Abstract
There are ∼1031 phage in the biosphere, making them the most abundant biological entities on the planet. Despite their great numbers and ubiquitous presence, very little is known about phage biodiversity, biogeography, or phylogeny. Information is limited, in part, because the current ICTV taxonomical system is based on culturing phage and measuring physical parameters of the free virion. No sequence-based taxonomic systems have previously been established for phage. We present here the “Phage Proteomic Tree,” which is based on the overall similarity of 105 completely sequenced phage genomes. The Phage Proteomic Tree places phage relative to both their near neighbors and all other phage included in the analysis. This method groups phage into taxa that predicts several aspects of phage biology and highlights genetic markers that can be used for monitoring phage biodiversity. We propose that the Phage Proteomic Tree be used as the basis of a genome-based taxonomical system for phage.
Phage, viruses that infect prokaryotes, were first described in the early 1900s (19, 63). Studies of phage model systems revolutionized biology and established the field of molecular biology (12). Only recently have the enormous influences of phage on ecosystems been realized (26, 67). Phage are extremely common in the environment: there are ∼1010 phage per liter of surface seawater (6) and 107 to 109 per g of sediment or topsoil (16, 17, 32; David Lipson, unpublished data). In the ocean, phage are major predators of bacteria and significant sinks of essential nutrients (e.g., nitrogen and phosphorus) (65). Phage are also major conduits of genetic exchange, transducing an estimated 1025 to 1028 bp of DNA per year in the world's oceans (34, 50).
Historically, phage have been characterized by their host range and the physical characteristics of the free virion, including capsid size, shape, resistance to organic solvents, and structure, as well as genome size and type (e.g., single-stranded RNA [ssRNA], ssDNA, double-stranded RNA [dsRNA], and dsDNA). The resulting taxonomic system is regularly approved and updated by the International Committee on the Taxonomy of Viruses (ICTV) (45; see also the review by Ackermann [1]). The ICTV taxonomic system requires visualization of the phage particles by electron microscopy to determine capsid morphology. Many investigators, however, do not routinely perform this procedure, as illustrated by the fact that many of the completely sequenced phage in GenBank (25 of 105 [5]) have not been formally classified by the ICTV system. Visualization also cannot be used to classify the numerous prophage genomes that are found within sequenced microbial genomes. As the rate of genomic sequencing increases, the proportion of phage considered “unclassified” will also increase, resulting in major discrepancies between the official taxonomy and the available data.
Analyses of the ribosomal DNA (rDNA) sequences revolutionized the taxonomic characterization of the major forms of life and identified Archaea as the third domain of life (25). Subsequent sequencing of uncultured 16S rDNAs dramatically changed our understanding of microbial diversity (38, 47). Phage do not contain a ribosomal sequence that allows them to be placed on the universal tree of life and, to date, have not benefited from their own gene-based taxonomic system. Previous attempts to classify and measure phage biodiversity based on genetic markers have met with limited success. Although structural proteins (e.g., capsids) could hypothetically serve as a basis for phage taxonomy (27, 29, 41, 60), they are highly diverse and, unlike rDNAs, do not contain conserved regions that allow them to be easily identified. This limits the usefulness of these proteins as markers for biodiversity studies.
We show here that no single gene is found in all phage that can be used as the basis for a classification system. As an alternative, we present a new taxonomic system based on the predicted phage proteome. The resulting taxonomy is compatible with the ICTV system, is supported by our knowledge of phage biology, and allows phage taxonomy to enter the postgenomic era. Using the proposed system, we identified a number of phage classes that are underrepresented in the databases and collections, as well as “signature genes” associated with many phage groups. We believe that the “Phage Proteomic Tree,” described here, offers a necessary platform for classifying phage based on their genomic sequences.
MATERIALS AND METHODS
Sequences.
