. 27:2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 8.Choi, S. H., and E. P. Greenberg. 1992. Genetic evidence for multimerization of LuxR, the transcriptional activator of Vibrio fischeri luminescence. Mol. Mar. Biol. Biotechnol. 1:408-413. [Google Scholar]
- 9.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. USA 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 11.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 12.Egland, K. A., and E. P. Greenberg. 2000. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 182:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 14.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 95:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrem, S. T., T. Gaal, W. Ross, Z. W. S. Chen, W. Niu, R. H. Ebright, and R. L. Gourse. 1999. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev. 13:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 18.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 19.Gourse, R. L., W. Ross, and T. Gaal. 2000. Ups and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg, E. P. 1997. Quorum sensing in gram-negative bacteria. ASM News 63:371-377. [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Igarashi, K., and A. Ishihama. 1991. Bipartite functional map of the E. coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 32:319-325. [DOI] [PubMed] [Google Scholar]
- 23.Ishihama, A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon, Y. H., T. Negishi, M. Shirakawa, T. Yamazaki, N. Fujita, A. Ishihama, and Y. Kyogoku. 1995. Solution structure of the activator contact domain of the RNA polymerase α subunit. Science 270:1495-1497. [DOI] [PubMed] [Google Scholar]
- 25.Kainz, M., and R. L. Gourse. 1998. The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J. Mol. Biol. 284:1379-1390. [DOI] [PubMed] [Google Scholar]
- 26.Kolb, A., K. Igarashi, A. Ishihama, M. Lavigne, M. Buckle, and H. Buc. 1993. E. coli RNA polymerase deleted in the C-terminal part of its α-subunit interacts differently with the cAMP-CRP complex at the lacP1 and galP1 promoter. Nucleic Acids Res. 21:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombardo, M., D. Bagga, and C. G. Miller. 1991. Mutations in rpoA affect expression of anaerobically regulated genes in Salmonella typhimurium. J. Bacteriol. 173:7511-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLeod, S. M., S. E. Aiyar, R. L. Gourse, and R. C. Johnson. 2002. The C-terminal domains of the RNA polymerase subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 316:517-529. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, K., N. Fujita, and A. Ishihama. 1996. Transcription factor recognition surface on the RNA polymerase α subunit is involved in contact with the DNA enhancer element. EMBO J. 15:4358-4367. [PMC free article] [PubMed] [Google Scholar]
- 30.Naryshkin, N., A. Revyakin, Y. Kim, V. Mekler and, R. H. Ebright. 2000. Structural organization of the RNA polymerase-promoter open complex. Cell 101:601-611. [DOI] [PubMed] [Google Scholar]
- 31.Niu, W., Y. Kim, G. Tau, T. Heyduk, and R. H. Ebright. 1996. Transcription activation at Class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell 87:1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 33.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strength: modular structure of bacterial promoters. J. Bacteriol. 180:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross, W., A. Ernst, and R. L. Gourse. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 15:491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosson, R. A., and K. H. Nealson. 1981. Autoinduction of bacterial bioluminescence in a carbon limited chemostat. Arch. Microbiol. 129:299-304. [Google Scholar]
- 36.Savery, N. J., G. S. Lloyd, M. Kainz, T. Gaal, W. Ross, R. H. Ebright, and R. L. Gourse. 1998. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase α subunit. EMBO J. 17:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savery, N. J., G. S. Lloyd, S. J. W. Busby, M. S. Thomas, R. H. Ebright, and R. L. Gourse. 2002. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase α subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 184:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens, A. M., and E. P. Greenberg. 1997. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 179:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens, A. M., N. Fujita, A. Ishihama, and E. P. Greenberg. 1999. Involvement of the α subunit of RNA polymerase in LuxR-dependent activation of the luminescence genes during quorum sensing. J. Bacteriol. 181:4704-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, A. M., and E. P. Greenberg. 1999. Transcriptional activation by LuxR, p. 231-242. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 42.Swift, S., J. P. Throup, P. Williams, G. P. C. Salmond, and G. S. A. B. Stewart. 1996. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214-219. [PubMed] [Google Scholar]
- 43.Tang, H., K. Severinov, A. Goldfarb, D. Fenyo, B. Chait, and R. H. Ebright. 1994. Location, structure, and function of the target of a transcriptional activator protein. Genes Dev. 8:3058-3067. [DOI] [PubMed] [Google Scholar]
- 44.Tang, H., K. Severinov, A. Goldfarb, and R. H. Ebright. 1995. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 92:4902-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood, L. F., N. Y. Tszine, and G. E. Christie. 1997. Activation of P2 late transcription of P2 Ogr protein requires a discrete contact site on the C-terminus of the α subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 274:1-7. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 48.Yasuno, K., T. Yamazaki, Y. Tonaka, T. S. Kodama, A. Matsugami, M. Katahira, A. Ishihama, and Y. Kyogoku. 2001. Interaction of the C-terminal domain of the E. coli RNA polymerase α subunit with the UP element: recognizing the backbone structure in the minor groove surface. J. Mol. Biol. 306:213-225. [DOI] [PubMed] [Google Scholar]
- 49.Zou, C., N. Fujita, K. Igarashi, and A. Ishihama. 1992. Mapping the cAMP receptor protein contact site on the α subunit of Escherichia coli RNA polymerase. Mol. Microbiol. 6:2599-2605. [DOI] [PubMed] [Google Scholar]