Abstract
The RpoS sigma factor (also called σS or σ38) is known to regulate at least 50 genes in response to environmental sources of stress or during entry into stationary phase. Regulation of RpoS abundance and activity is complex, with many factors participating at multiple levels. One factor is the nutritional stress signal ppGpp. The absence of ppGpp blocks or delays the induction of rpoS during entry into stationary phase. Artificially inducing ppGpp, without starvation, is known to induce rpoS during the log phase 25- to 50-fold. Induction of ppGpp is found to have only minor effects on rpoS transcript abundance or on RpoS protein stability; instead, the efficiency of rpoS mRNA translation is increased by ppGpp as judged by both RpoS pulse-labeling and promoter-independent effects on lacZ fusions. DksA is found to affect RpoS abundance in a manner related to ppGpp. Deleting dksA blocks rpoS induction by ppGpp. Overproduction of DksA induces rpoS but not ppGpp. Deleting dksA neither alters regulation of ppGpp in response to amino acid starvation nor nullifies the inhibitory effects of ppGpp on stable RNA synthesis. Although this suggests that dksA is epistatic to ppGpp, inducing ppGpp does not induce DksA. A dksA deletion does display a subset of the same multiple-amino-acid requirements found for ppGpp0 mutants, but overproducing DksA does not satisfy ppGpp0 requirements. Sequenced spontaneous extragenic suppressors of dksA polyauxotrophy are frequently the same T563P rpoB allele that suppresses a ppGpp0 phenotype. We propose that DksA functions downstream of ppGpp but indirectly regulates rpoS induction.
Eubacteria have developed complex regulatory networks that recognize and respond to a variety of environmental sources of physiological stress. One element common to many such networks in gram-negative bacteria is RpoS (29), a regulator defined by sequence and functional studies as an alternative sigma subunit of RNA polymerase (38). Over the past decade, it has come to be appreciated that RpoS participates in the regulation of at least 50 genes and that RpoS is itself regulated by nearly half as many factors. RpoS has been referred to as “the master regulator of the general stress response in Escherichia coli” (6).
Regulation of RpoS itself is arguably the most complicated system in bacteria. Regulation of RpoS involves transcription, mRNA turnover, translation initiation, and proteolysis. Reported transcription regulators include BarA (37), cyclic AMP/cyclic AMP receptor protein (30), and ppGpp (27). Leader mRNA is a regulatory target affecting efficiency of translation initiation in different ways. With the rpoS transcript originating in nlpD and the initiating AUG at +565 (50), a structure extending from +458 to +565 sequesters the ribosomal binding sequence through a much smaller cis-acting antisense element. One hypothesis is that translation initiation is positively regulated by hfq (7, 36) with HF-1 binding to leader RNA changing antisense element conformation (8). A small RNA, called DsrA, is normally made only at low temperatures where it functions as an antiantisense element (34), but RNA binding also stabilizes rpoS mRNA turnover, whereas binding to hns mRNA makes it more labile (30). Turnover of DsrA is itself stabilized by HF-1 binding (48). Another small RNA, called RprA, when present in multicopy can stimulate translation in dsrA mutants, but RprA is not complementary to the antisense element, and its site of action is uncertain (33). DNA binding proteins are also involved. The HU protein binds the rpoS leader region and stimulates translation (2), while H-NS inhibits RpoS induction (3) in a manner antagonized by DsrA (31). OxyS is a third small RNA inhibiting rpoS translation by binding HF-1 in competition with rpoS leader binding (56). Turnover of RpoS protein by the ClpXP protease is strikingly limited during entry into stationary phase (47, 55), which is sensitized by phosphorylation of the RssB protein affecting a specific interaction with a region of the RpoS protein (5, 57).
Examples of stress that trigger RpoS regulation include (i) starvation for sources of carbon, nitrogen, or phosphate; (ii) steady-state growth on secondary metabolites; (iii) the transition from exponential growth in rich media into stationary phase accompanying complex-nutrient exhaustion; (iv) hyperosmotic shock; (v) cold shock; (vi) acid shock; (vii) heat shock; and (viii) oxidative damage (for reviews, see references 21 and 32). Still at issue is a clear understanding of exactly how many sources of stress are linked to RpoS regulation through the factors cited above. A plausible participant in this regard is ppGpp, because cellular levels of ppGpp increase under the first five stress conditions listed above with weak or otherwise qualified responses to the last three (for a review, see reference 10). Sources of stress for induction of ppGpp are now thought equivalent to those provoking induction of the “universal stress protein” UspA (16, 42). Measurements of RpoS abundance by immunoblots revealed a 25- to 50-fold increase in RpoS when ppGpp was induced without starvation and showed that a complete ppGpp0 deficiency blocks RpoS induction during starvation; furthermore, rpoD suppressors of a ppGpp0 deficiency phenotype are found to suppress this latter defect in RpoS accumulation (18, 22). It should be pointed out that an early report concluded that negative effects of a ppGpp deficiency on lacZ reporters for rpoS expression are exerted at the level of transcription (27). From the work presented here, it is concluded that the major effects of ppGpp induction are not exerted on rpoS mRNA abundance or on protein turnover but instead are exerted on translational efficiency. A report (51) stating that dksA is required for RpoS induction during stationary phase entry and acid shock by affecting translational efficiency at a site distinct from the antisense stem has led us to find parallels between dksA effects and ppGpp induction of RpoS. The mnemonic for the dks gene is dnaK suppressor, because it is a dosage-dependent suppressor of a dnaK deletion (25); it was subsequently found that dksA has activities in addition to its effects on rpoS (see Discussion).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains, plasmids, and oligonucleotide primers are shown in Table 1. We chose to use strain MG1655 instead of MC4100 (commonly used to study rpoS) because we have observed that MG1655 is more tolerant to high levels of ppGpp. We found that the weak uracil requirement in MG1655 due to an rph frameshift mutation (24) is somewhat enhanced by the dksA mutation; therefore, uracil was always added for growth of these mutants. The enriched minimal medium used for immunoprecipitation experiments consisted of morpholinepropanesulfonic acid (MOPS) medium (40) with 0.4% glucose and supplements at recommended concentrations (13). These are biotin, diaminopimelic acid, nicotinic acid, pantothenic acid, pyridoxine, thiamine, adenine, guanine, cytosine, uracil, and thymine as well as all amino acids except methionine and cysteine. Cultures were grown at 37°C with ampicillin present (100 μg/ml) for plasmid maintenance. Overnight cultures were diluted 1/2,000 into prewarmed medium and were grown to an A600 of at least 0.5 and were then diluted with prewarmed medium to an A600 of approximately 0.1 and grown to an A600 of about 0.5. This procedure assured that the cultures were in exponential phase at the start of the experiment and that ppGpp basal levels were at the lowest levels supported by a given medium. In labeling experiments using ppGpp0 strains, the chance of reversion was minimized by resuspending a few colonies from an overnight plate into a small volume of fresh medium and diluting to an A600 of about 0.005. These cultures were grown to an A600 of at least 0.5 and were then diluted to an A600 of about 0.1 and grown again to an A600 of about 0.5.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Phenotype, description or sequence | Source, reference, or description |