Most of the genome sequences used in these analyses were obtained from the phage genome page at http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/phg.html. An additional 13 phage genomes, also deposited in GenBank but not located at the above URL, were included: bacteriophage φ105 (accession number AB016282), bacteriophage M11 (AF052431), Haemophilus influenzae phage HP2 (AY027935), Vibrio cholerae O139 fs1 phage (D89074), bacteriophage L5 (L06183), Spiroplasma virus SPV4 (M17988), Spiroplasma virus SpV1 (NC001365), bacteriophage r1t (U38906), Streptococcus thermophilus temperate bacteriophage O1205 (U88974), coliphage M13 (V00604), bacteriophage SP (X07489), bacteriophage φg1e (X98106), and bacteriophage B103 (X99260). Vibrio parahaemolyticus φTB16 was also included (35; V. Seguritan, I. Feng, F. Rohwer, and A. Segall, unpublished data). Wherever possible, the open reading frame (ORF) sequences described by the submitting authors were used in these analyses. Three genomes either had none or only a few ORFs identified in the GenBank sequence (Xanthomonas campestris φCf1c, Spiroplasma φSPV4, and Leuconostoc oenos φL5). For these genomes, ORFs were identified by using Artemis (55) and annotated by comparison with proteins in the SWISS database (4). Together, these 105 genomes had a total of 3,981 predicted proteins. Prophage within completed bacterial genomes, but not deposited independently into GenBank, were not included because of potential problems with misidentifying the phage ends.
Computer analyses and web access.
All computer analyses were performed on Intel-based PC's with the RedHat GNU/Linux 7.2 operating system (RedHat, Durham, N.C.). All of the programs were written with PERL 5.6 (http://www.perl.com/) and are available online at http://salmonella.utmem.edu/phage/tree/. The website also contains tables describing many additional analyses that were performed on the phage genomes, as well as information on the natural history of the phage used in this study.
Calculating BLASTP distances.
All predicted phage protein sequences were compared against all other predicted phage protein sequences by using the BLASTP program (2, 3) and the BLOSUM62 matrix. The data was parsed by using E values of <0.1, <0.01, or <0.001 as cutoffs. The BLASTP distance between two genomes was calculated as the fraction of the genes with significant hits to each other in each of the genomes by using the following equation: 1 − (the number of significant hits between the two genomes/total number of genes in the two genomes). The resulting distance matrix was written to a PHYLIP format file (22).
Calculation of proteomic distances.
All predicted phage protein sequences were compared against all other predicted phage protein sequences by using BLASTP and the BLOSUM62 matrix. All sequences in each comparison with a BLASTP E value of <0.1 were aligned by using the CLUSTALW program with a gap opening penalty of 10.00 and a gap extension penalty of 0.20 (61). Output from the CLUSTALW program was written in PHYLIP format and PROTDIST (part of the PHYLIP package [22]) was used to estimate protein distance scores by using the Dayhoff-PAM matrix. Protein distance scores of >5 were considered not significant (Gary Olsen, unpublished data). If PROTDIST was completely unable to identify any similarity between two proteins the program applied a score of −1. Therefore, in our analyses, any protein distance score of >5 or equal to −1 was treated as a nonsignificant match. To compensate for different predicted protein lengths, each protein distance score was multiplied by the average length of the two proteins.
If two proteomes did not share any proteins, they did not received a pairwise score. Because a zero score indicates a high similarity, it was necessary to add a penalty for each protein that was present in one genome but absent in another. We tested the effect of different penalties (including no penalty) for every protein for which there was no significant match between two genomes. This penalty value was determined empirically and is discussed below. Since there is no evidence that suggests large or small insertions or deletions are more likely, no length factor was included with this penalty.
The proteomic distance score was calculated from the sums of the length-corrected protein distance scores and the penalties and then divided by the total average lengths and the number of missing proteins. These data were written as a distance matrix in the PHYLIP format.
Generation of the trees.
FITCH (part of the PHYLIP package) was used to produce a tree from each PHYLIP format distance matrix. The input data was randomized by using the jumble feature of FITCH and, after generation of the tree, it was globally rearranged to optimize the resulting tree. During our analysis of the phage genomes, we generated a wide diversity of trees based on individual proteins, groups of proteins, phage genomes, families of genomes, and by using different tree generation methods (e.g., neighbor-joining). We also generated an interactive website that provides both graphical and textual comparisons of phage genomes. These trees, images, and additional supporting data are presented online at http://salmonella.utmem.edu/phage/tree/.
RESULTS AND DISCUSSION
The primary goal of this study was to construct a genome-based taxonomy for phage. Such a system has become a necessity due to the rapid accumulation of phage genomic data in the absence of morphology and culturing data required for classification by the current ICTV system. Ideally, a new phage taxonomical system would predict aspects of phage biology, provide tools for measuring uncultured phage biodiversity, and resolve a number of counterintuitive assignments of phage by using the ICTV system (e.g., P22 grouped with T7 as discussed below).
Phage proteome analyses.
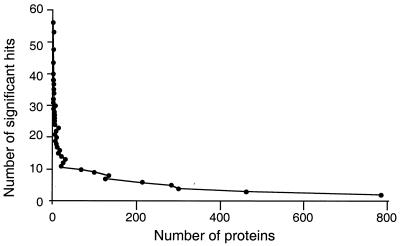
A taxonomic system based on a single locus analogous to the 16S rDNA in bacteria would be the most straightforward method for classifying phage based on their genomic sequence. It has been generally believed for many years that phage do not contain a single genetic marker present in all genomes. To confirm this, we compared all of the predicted phage encoded proteins (n = 3,981) from 105 completed genomes to each other by using BLASTP. Figure 1 shows a rank abundance curve from this analysis. There were 21,153 significant hits (E value of <0.1) between the predicted phage proteins. However, no single protein was found in all 105 genomes. YomI, a putative transglycosylase from Bacillus subtilis φSPBc2, had the greatest number of significant hits (56) to other proteins. However, those 56 similarities were distributed among only 45 genomes. Closer analysis of the YomI BLASTP results showed that most were due to a highly conserved transglycosylase domain that appears multiple times in some phage genomes, either in the same or different proteins. The similarities reported by BLASTP ranged from 16 to 63% identity over short regions of the YomI protein: >60% of the similarities span less than half the YomI protein. These results show that there is no single protein marker that is conserved even in the majority of phage genomes, effectively ruling out a taxonomy system based on a single genetic locus. In addition, a separate analysis showed that no DNA sequence motif is conserved throughout all phage genomes that could serve as a locus for biodiversity analyses (data not shown).
FIG. 1.
Rank abundance curve showing the number of significant similarities between each predicted phage protein against all other predicted phage proteins, as determined by BLASTP with an E-value cutoff of <0.1.
Compatibility analyses.
Since there is not a phage equivalent of the 16S rDNA, different compatibility approaches were investigated (43). In a compatibility taxonomic scheme, the more characters that two organisms share, the more closely related they are. The predicted protein sequences in the genomes were the most obvious characters to use in this analysis, with the expectation that related phage would have a similar complement of proteins. With this approach, it does not matter if the common protein pool arises from a common ancestor or via lateral transfer. The main challenge was deciding which method best predicts which proteins are related. Different approaches (i.e., BLASTP and protein distances) were tested, and the resulting trees were evaluated by using the criteria listed in Table 1. These criteria were extracted from the literature and were chosen because they discriminate between different possible relationships among phage. Both the BLASTP distance and proteomic distance methods produced groups that matched most of the criteria in Table 1. Interestingly, both methods also predicted phage groups similar to those of the ICTV system. These results strongly suggest that a genome-based compatibility approach should be appropriate for classifying phage.
TABLE 1.
Criteria extracted from the literature that were used to evaluate the Phage Proteomic Treesa
| Criteriab | BLASTP result with E-value cutoff of:
|
Protein distance result with penalty value of:
|
|||||
|---|---|---|---|---|---|---|---|
| 0.001 | 0.01 | 0.1 | 0 | 5 | 10 | 100 | |
| dsDNA, ssDNA, and ssRNA phage are fundamentally different | Nc | Nc | Nc | Nd | Y | Y | Y |
| ssRNA phage fall into two groups: (i) leviviruses (fr, MS2, and GA) and (ii) alloleviviruses (SP, NL95, M11, and MX1); PP7 is an out group (7) | N | N | N | Y | Y | Y | Y |
| Leviviruses consist of two groups: group I (fr and MS2) and group II (GA and KU1) (28) | N | N | Y | Y | Y | Y | Y |
| φCPG1, φAR39, and Chp2 are more closely related to each other than to the avian Chlamydia psittaci φChp1 (53) | Y | N | N | Y | Y | Y | Y |