|---|---|---|
| Strains | ||
| CF1648 | MG1655 wild type | 52 |
| CF1652 | MG1655 ΔrelA251::kan | 52 |
| CF1693 | MG1655 ΔrelA251::kan ΔspoT207::cat | 52 |
| CF7968 | MG1655 Δlac (rph+) | Cashel lab |
| CF9084 | CF7968 ΔrelA255::cat | Cashel lab |
| CF9992 | CF9084 ΔrelA255::cat ΔdksA::tet | This work |
| TE8114 | MG1655 ΔdksA::tet | This work |
| PK201 | MG1655 ΔdksA::kan | 25 |
| TE6406 | MG1655 ΔlacX74 trp::put::kanR-rpoS-lac (fusion A) | 11 |
| TE2680 | F− λ− IN(rrnD-rrnE)1 Δ(lac)X74 rpsL galK2 recD1903::Tn10d-Tet trpDC700:: putPA1303::(Kans-Camr-lac) | 14 |
| TE6608 | TE2680 trp::put::kan Ptac-rpoS-lacΔ2 (fusion J) | 14 |
| TE6798 | TE2680 trp::put::kan Ptac-rpoS-lac (fusion F) | 14 |
| TE6987 | TE2680 trp::put::kan PlacUV5c-rpoS-lac (fusion K) | 14 |
| TE6989 | TE2680 trp::put::kan PlacUV5-rpoS-lacΔ2 (fusion M) | 14 |
| CF9993/CF10003 | CF9084/CF9992 trp::put::kan-rpoS-lac (fusion A)/ΔdksA | This work |
| CF9995/CF10005 | CF9084/CF9992 trp::put::kan Ptac-rpoS-lacΔ2 (fusion J)/ΔdksA | This work |
| CF9997/CF10007 | CF9084/CF9992 trp::put::kan Ptac-rpoS-lac (fusion F)/ΔdksA | This work |
| CF9999/CF10009 | CF9084/CF9992 trp::put::kan PlacUV5c-rpoS-lac (fusion K)/ΔdksA | This work |
| CF10001/CF10011 | CF9084/CF9992 trp::put::kan PlacUV5c-rpoS-lacΔ2 (fusion M)/ΔdksA | This work |
| CF10013/CF10018 | CF9084/CF9992 trp::put::kan rpoS-lac (fusion A) pLB8/ΔdksA | This work |
| CF10014/CF10019 | CF9084/CF9992 trp::put::kan Ptac-rpoS-lacΔ2 (fusion J) pLB8/ΔdksA | This work |
| CF10015/CF10020 | CF9084/CF9992 trp::put::kan Ptac-rpoS-lac (fusion F) pLB8/ΔdksA | This work |
| CF10016/CF10021 | CF9084/CF9992 trp::put::kan PlacUV5c-rpoS-lac (fusion K) pLB8/ΔdksA | This work |
| CF10107/CF10022 | CF9084/CF9992 trp::put::kan PlacUV5c-rpoS-lacΔ2 (fusion M) pLB8/ΔdksA | This work |
| Plasmids | ||
| pALS13 | Ptac-relA′ (RelA 1-455) Apr | 49 |
| pLB8 | PBAD-relA′ (RelA 1-394) Apr | This work |
| pJK537 | dksA in pBR322 Apr | 25 |
| pMMKatF3 | rpoS | 39 |
| pGN66 | bolA promoter plasmid | 41 |
| Primers | ||
| DG62 | GGGTAGGAGCCACCTTATGAGTCAGAATAC | rpoS probe, −16 to +14 with AUG +1 |
| DG63 | GGATCCTAATACGACTCACTATAGGGAGGGGGTAAAGCGAGTC GCGTCCAACACACGCT | rpoS probe, 154 to 183 with AUG +1, pT7 underlined |
| DG22 | GCGGGATCCTGCTGTGGCAGT | bolA p2 probe, −364 to −384 with AUG +1 |
| DG71 | GGATCCTAATACGACTCACTATAGGGAGGGCGATCGCTGACAG ACAAC | bolA p2 probe, 153 to 174 with AUG +1, pT7 underlined |
A ΔdksA::tet insertion was constructed by amplifying the Tn10 tetAR genes from pWM7 (12) by PCR using the E. coli dksA tet up and E. coli dksA tet down primers described in Table 1. The resulting PCR product was recombined into the E. coli chromosome (54).
The pLB8 plasmid was used instead of pALS13 to induce rpoS-lac fusion strains, because even uninduced levels of ppGpp from pALS13 inhibited growth of these strains. Plasmid pLB8 was constructed from pALS10 to contain the N-terminal 394 residues of RelA. This RelA′ fragment is constitutively active, similar to pALS13 with 455 residues of RelA (49) but under the control of the PBAD arabinose-inducible promoter from plasmid pBAD22A (20). The ΔrelA255::cat allele was constructed by deleting sequences between the AUG and TAA codons of relA and substituting a chloramphenicol resistance element.
For experiments where RpoS or rpoS-lac expression was measured during growth into stationary phase, overnight cultures were diluted 1/200 into either Luria broth (LB) medium or MOPS minimal medium containing 0.4% glucose, each of the 20 amino acids at 40 μg/ml, and 20 μg each of adenosine, cytidine, guanosine, uridine, and thymidine per ml. Cultures were grown to an A600 of approximately 0.2 and were then diluted to an A600 of 0.02 and sampled when cultures reached an A600 of 0.2 to 0.3.
For experiments where ppGpp was overexpressed from either pALS13 (49) or pLB8, overnight cultures were diluted 1/200 into LB medium, grown to an A600 of approximately 0.2, and diluted again to an A600 of 0.02. At an A600 of approximately 0.2, ppGpp was induced by adding either 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (pALS13) or 0.2% arabinose (pLB8). Samples were taken at various times following induction and analyzed by either Western blot or β-galactosidase assay.
RNA isolation and RNase protection assays.
Isolation of RNA at different points during culture growth of CF1648 bearing pALS13 was by a protocol using an acid phenol/guanidinium thiocyanate solution (TriReagent; Molecular Research Center, Inc.), pelleting cells from 1.0- to 5.0-ml aliquots, and resuspending in 1.0 ml of TriReagent. The suspension was incubated for 5.0 min at 65°C, cooled to room temperature, extracted with 200 μl of chloroform, and RNA precipitated from the aqueous phase by the addition of 1 volume of isopropanol. Precipitated RNA was washed with 70% ethanol, dried, and resuspended in 10 μl of hybridization buffer (Ambion). In most cases, RNA was isolated from E. coli liquid cultures using the boiling sodium dodecyl sulfate (SDS) method (15). Where appropriate, an external standard of 5.0 μg of mouse liver RNA (Ambion) was added to each sample after boiling in SDS.
RNase protection assays were performed using RNA probes synthesized using T7 RNA polymerase and templates generated by PCR using primers indicated in Table 1. Primers were designed such that a T7 RNA polymerase promoter was included in the PCR fragment allowing for transcription of a cRNA. The bolA p2 upstream probe was synthesized using T7 RNA polymerase and DraI-digested pGN66 (41). RNA probes were synthesized using a kit from Ambion as directed; an enclosed template for the mouse actin gene was used to synthesize the external standard. Synthesized RNA probes were purified from unincorporated nucleotides and RNase protection assays were performed using the RPAII kit (Ambion) as directed by the manufacturer, except that the RNAs were not precipitated. Digested RNAs were separated on 8% polyacrylamide urea gels. Bands corresponding to rpoS transcripts were visualized by autoradiography and quantitated by scanning. For most experiments the level of rpoS-specific transcripts was normalized to the external standard. Assays were performed in duplicate and generally varied by less than 5%. When transcripts were normalized to total RNA, 2.5 μg of total RNA was used in each assay. Pilot experiments revealed that the assay was linear from less than 1.0 μg up to 10 μg of total RNA.
Labeling cellular proteins.
For determining protein stability and rates of translation, cultures of CF1648 bearing pALS13 were labeled with a mixture of [35S]methionine and cysteine ([35S]Met-Cys mixture; Trans [35S] Label; ICN). RpoS decay rates were measured in log-phase cultures grown in supplemented MOPS medium, labeled with 100 μCi of [35S]Met/Cys per ml for 2.0 min, and were then chased with unlabeled methionine and cysteine (10 mM each). Samples were precipitated with 5% trichloroacetic acid (TCA), immunoprecipitated, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Translation rates were measured using log-phase cultures grown in supplemented MOPS media. Samples were removed at various times before or after IPTG addition, pulse-labeled with [35S]Met/Cys for 30 or 60 s, immunoprecipitated (see below), and similarly processed by SDS-PAGE. The effect of adding IPTG on total protein synthesis was measured as radioactivity that was obtained after similar pulse-labeling but was recovered on nitrocellulose filters after TCA precipitation.
Immunoprecipitation.
Appropriately labeled samples were TCA precipitated and resuspended in 75 μl of 50 mM Tris-acetate, 5 mM EDTA, and 1% SDS, pH 8.0. Immunoprecipitation (23) was by adding 50 μl of a 50% suspension of Staph A cells (Boehringer Mannheim) and mixing the tubes end over end for 1 to 2 h (25°C). The precipitates were washed five times with immunoprecipitation buffer (50 mM Tris-acetate, 0.15 M NaCl, 0.1 mM EDTA, 2% Triton X-100, and 0.2% sodium deoxycholate), once with PBS (137 mM NaCl, 2.7 mM KCl, 4.2 mM Na2HPO4, and 1.4 mM KH2PO4, pH 7.1) and were washed once with PBS + 1.0 M NaCl, and a final wash with PBS was performed. The immunocomplexes were dissolved in 100 μl of SDS sample buffer (3% SDS, 5% 2-mercaptoethanol, 10% glycerol, and 63 mM Tris-HCl, pH 6.8), boiled for 2.0 min, and centrifuged for 2.0 min. Labeled proteins were then separated by SDS-PAGE, fixed with 5% TCA, and dried, and activity was measured by scanning.
Sample preparation for Western analysis.
Protein samples from liquid cultures (see Fig. 4, 5, and 7) were prepared by removing 1.0 ml of liquid culture that was centrifuged and immediately resuspended at a concentration of 0.01 A600 units/μl in SDS sample buffer (3% SDS, 5% 2-mercaptoethanol, 10% glycerol, and 63 mM Tris-HCl, pH 6.8), boiled for 2 min, and applied to SDS-9 or 15% polyacrylamide gels. The resolved proteins were electroblotted to polyvinylidene difluoride membranes and probed. RpoS was detected using the 1RS1 monoclonal antibody (41), DksA was detected using a polyclonal antibody from Diana Downs, and RelA was detected using a polyclonal antibody made in this laboratory. An anti-mouse horseradish peroxidase-conjugated secondary antibody was used with the supersignal chemiluminescent substrate (Pierce). Each experiment was repeated twice with independent cultures, and samples within a given experiment were run twice.
FIG. 4.
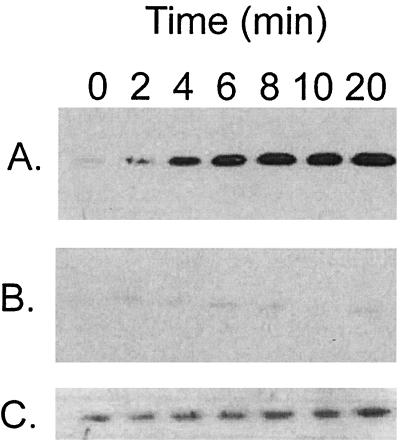
A dksA mutation impairs induction of RpoS by overproduction of ppGpp in exponential phase. Cultures were grown in LB medium to an A600 of 0.3 when IPTG was added to induce ppGpp (time zero), and timed samples were assayed for RpoS by Western blotting. (A) CF1648 (wild type/pALS13). (B) TE8114 (ΔdksA/pALS13). (C) Overexposure of the blot in panel B in order to visualize the small amount of RpoS detected.
Measurement of ppGpp.
Assays for ppGpp (9) involved cells uniformly labeled with 100 to 200 μCi of H332PO4 per ml of MOPS medium supplemented as before but containing 0.4 mM phosphate and 40 μg each of 19 amino acids per ml, omitting serine. Serine hydroxamate (1 mg/ml) was added at an A600 of about 0.3, and samples were taken at the indicated times by diluting into an equal volume of 13 M formic acid. They were then frozen and thawed three times and centrifuged, and duplicate samples of the supernatant were spotted onto polyethyleneimine-cellulose thin-layer plates. Nucleotides were separated by chromatography in 1.5 M KPi (pH 3.4). Radiolabeled nucleotides were visualized by exposure to X-ray film and were quantitated by scanning on a PhosphorImager. These experiments were repeated twice.
Measurement of RNA accumulation.
The accumulation of RNA in wild-type, relA, and dksA strains following starvation for serine was determined as the alkali-labile fraction of TCA-precipitable [32P] activity from uniformly labeled cultures (17) quantitated on a PhosphorImager. The alkali-labile activity is the difference in duplicate TCA-precipitable activities before and after hydrolysis in 0.33 M NaOH (17).
β-Galactosidase assays.
Overnight cultures were diluted three times in LB (see above) to insure exponential growth and were induced with 0.2% arabinose at an optical density at 600 nm of 0.2 to 0.3 and with sampling thereafter. Enzyme activity assays were measured at 30°C with chloroform-permeabilized cells in Z buffer containing 50 mM β-mercaptoethanol by a kinetic method using an automated plate reader. Specific activities of duplicate samples, each assayed twice, were calculated as averages of the change in A420 per minute normalized to the cell density (A650) of the actual sample and with correction for controls lacking enzyme (35). These experiments were repeated in their entirety at least three times with similar results.
RESULTS
Overproduction of ppGpp does not lead to increased transcription of rpoS.