| α3 and φK ssDNA phage are more closely related to each other than to φX174 and G4 (36) | N | Y | N | Y | Y | Y | Y |
| ssDNA phages I2-2 and Ike are more closely related to each other than to F1 (58) | N | N | N | Y | Y | Y | Y |
| ssDNA phage Pf3 is only distantly related to M13, f1, and fd (42) | Y | Y | Y | Y | Y | Y | Y |
| Fs-2 is similar to f1, fd, M13, Ike, and Pf3 (33) | Y | Y | Y | Y | Y | Y | Y |
| ssDNA phage SVTS2 is related to Spiroplasma citri φSpVI (56) | Y | Y | Y | Y | Y | Y | Y |
| Among the podoviruses, PZA is more closely related to B103 than to GA1 (51) | Y | Y | Y | Y | Y | Y | N |
| Mycoplasma φP1 is related to other terminal protein-containing phage (e.g., φ29) (62) | Y | Y | Y | N | N | Y | Y |
| Podoviruses SIO1, T7, YeO3-12 are related (48, 49, 54) | Y | Y | Y | Y | Y | Y | Y |
| Lambdoid phage include lambda, 933W, N15, HK022, HK97, VT2-Sa, P22, D3, APSE-1, and HK620 (8, 13, 15, 37, 59, 64) | Y | Y | Y | N | Y | Y | Ne |
| D29, L5, and Bxb are closely related (24, 44) | Y | Y | Y | Y | Y | Y | Y |
| Relative relationships of Sfi21 > adh > PVL ≥ φ105 (18) | Y | Y | N | N | Y | Y | Y |
| Methanobacterium φM2 is closely related to Methanobacterium wolfeii prophage ψM100 (52) | Y | Y | Y | Y | Y | Y | Y |
| HP1, P2, and 186 are related (21) | Y | Y | Y | Y | Y | Y | Y |
“No” (N) indicates that the particular criteria were not met by the tree in question. A tree using the protein distance method with a penalty of 10 met all of the criteria (i.e., Yes [Y]) and is shown in Fig. 2.
Source references are indicated in parentheses.
dsDNA Fuselloviridae and Corticoviridae phage group with ssDNA phages due to one similar protein.
Inoviridae MV-L1 groups with Siphoviridae BK5-T.
T4 is forced into lambdoid phage by the penalty.
While both the BLASTP distance and protein distance methods resolved many of the relationships predicted by the literature-derived criteria, there were a number of exceptions (Table 1). For example, the BLASTP method was unable to resolve the relationships among the Leviviridae, Inoviridae, and Microviridae, even when the different E-value cutoffs were used. Using strict E-value cutoffs ignores potential information about the relative distance between the protein sequences. In contrast, protein distances, which are an estimate of the number of changes from one protein to another, resolve the finer relationships of these phage groups (Table 1).
Penalty scores.
An apparent problem with both methods was the strong attraction of genomes that are very different from everything in the current database except for one or two proteins, because the similar proteins draw the genomes together. Therefore, the effect of imposing a penalty for every ORF that two genomes do not share with each other was tested in conjunction with the protein distance method. For each ORF not shared between two genomes, penalty values of 5, 10, or 100 were tested. A penalty of 5 produced a tree that matched all of the criteria except that Mycoplasma spp. φP1 was not included in the same group as those phage with terminal protein primers (Table 1). A penalty of 100 was found to force unrelated genomes together based on the penalty alone. A tree that incorporated a penalty of 10, however, matched all of the literature-derived criteria.
Proposed taxonomy scheme.
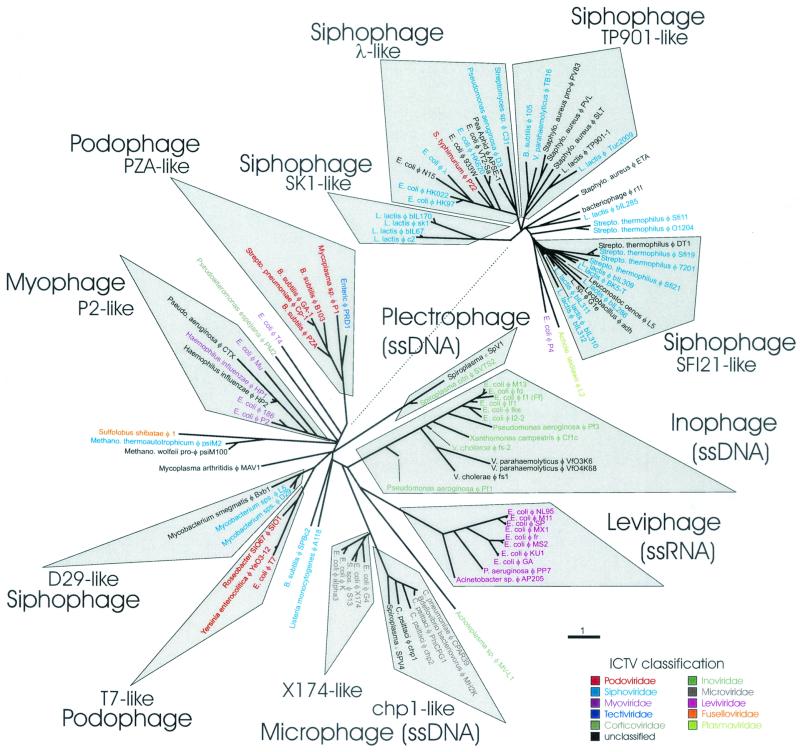