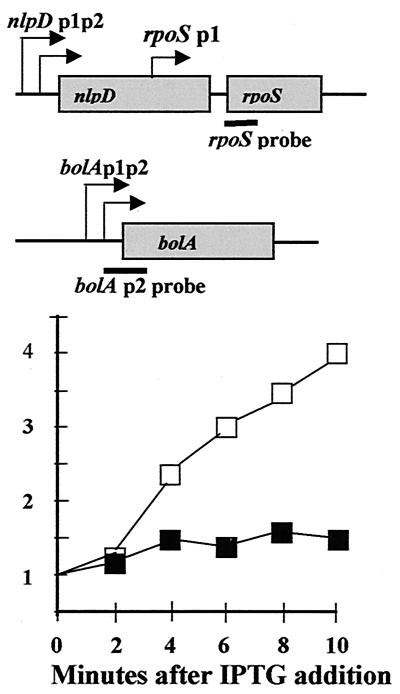
The starting point of this work is to understand how gratuitous induction of ppGpp by IPTG in cultures growing exponentially in LB leads to about a 50-fold increase in abundance of RpoS protein within 10 min that is virtually coincident with ppGpp accumulation (19). Given that ppGpp is a known regulator of transcription (10) and was previously reported to induce rpoS-lac operon fusion activity (27), we first measured induction effects of ppGpp on transcription by assaying rpoS mRNA content as RNase-resistant hybrids (Fig. 1, filled symbols). Since RpoS protein induction could not be verified in these same RNA samples, hybrids to bolA transcripts were used as an internal control to document induction (Fig. 1, open symbols). The figure shows that, 10 min after IPTG addition, only a 50% increase in the abundance of rpoS transcripts occurs while bolA transcript levels increase about fourfold. The increase in bolA transcripts is taken to indicate that induction of RpoS occurs, very probably accompanied by direct stimulatory effects of ppGpp (1, 26).
FIG. 1.
Overproduction of ppGpp does not stimulate rpoS transcription. Upper panel, the rpoS and bolA operons are shown schematically with black bars indicating mRNA regions hybridizing to RNA probes. Graph, activities measured at various times in minutes are normalized to an actin RNA external standard. Filled symbols = rpoS probe hybrids. Open symbols = bolA probe hybrids. Induction of ppGpp was achieved by adding IPTG to exponentially growing cells bearing plasmid pALS13 to overexpress a fragment of the RelA protein that is constitutive for ppGpp synthetic activity.
Induction of ppGpp does not alter RpoS protein stability.
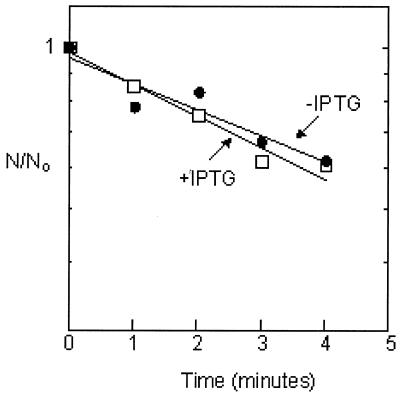
Since entry into the stationary phase increases RpoS levels by stabilizing the RpoS protein (see the introduction), it is plausible that ppGpp induction might act similarly. We estimated the metabolic stability of RpoS before and after ppGpp induction as the rate of decay of activity immunoprecipitated with anti-RpoS antibody and resolved by SDS-PAGE during a chase of [35S]Met/Cys pulse-labeled RpoS. In order to obtain the desired specific activities, it was necessary to perform the induction using supplemented MOPS media lacking methionine or cysteine instead of LB (see Materials and Methods). Figure 2 reveals that ppGpp overproduction had little effect on RpoS degradation rates: a half-life for RpoS of about 5 min before induction was found to increase to 6 min after exposure to IPTG for 10 min.
FIG. 2.
Overproduction of ppGpp does not affect RpoS stability. Metabolic decay of pulse-labeled RpoS was measured during a chase imposed either before (open squares) or 10 min after (closed circles) induction of ppGpp. The apparent rates of exponential decay of activities lead to estimates of the RpoS half-life as about 5 min before and 6 min after IPTG induction.
Induction of ppGpp specifically increases the rate of RpoS translation.
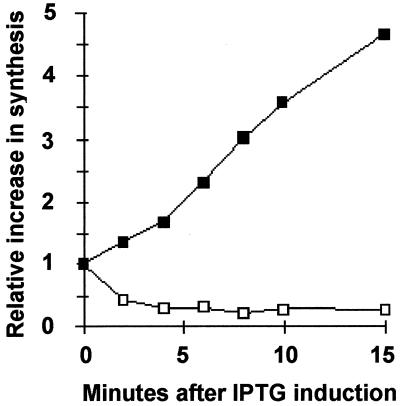
Since regulation of RpoS at a posttranscriptional level has been observed repeatedly (see the introduction), we next asked whether ppGpp induction under these growth conditions might alter the instantaneous rate of rpoS mRNA translation, again using enriched MOPS media. For these estimates, for each IPTG induction time point, two culture samples were labeled with [35S]Met/Cys, one for 30 and the other for 60 s. Samples were then TCA precipitated and immunoprecipitated with anti-RpoS antibody, and the labeled RpoS protein was resolved by SDS-PAGE and activities were scanned. Initial rates of RpoS labeling as a function of IPTG induction times are shown in Fig. 3. The rate of RpoS labeling apparently increases linearly for at least 15 min after IPTG induction; after 10 min the induced rate is about 3.5-fold more than the uninduced rate (Fig. 3, filled symbols). As previously reported (49), ppGpp overproduction by IPTG induction is again found here to inhibit the instantaneous total protein synthesis rate as judged by TCA-precipitable activities of cells pulse-labeled with [35S]Met/Cys mixtures (Fig. 3, open symbols). Since mRNA levels and RpoS metabolic turnover are each modestly affected by ppGpp overproduction (Fig. 1 and 2), the differential effects of selective activation of RpoS translation coupled with severe inhibition of overall translation seem to largely account for the previously observed dramatic elevation of RpoS protein levels by ppGpp (19).
FIG. 3.
Overproduction of ppGpp increases specific rates of RpoS synthesis while decreasing overall rates of protein synthesis. At times indicated after IPTG addition, samples were pulse-labeled with [35S]Met/Cys for 30 and 60 s and initial rates of synthesis (plotted) were determined from the slopes. Activities associated with RpoS by SDS-PAGE of immunoprecipitates were used to estimate a rate of RpoS synthesis for each induction time normalized to rates of synthesis before IPTG addition (closed squares). Total protein synthesis rates (open squares) were estimated from the slope obtained for total TCA-precipitable activity recovered on filters after both pulses without immunoprecipitation, normalized to rates before IPTG addition.
A dksA deletion mutation blocks induction of rpoS by ppGpp.
Although ppGpp apparently provokes significant differential activation of rpoS mRNA translation, reports of ppGpp regulatory effects on the many factors known to alter translational efficiency of RpoS have not appeared in the literature (see the introduction). This has led us to search for components involved in ppGpp induction of RpoS. One candidate is DksA, found in Salmonella enterica serovar Typhimurium to be required for the induction of rpoS in stationary phase (51). The effect of a dksA deletion on the ability of ppGpp to induce RpoS was examined when ppGpp is overproduced from pALS13 by IPTG addition to exponentially growing cells. The ΔdksA mutant does show an induction defect (Fig. 4B) in comparison to wild-type strain behavior (Fig. 4A). The induction defect may be even more severe than apparent in Fig. 4B, because the trace of RpoS present in the dksA deletion strain hardly increases during induction when the blot is overexposed (Fig. 4C).
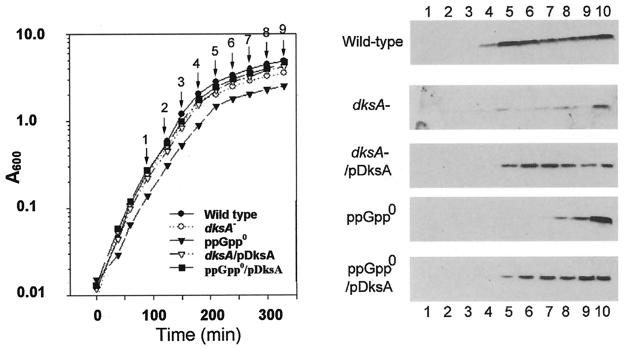
We next examined the effects of a dksA mutation on accumulation of RpoS during the early transition between the exponential growth phrase in LB and stationary phase. Induction of RpoS under this condition is known to require the presence of ppGpp (19), and this dependence is again verified here: compare the wild-type and ppGpp0 blots in the right panel of Fig. 5. We also verify that accumulation of RpoS occurs even in a ppGpp-deficient host late in stationary phase and in overnight cultures. Comparing the two uppermost blots in Fig. 5 reveals that the ΔdksA E. coli mutant impairs RpoS accumulation during entry into stationary phase, as it does in S. enterica serovar Typhimurium (49). It is notable that RpoS levels are not as severely affected in mutant overnight cultures (Fig. 5, sample 10). The middle blot of Fig. 5 reveals that the RpoS accumulation defect of the deletion mutant is almost fully complemented by a pBR322-derived plasmid expressing DksA (pJK537), which indicates the absence of possible polar effects of the insertion allele. The two lower blots in the figure indicate that multicopy expression of DksA also largely complements the defect in RpoS accumulation occasioned by a complete deficiency of ppGpp. Growth of all strains shown in the left panel of Fig. 5 is taken to be comparable.
FIG. 5.
RpoS induction during growth into stationary phase is affected by DksA. Left, LB culture growth. Samples 1 to 9 were taken at the points indicated by the arrows, while sample 10 represents an overnight culture. Right, RpoS Western blots of cultures from top to bottom: wild type (MG1655); dksA− (TE8114 = MG1655 but dksA::tet); dksA−/pdksA (TE8114/pJK537); ppGpp0 (CF1693 = MG1655 but ΔrelA ΔspoT); and ppGpp0/pdksA (CF1693/pJK537).
The dksA deletion mutant does not impair either relA expression, ppGpp regulation, or RNA control during the stringent response.
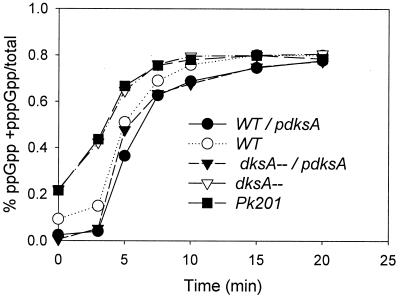
Since dksA in multicopy can suppress a variety of mutations (see Discussion), we tested whether the ΔdksA strain might lower expression of relA. The same blots shown in Fig. 5 were stripped and reprobed with antibody to RelA. The amount of RelA was found to be similar for the wild type and the dksA mutant strain (data not shown). We also measured the accumulation of ppGpp during a stringent response, due to serine hydroxamate addition, in the wild type and a ΔdksA strain (Fig. 6). Cultures were uniformly labeled with 32Pi, and levels of ppGpp were determined by polyethyleneimine-cellulose thin-layer chromatography. Both the wild-type and the dksA mutant cultures show similar patterns of ppGpp induction. One difference found is that the basal level of ppGpp is significantly elevated in dksA mutants before triggering the stringent response (Fig. 6). This effect on basal levels is the opposite of what one would expect if the defective RpoS accumulation of the dksA mutant were due to impaired ppGpp accumulation. Such a change might be imagined as an adaptation in response to putative dksA mutant impairments of either positive or negative control by ppGpp. We next asked whether negative regulation of stable RNA accumulation during the stringent response was affected by the dksA deletion. Inhibition of stable RNA accumulation during the stringent response was measured as alkali-soluble, TCA-precipitable radioactivity from wild-type and dksA strains after the addition of serine hydroxamate. No significant difference between the wild type and an otherwise isogenic ΔdksA mutant was found for the inhibition of RNA accumulation during the stringent response (data not shown). We conclude that negative control of RNA by ppGpp is unaffected in a dksA mutant.
FIG. 6.
DksA does not alter ppGpp regulation during the stringent response. Cultures were grown and uniformly labeled with 32Pi in MOPS minimal glucose medium, five nucleosides, and all amino acids except serine. At time zero, 1 mg of serine hydroxamate/ml was added and the culture was assayed for guanine nucleotide content at various times thereafter. The amount of ppGpp is normalized to the sum of GTP, ppGpp, and pppGpp.
Accumulation of DksA protein with single-copy and multicopy dksA is unaltered by a ppGpp deficiency.
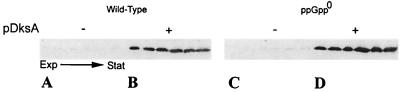
Since expression of dksA from a multicopy plasmid can complement the ppGpp0 defect for RpoS induction during the transition into stationary phase, it seemed possible that ppGpp could be a positive regulator of DksA and thereby induce RpoS. We attempted to determine the effect of a ppGpp deficiency on DksA expressed from a single chromosomal gene as well as expressed from its natural promoter in multicopy from pJK537, a pBR322 derivative bearing dksA (25). Marginally detectable levels of DksA were found by Western blots of extracts of wild-type cells undergoing the transition from exponential growth phrase to stationary phase (Fig. 7A). With multicopy dksA present in the same wild-type strain, DksA was easily visualized, but the levels did not change during the transition (Fig. 7B). No differences in DksA levels were found for the same comparisons in the ppGpp0 host CF1693 (Fig. 7C and D). These results suggest that ppGpp is probably not a positive regulator of dksA expression. The possibility that ppGpp might enhance a relevant specific activity of DksA without changing dksA expression cannot be ruled out.
FIG. 7.
The presence or absence of ppGpp does not alter the DksA accumulation. Cultures of strains A to D were grown in LB medium, and in each case six samples were taken representing the growth transition to stationary phase as in Fig. 5. The DksA content was analyzed by Western blots. (A) CF1648 = MG1655 (wild type); (B) CF1648/pdksA; (C) CF1693 ΔrelA ΔspoT ppGpp0; and (D) CF1693/pdksA. The presence of the pdksA plasmid is indicated by a plus at the top of sample series B and D.
RpoS-Lac protein fusion evidence that the dksA requirement for ppGpp induction of RpoS depends on the mRNA far upstream of the leader region but not on PrpoS or RpoS protein turnover.
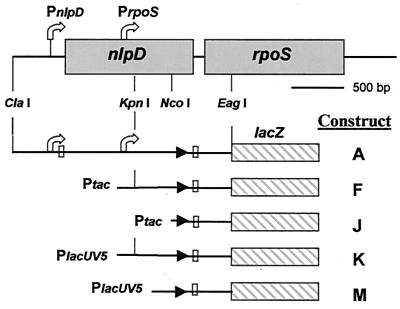
We have measured the effects of deleting dksA on RpoS induction using a previously constructed set (Fig. 8) of single-copy rpoS-lac protein fusions insensitive to degradation by the ClpX proteolytic pathway (11). Construct F simply substitutes the Ptac promoter for the major rpoS promoter within nlpD and removes all upstream sequences. Construct K substitutes the PlacUV5 promoter for the preceding promoter as well as deleting 72 nucleotides (nt) of the far upstream leader to the DNA KpnI site. The remaining members of the fusion set (constructs J and M) have deleted 454 nt of the leader, leaving only about 110 nt upstream of the intact sense-antisense region and the rpoS AUG codon. The effects of a dksA deletion on reporter activities have been measured during growth into stationary phase (Fig. 9) as well as when ppGpp was induced with arabinose from a multicopy plasmid (Fig. 10).