Using protein distances and a penalty of 10, we propose the Phage Proteomic Tree in Fig. 2 most closely describes the relationships of different phage to each other and can serve as a genome-based classification system for phage. Unlike the ICTV system, this classification does not require direct visualization of the free virion, information about host range, or lifestyle information about the phage. The Phage Proteomic Tree shows that a genotype-based taxonomy recapitulates many aspects of the morphology-based ICTV classification. Because of this overlap between the two systems, we suggest that the Phage Genomic Tree taxa be named after the most common ICTV morphology within the proposed groups. To avoid confusion between computational and morphological classifications, we propose that the suffix “-phage” replace “-viridae” when the genome is used to classify the phage. When a group is broken into subgroups, the subgroup should be named after the most widely studied phage in the group followed by “-like” (e.g., φ X174-like microphage). We also noted a number of phage with very similar names that were unrelated to each other (e.g., there are ssDNA and ssRNA phages that infect Escherichia coli that are both called Ff, and there are two dsDNA L5 phages: one associated with Mycobacterium spp. and the other associated with Leuconostoc oenos). Therefore, we suggest that future phage genotypes be named by using the full genus species of the first characterized host, followed by φ (phi for phage), and then the designation of the phage in the first known reference to the phage (e.g., E. coli φ X174 [as shown in Fig. 2]). After the initial naming of the phage within a communication, the designation should be sufficient thereafter. For newly named phage, this designation should include some reference to the geographical location where the phage was isolated (e.g., HK97 = Hong Kong; SIO1 = Scripps Institution of Oceanography). In the future, when uncultured phage will be sequenced, “unknown” will take the place of the host name. The goal of these suggestions is to provide the most information possible to the reader without making any significant changes to current conventions.
FIG. 2.
The Phage Proteomic Tree was constructed from 105 completely sequenced phage genomes. The tree was generated from length-corrected protein distance scores with a penalty of 10 for missing proteins as described in the text. Each phage genome is colored according to its ICTV classification as shown in the key. To make the figure easier to read, the large group of siphophage has been manually shifted away from the other phage groups. The dotted line indicates where the two groups would connect.
The Phage Proteomic Tree resolves a number of classification anomalies associated with the ICTV system. Most conspicuous among these is the ICTV classification of Salmonella φP22 as a podovirus, because of its morphology (i.e., it has a short tail). Botstein and Herskowitz (8) showed that P22 recombines with lambda to produce functional hybrids. Eventually, lambdoid phage were defined as phage whose genomes form viable hybrids with lambda (13). Phage that fulfill this criteria include 933W, N15, HK022, HK97, VT2-Sa, P22, and S. flexneri φSF6, as well as the prophage RAC, DLP12, and VT2-Sakai. Clark et al. (15) suggested that phage APSE-1 and HK620 share genes from a common pool and should also be considered lambdoid phage. In contrast, there is no known genetic relationships between P22 and T7, and grouping of these phage together based on the length of the tail provides very little insight into the biology of these phage. In the Phage Proteomic Tree, P22 groups with the other lambdoid phage, which more accurately reflects its biology.
The remaining Podoviridae separate into two distinct groups in the Phage Proteomic Tree. One group contains the T7, Roseobacter SIO67 φSIO1 and the Yersinia enterocolitica φYeO3-12. The latter two phage have been previously shown to be similar to T7 (48, 49, 54), and therefore we propose that this group be called the T7-like podophage. The second group of podophage, PZA-like, contains six phage, including the enteric φPRD1. Although PRD1 is classified as belonging to the Tectiviridae based on its morphological characteristics, it shares a DNA replication machinery based on protein primers with all members of this group. These two groups of podophage also have very different genome sizes (Table 2).
TABLE 2.