FIG. 8.
Schematics of rpoS-lac fusions. The top line shows diagrams of the nlpD and rpoS genes of E. coli. Native rpoS is transcribed from two promoters indicated by the bent arrows. The lower lines represent the different protein fusions made by Cunning et al. (11) that were transduced into an rph+ MG1655 derivative with an internal lacIZ deletion (CF7968) for analysis. The AUG start codons are indicated by open squares, and stop codons are indicated by arrowheads.
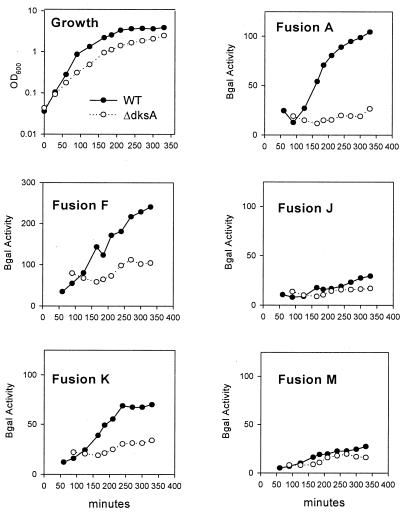
FIG. 9.
β-Galactosidase assays of rpoS-lac fusions in a dksA deletion mutant during growth into stationary phase. (A) Cultures were grown in MOPS minimal media with glucose, all amino acids, and the five nucleosides. Upper left panel (Growth) shows typical growth for each of the different fusions. Remaining labeled panels show β-galactosidase-specific activities for each fusion construct assayed. OD600, optical density at 600 nm.
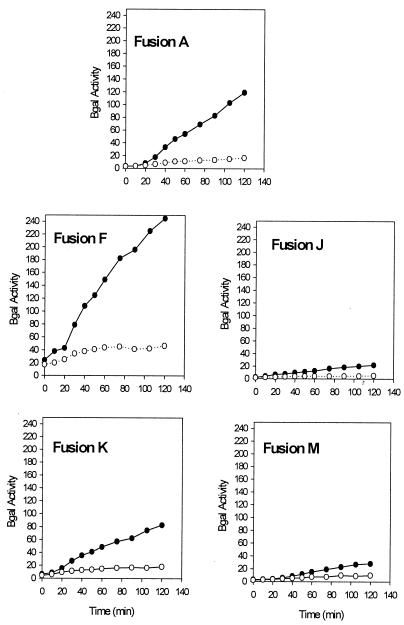
FIG. 10.
β-Galactosidase assays of rpoS-lac fusions after induction of ppGpp. Cultures were grown in LB medium, and 0.2% arabinose was added at an A600 of 0.3 (time zero) to induce relA expression from pRelA. Samples thereafter were assayed for β-galactosidase. Specific activities for each fusion are shown. Filled circles, wild type; and open circles, ΔdksA.
Reporter activities from fusion A indicate that a dksA deletion severely impairs RpoS induction under both sets of growth conditions. This is consistent with Western blots already presented, as well as with our estimates from pulse-chase experiments that RpoS turnover does not appreciably change during ppGpp induction.
When the major rpoS promoter is replaced without changing the initiation site for the Ptac construct (Fig. 9 and 10, fusions A and F), RpoS is still apparently induced in a ppGpp- and dksA-dependent manner. The PlacUV5 promoter construct K deletes the first 72 nt of the rpoS leader, and quantitative differences are evident (see Discussion). Somewhat more marked dependences on dksA are displayed during induction of ppGpp than during entry into stationary phase for fusions F and K (compare Fig. 9 and 10).
With fusions J and M, the Ptac and PlacUV5 promoters are moved closer to the initiation codon for RpoS translation but with equal-length transcripts that still allow folding of leader region sense and antisense elements that limit translation initiation. Reporter activities are very low for fusions J and M, as if RpoS induction is largely blocked both during growth into stationary phase as well as during ppGpp induction (Fig. 9 and 10, fusions J and M). The degree of dependence on dksA seems weak, but it is difficult to assess because of the low reporter activities of these fusions.
Growth phenotypes of dksA mutants have similarities to those of ppGpp0 strains.
Strain MG1655 with a ΔdksA::kan allele (strain PK201) was reported to grow on LB but not on M9 minimal medium containing all 20 amino acids and vitamins despite testing of several carbon sources (25). The deletion in PK201 extends into portions of the sfsA and yadB genes that flank dksA and comprise a possible operon. However, the dksA phenotype is probably not due to polar effects, since complementation occurs with plasmid pJK537, a pBR322 derivative bearing dksA as well as only small portions of sfsA and yadB (25). We found that supplementing either the ΔdksA::tet or the ΔdksA::kan mutant derivative of MG1655 with uracil (24) did allow growth on complete amino acid mixtures but not on single amino acids. The individual requirements for strains PK201 as well as TE8114 then were found to be leucine, valine, glycine, phenylalanine, and threonine using amino acid dropout plates consisting of M9 mininal glucose plates containing uracil and all amino acids except one (Table 2). No growth was found with 19 amino acids minus isoleucine, leucine, valine, glycine, phenylalanine, or threonine. The apparent isoleucine requirement is expected on dropout plates, since E. coli K12 derivatives are sensitive to valine in the absence of isoleucine. Rapid growth was not obtained by supplementation with only these five amino acids (L, V, G, F, and T) whether or not isoleucine was present as a sixth amino acid. Strain MG1655 with a deletion of relA and spoT on similar dropout plates requires the same five amino acids in addition to requiring arginine, histidine, and serine (52). Thus, in the MG1655 strain background, requirements for six of the nine amino acids by ppGpp0 derivatives are shared by dksA mutants. The dksA mutation also abolishes aminotriazole resistance but without generating a histidine requirement, as if positive regulation of the histidine operon was compromised despite the presence of wild-type relA and spoT genes (46). Although dksA in multicopy complements the amino acid requirements of dksA mutants, it does not allow growth of the ppGpp0 strain CF1693 on glucose minimal medium (Table 2). The presence of the pJK537 (dksA) plasmid does suppress the aminotriazole sensitivity of a dksA mutant but does not suppress the aminotriazole sensitivity of ppGpp0 strain CF1693 (Table 2).
TABLE 2.
Multicopy dksA suppresses a ΔdksA deletion phenotype and some features of a ppGpp-dependent phenotypea
| Strain | AT sensitivity | Minimum medium growthb | Glycogen stain |
|---|---|---|---|
| MG1655 (wt) | R | + | Brown |
| MG1655/pdksA | R | + | Brown |
| CF1652 (ΔrelA) | S | + | White |
| CF1652/pdksA | S | + | Brown |
| CF1693 (ΔrelA ΔspoT) | S | −c | White |
| CF1693/pdksA | S | − | Brown |
| PK2C1 (ΔdksA::kan) | S | −d | ND |
| TE8114 (ΔdksA::tet) | S | −d | White |
| TE8114/pdksA | R | + | Brown |
| TE8114/rpoB (T563P) | R | + | Brown |
AT, aminotriazole; pdksA, pJK537; ND, not determined; S, sensitive; R, resistant; wt, wild type.
Growth on M9 glucose minimal medium + uracil, +, growth; −, no growth.
Requirements on dropout plates (uracil) + L, V, F, T, S, R, G, H, and probably I.
Requirements on dropout plates (uracil) + L, V, G, F, T, (weak), and probably I.
Limited accumulation of glycogen in cells grown to stationary phase in the presence of glucose is a phenotypic trait of relA as well as of rpoS mutants and is easily screened by staining colonies with iodine vapors (29, 45). Table 2 indicates that multicopy dksA restores iodine staining to ΔrelA, ppGpp0 cells, and a dksA mutant, consistent with the RpoS Western analysis and known dependence of glycogen accumulation (iodine staining) on RpoS function.
Extragenic suppressors of dksA multiple-amino-acid auxotrophy can be identical to ppGpp0 suppressors.
Colonies appeared after incubating dksA mutants on minimal glucose plates for several days. Suppressors were also found on plates containing all amino acids lacking Thr, Phe, or Gly as single omissions, lacking all three binary combinations of Thr, Phe, and Gly as double omissions, and lacking triple omissions of Ile, Leu, and Val or Thr, Phe, and Gly. However, rather than displaying prototrophy specific for the amino acid omissions used for selection of growth, all 10 suppressors tested instead suppressed the parental requirements for all amino acids. Fourteen independent suppressor mutations were isolated from a ΔdksA::kan parent at 37°C. All 14 extragenic suppressors were mapped to the rpoBC region by linkage to btuB::Tn10 with phage P1 transduction, and several were found to be resistant to rifampin. Sequencing of the regions associated with rifampin resistance revealed several identical isolates of the rpoB3370 (T563P) allele, a frequently encountered suppressor of the multiple-amino-acid auxotrophy phenotype of ppGpp0 strains (10). This RNA polymerase mutation restored induction of RpoS in a dksA mutant background during growth into stationary phase as measured by a rpoS-lac fusion (data not shown), as well as suppressing the aminotriazole sensitivity and the iodine-staining defect of a ΔdksA mutant (Table 2).
DISCUSSION
In wild-type E. coli cells, the abundance of RpoS protein increases dramatically during the transition from exponential growth phrase to stationary phase when ppGpp levels are expected to be elevated due to nutrient exhaustion (19). Previous work also showed that RpoS is also induced from the very low basal levels characteristic of exponential growth in response to gratuitously elevating ppGpp levels even a few times above normal (19). The data presented here lead us to propose that positive regulation of RpoS during ppGpp induction occurs by enhancing the efficiency of rpoS expression at the posttranscriptional level. This is based on the finding that ppGpp induction increases initial rates of RpoS synthesis by pulse-labeling (Fig. 3) coupled with a failure to document major effects of ppGpp induction on rpoS mRNA abundance by RNase protection assays (Fig. 1) or on rates of RpoS metabolic turnover (Fig. 2). A possible caveat is that estimates of transcription were performed on LB-grown cells while measurements of RpoS protein synthesis and turnover were made with cells grown on enriched MOPS media to allow high specific activities of labeling with [35S]. The conclusion of posttranslational effects is consistent with the findings of Lange and Hengge-Aronis (28), who showed that the rate of rpoS translation increased at the onset of stationary phase. That ppGpp affects translation is also consistent with promoter substitution experiments in which the rpoS promoter can be replaced by Ptac or PlacUV5 without altering regulation of a RpoS-LacZ reporter while being in growth-enriched MOPS media up to stationary phase or during ppGpp induction in exponentially growing cells in LB (Fig. 8 to 10). Our LacZ reporter experiments (Fig. 10) use a protein fusion that is insensitive to ClpXP degradation, and they therefore indirectly verify that ppGpp induction does not work through effects on the metabolic stability of the RpoS protein. We have searched unsuccessfully for an effect of ppGpp on rpoS translation in vitro using S30 extracts of exponentially growing E. coli, either when translation was coupled with transcription (by adding rpoS encoding plasmid DNA) or when synthetic rpoS mRNA was added directly to extracts (data not shown). It therefore seems unlikely that ppGpp interacts directly with translating ribosomes to specifically enhance the efficiency of rpoS translation. We propose instead that ppGpp indirectly regulates one or more additional factors specifically required for rpoS translation.
There are many candidates for this intermediary factor, including a number of proteins and small RNAs already found to affect RpoS translational efficiency (see the introduction). Among these, our attention was drawn to the pleiotropic regulator DksA (51), because mutants defective in dksA show phenotypes similar to those of ppGpp0 mutants (Table 2). Unlike mutants lacking other factors found to regulate rpoS translation, dksA mutants show multiple auxotrophic requirements (Leu, Val, Thr, Gly, Phe, and probably Ile). These five amino acids are among the nine required due to a deficiency of ppGpp in the same host (52). Aminotriazole sensitivity and impaired glycogen accumulation are also displayed by both dksA and relA mutants. There are some differences: multicopy dksA will suppress the iodine-staining defect of a relA mutant (45) but not the aminotriazole sensitivity of a relA mutant. Multicopy dksA does not enable a ppGpp0 strain to grow on minimal medium but will reverse requirements for ILV on plates containing 17 amino acids and for H on plates containing 19 amino acids. Prototrophic suppressors of dksA deletions were isolated; most of these carry the same T563P allele of rpoB that restores growth of ppGpp0 strains on minimal medium (Table 2). Perhaps the only mutations that can simultaneously suppress the requirement for multiple amino acids made from unrelated biosynthetic pathways are those with global effects on transcription. Far more intriguing is the possibility that these mutations indicate a direct interaction between the DksA protein and RNA polymerase that shares functional similarity with the interaction between RNA polymerase and ppGpp.
Mutants of S. enterica serovar Typhimurium defective in dksA were recovered in a screen for impaired RpoS function in stationary phase (51). This RpoS accumulation defect was verified in E. coli by immunoblots of extracts of LB-grown cells entering stationary phase (Fig. 5). A new finding of this work is that a dksA mutation can block the ability of ppGpp to induce RpoS in exponentially growing cells (Fig. 4). Since a dksA mutant might have a defect in the production of ppGpp, we asked if a dksA mutant produces a similar amount of ppGpp during the stringent response to serine hydroxamate addition on enriched MOPS minimal medium lacking serine. No difference was found, although the dksA mutation significantly elevates, and multicopy dksA lowers, basal levels of ppGpp (Fig. 6). The lack of an effect of the dksA mutation on ppGpp induction during the stringent response is also consistent with the lack of mutant effects on RelA protein abundance during entry into stationary phase when the Western blots shown in Fig. 5 were stripped and reprobed with anti-RelA antibody (data not shown). However, elevation of ppGpp by SpoT activity can account for RpoS induction even in a strain for which relA has been deleted under these conditions (19). Since dksA alters positive regulation of rpoS by ppGpp, we also tested for negative control of stable RNA accumulation by ppGpp under the same conditions without seeing differences between the wild type and a dksA mutant (data not shown).