Signature genes and genome size range for the proposed phage groupsa
| Phage group | Genetic material | Subgroup | No. of:
|
Genome size range (kb) | |
|---|---|---|---|---|---|
| Genomes in subgroup | Signature genes | ||||
| Leviphage | ssRNA | None | 10 | 2 | 3.5-4.3 |
| Inophage | ssDNA | None | 13 | 0 | 5.8-8.8 |
| Plectrophage | ssDNA | None | 2 | 6 | 6.8-8.3 |
| Microphage | ssDNA | X174-like | 5 | 10 | 5.4-6.1 |
| Chp1-like | 6 | 5 | 4.4-4.8 | ||
| Podophage | dsDNA | PZA-like | 6 | 1 | 11.7-21.1 |
| T7-like | 3 | 7 | 39.6-39.9 | ||
| Siphophage | dsDNA | Lambda-like | 11 | 0 | 36.5-61.7 |
| D29-like | 3 | 53 | 49.1-52.3 | ||
| SK1-like | 4 | 15 | 22.2-31.8 | ||
| TP901-like | 7 | 9 | 37.7-49.7 | ||
| SFI21 like | 13 | 0 | 14.5-52.2b | ||
| Myophage | dsDNA | P2-like | 6 | 2 | 30.6-35.6 |
Signature genes were described as loci found in all of the genomes within the proposed group and that had a BLASTP E value of <0.1 between all members. Details about the signature genes, links to alignments, sequences, and more information can be found at http://salmonella.utmem.edu/phage/tree/signature.html.
Leuconostoc oenos φL5 was not included in this analysis.
The siphophages present the most problems for the proposed taxonomy system, probably because of rampant lateral gene transfer (31). In the Phage Proteomic Tree, the ICTV Siphoviridae are spread into at least five separate groups. As mentioned above, the lambda-like siphophage group together. The D29-like siphophages infect Mycobacterium spp. and appear to be more closely related to the T7-like podophages than to other siphophages. The SK1-like and TP901-like siphophage groups also form monophyletic taxa. SK1-like siphophages include the 936 and c6A groups proposed by Chopin et al. (14). The rest of the siphophage, however, either fall away from all of the other genomes (see below) or into a polyphyletic group designated the SFI21-like siphophage. The stability of the SFI21-like siphophage group is weak and will almost assuredly change as more genomes are added to the database. Interestingly, the siphophage that infect gram-positive versus gram-negative bacteria generally occupy different groups on the tree.
Similarly, P22, N15, and lambda all group together. Although P22 and lambda are similar, and N15 and lambda are similar, P22 and N15 are only distantly related. However, P22 and N15 are more similar to each other than they are to other phage that were considered, and therefore their close association is not only because of their relationship with each other and with lambda but also because of their relative similarity to the other phage in the tree.
The ICTV Myoviridae group together with the exception of the ICTV type coliphage T4 or P4. Both P4 and T4 appear to represent their own groups, of which is there is currently only one sequenced representative. Similarly, ∼15% of the genomes did not group with other phage on the Phage Proteomic Tree, suggesting that there is only one representative of this group in the database. We expect that as more phage genomes are sequenced and included in the analyses, these discrepancies will be resolved and allow an accurate classification of these phage by using the Phage Proteomic Tree.
The microphage separate into two distinct groups: the φX174-like phage consist of phage that primarily infect enteric bacteria, whereas the chp1-like phage primarily infect Chlamydia species. These groups are also distinguished by the sizes of their genomes (Table 2). Similarly, the inophage divide into two groups. Because one of these groups includes two phage genotypes, Spiroplasma φSpV1 and Spiroplasma citri φSVTS2, that fit the definition of plectroviruses, we have split them into a separate group called plectrophage. Two of the ICTV Inoviridae, Acholeplasma φMVL-1 and A. laidlawii φL2, each appear to represent their own proteomic groups.
Using the taxa shown in Fig. 2, we identified the proteins that are conserved in every member of each group. These ORFs can be considered genetic markers for their respective groups and may be used in the future to identify the presence of uncultured representatives in the environment (Table 2). We call these loci signature genes. Not all groups have signature genes (e.g., lambda-like siphophages). Details and alignments of the signature genes can be found at http://salmonella.utmem.edu/phage/tree/signature.html. The website has an interactive page that allows all of the signature genes within a selection of genomes to be visualized.
Does the Phage Proteomic Tree reflect an evolutionary history?