Another new finding is that multicopy dksA nearly completely restores wild-type accumulation of RpoS in a ppGpp0 host during entry into stationary phase but without inducing RpoS during exponential growth in either a ppGpp0 or an otherwise wild-type strain (Fig. 5). This might suggest that, during the transition into stationary phase, ppGpp induces dksA, which in turn induces rpoS. However, Western blot analysis reveals that the amount of DksA is similar during this transition in wild-type and ppGpp0 strains. This is true regardless of whether DksA levels are low or high corresponding to dksA in single copy or multicopy, respectively (Fig. 7). It may be that ppGpp alters an activity of DksA without changing its concentration and that a threshold of this activity is reached only with high levels of the protein expressed from multicopy dksA. In any event, during the growth transition but not during exponential growth, high levels of DksA seem necessary but not sufficient to induce RpoS, implying that ppGpp-independent factors are required. Other examples exist in which dksA has been identified as a multicopy suppressor but has little effect in single copy. Examples are suppression of mutations of dnaK, dnaJ, and grpE heat shock chaperones (25), of the prc periplasmic protease (4), of the chromosome-partitioning gene mukB (53), and near the pSC101 ori (51). There is also evidence that dnaK can affect RpoS proteolysis during carbon source starvation: an excess of dnaK elevates RpoS twofold, and a deficiency of dnaK lowers RpoS threefold (44). Even if dksA could modulate dnaK function as dramatically as deleting or overproducing DnaK, the mild effects predicted on RpoS seem insufficient to account for the regulation observed here. It is noteworthy that ppGpp, dksA, and rpoS are themselves pleiotropic regulators with incompletely understood properties, not to mention their putative interactions.
In addition to ppGpp and DksA, there are several other stimuli or protein effectors thought to regulate RpoS at the translational level, but so far the only well-understood mechanism involves sequestration of the rpoS ribosome binding site in an inhibitory RNA secondary structure (8). The antisense element (nt 458 to 477) lies about 100 nt upstream of the rpoS AUG initiation codon (nt 565; see the introduction). The antisense element's inhibitory function can be neutralized by competitive binding to a small untranslated RNA, DsrA (34). In contrast, osmotic induction and the response to hfq are not well understood; these also require sequences far upstream in the untranslated leader of rpoS mRNA, but they are independent of the native promoter (11). We investigated the promoter dependence and requirement for upstream sequences in the response of rpoS to stationary phase and ppGpp. Constructs with rpoS-lac driven by Ptac (fusion F) or PlacUV5 (fusion K) retain both stationary-phase induction (Fig. 9) and the response to ppGpp overproduction (Fig. 10), compared to a construct driven by native promoters (fusion A). Derivatives of the promoter-substitution constructs that lack sequences upstream of nt 454, whether driven by Ptac (fusion J) or PlacUV5 (fusion M), are clearly reduced in their response to stationary phase and also appear less sensitive to ppGpp. Nevertheless, with both these deletion constructs, a portion of the remaining ppGpp response is dksA dependent. Assessing the extent of this dependence for the deletion derivatives would require accurate values for the very small slopes of the dksA mutant plots, which cannot be reliably determined from this data.
It seems unlikely that dksA interacts with dsrA, because overexpression of pdsrA induces rpoS in a dksA mutant and because induction of pRelA induces rpoS in a dsrA mutant, while we see similar effects of dksA at 37°C, a temperature where RpoS regulation by dsrA does not occur (43; data not shown). HF-I and OxyS remain possible candidates. Alternatively, DksA may interact with rpoS RNA directly, possibly though the C4-zinc binding motif in the dksA C terminus. We have no evidence that ppGpp directly interacts with DksA or that DksA directly interacts with RpoS. On two-dimensional gels, dksA mutations alter the expression of a number of proteins not affected by mutations in rpoS (51). Indirect effects of DksA seem a possibility through regulation of expression of other genes. As a speculation, DksA might thereby bridge the gap between the ppGpp transcription factor and the observed translational effects on rpoS. It remains to be seen whether ppGpp and dksA interact with each other, with rpoS, and possibly even with RNA polymerase.
Acknowledgments
Helen Murphy provided helpful and expert technical assistance in these experiments.
We gratefully acknowledge receiving the dksA mutant strain PK201 and the plasmid pJK537 from Elizabeth Craig, the anti-DksA polyclonal antibody from Diana Downs, and the 1RS1 RpoS monoclonal antibody from R. R. Burgess.
REFERENCES
- 1.Aldea, M., T. Garrido, C. Hernandez-Chico, M. Vicente, and S. R. Kushner. 1989. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 8:3923-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balandina, A., L. Claret, R. Hengge-Aronis, and J. Rouviere-Yaniv. 2001. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 39:1069-1079. [DOI] [PubMed] [Google Scholar]
- 3.Barth, M., C. Marschall, A. Muffler, D. Fischer, and R. Hengge-Aronis. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoA), DksA, and a truncated RlpA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for sigmaS proteolysis in Escherichia coli, can act like an anti-sigmaS factor. Mol. Microbiol. 35:657-666. [DOI] [PubMed] [Google Scholar]
- 7.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L., and T. Elliott. 1997. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cashel, M. 1994. Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants, p. 341-356. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 3, part A. Academic Press, New York, N.Y.
- 10.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 11.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson, J. W., and C. A. Gross. 1989. Identification of the Sigma-E subunit of Escherichia coli RNA polymerase: a second alternative sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 16.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 17.Gentry, D., H. Xiao, R. Burgess, and M. Cashel. 1991. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 173:3901-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19:1373-1384. [DOI] [PubMed] [Google Scholar]
- 19.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengge-Aronis, R. 1996. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez, V. J., and M. Cashel. 1995. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536-549. [DOI] [PubMed] [Google Scholar]
- 23.Ito, P., J. Bassford, and J. Beckwith. 1981. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell 24:707-717. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, P., and E. Craig. 1990. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 172:2055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 27.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 29.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 30.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 31.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 33.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 34.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 38.Mulvey, M. R., and P. C. Loewen. 1989. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 17:9979-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulvey, M. R., P. A. Sorby, B. L. Triggs-Raine, and P. C. Loewen. 1988. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene 73:337-345. [DOI] [PubMed] [Google Scholar]
- 40.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen, L. H., D. B. Jensen, N. E. Thompson, D. R. Gentry, and R. R. Burgess. 1993. In vitro functional characterization of overproduced Escherichia coli katF/rpoS gene product. Biochemistry 32:11112-11117. [DOI] [PubMed] [Google Scholar]
- 42.Nystrom, T., and F. C. Neidhardt. 1992. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6:3187-3198. [DOI] [PubMed] [Google Scholar]
- 43.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romeo, T., M. Gong, M. Y. Liu, and A.-M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudd, K. E., B. R. Bochner, M. Cashel, and J. R. Roth. 1985. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svitil, A. L., M. Cashel, and J. W. Zyskind. 1993. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268:2307-2311. [PubMed] [Google Scholar]
- 50.Takayanagi, Y., K. Tanaka, and H. Takahashi. 1994. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol. Gen. Genet. 243:525-531. [DOI] [PubMed] [Google Scholar]
- 51.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 53.Yamanaka, K., T. Mitani, T. Ogura, H. Niki, and S. Hiraga. 1994. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13:301-312. [DOI] [PubMed] [Google Scholar]
- 54.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zgurskaya, H. I., M. Keyhan, and A. Matin. 1997. The sigma S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]