The 16S rDNA locus has been used both as a predictor of evolutionary relationships and for measuring uncultured microbial diversity (66). Other studies have used FASTA3, Smith-Waterman, or BLAST searches to compare protein similarities to compare bacterial, archaeal, and eukaryotic genomes to the 16S rDNA phylogeny (10, 23, 57). In general, these trees agree with each other. However, lateral transfer of DNA makes the prediction of phenotype based on 16S rDNA a hazardous endeavor (20, 39, 40, 46). The case is much more problematic for phage and other viruses. In trying to infer history from sequence data, it should be considered that a proportional number of events must have occurred in the two sequences in a linear fashion. There is no reason to assume that this criterion is being met by the phage genomes. Additionally, a tree does not reflect evolutionarily history if horizontal transfer has occurred (10). It is well established that siphophage genomes are mosaics of genes from various sources including other phage and their hosts (11, 30, 31). Despite these caveats, there are two reasons to believe that the Phage Proteomic Tree provides important information about the evolutionary history of phage. First, if some phage groups genetically recombine and belong to a common gene pool as proposed by Hendrix et al. (31), then these relationships should be reflected in the Phage Proteomic Tree (e.g., the lambdoid phage versus gram-positive Siphophage). Second, if a group of phage evolves from a common ancestor, then there should also be evidence of these relationships in the Phage Proteomic Tree. Previous investigators have made detailed analyses of the evolutionary relationships among ssDNA and ssRNA phage (see references given in Table 1). The proposed method of proteomic analyses reproduced these relationships, suggesting that evolutionary relationships are reflected in the Phage Proteomic Tree.
In the future, there may be subtle improvements in the algorithms describing phage relationships. For example, we tested the possibility of weighting genes that move together more heavily in the Phage Proteomic Tree construction. Initial forays in this direction suggested that these analyses will have very little effect on the overall structure of the tree. For this reason, we believe that the proposed taxonomical system will not change significantly in the future and can be implemented now.
Acknowledgments
We thank Anca Segall, Mya Breitbart, Gary Olsen, and Stanley Maloy for helpful discussions and comments on the manuscript.
This work was funded by a grant to R.E. from the University of Tennessee's Center of Excellence in Genomics and Bioinformatics and by NSF SGER OCE01-16900 to F.R. and Farooq Azam (Scripps Institution of Oceanography).
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 7.Bollback, J. P., and J. P. Huelsenbeck. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J. Mol. Evol. 52:117-128. [DOI] [PubMed] [Google Scholar]
- 8.Botstein, D., and I. Herskowitz. 1974. Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature 251:585-589. [DOI] [PubMed] [Google Scholar]
- 9.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. R., and W. F. Doolittle. 1999. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Evol. 49:485-495. [DOI] [PubMed] [Google Scholar]
- 11.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 12.Cairns, J., G. S. Stent, and J. D. Watson. 1992. Phage and the origins of molecular biology, expanded ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 13.Campbell, A., and D. Botstein. 1983. Evolution of the lambdoid phage, p. 365-380. In R. E. A. Hendrix (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 14.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark, A. J., W. Inwood, T. Cloutier, and T. S. Dhillon. 2001. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Mol. Biol. 311:657-679. [DOI] [PubMed] [Google Scholar]
- 16.Danovaro, R., A. Dell'Anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danovaro, R., and M. Serresi. 2000. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 19.d'Herelle, F. 1917. Sur un microbe invisible antagoniste des bacilles dysentériques. C. R. Acad. Sci. Ser. D 165:373. [Google Scholar]
- 20.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2128. [DOI] [PubMed] [Google Scholar]
- 21.Esposito, D., W. P. Fitzmaurice, R. C. Benjamin, S. D. Goodman, A. S. Waldman, and J. J. Scocca. 1996. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Res. 24:2360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein, J. 1990. PHYLIP package, v3.3. Department of Genetics, University of Washington, Seattle.
- 23.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 25.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209:457-463. [DOI] [PubMed] [Google Scholar]
- 26.Fuhrman, J. A. 1999. Marine viruses: biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 27.Fuller, N. J., W. H. Wilson, I. R. Joint, and N. H. Mann. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groeneveld, H., F. Oudot, and J. Van Duin. 1996. RNA phage KU1 has an insertion of 18 nucleotides in the start codon of its lysis gene. Virology 218:141-147. [DOI] [PubMed] [Google Scholar]
- 29.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:499-500. [DOI] [PubMed] [Google Scholar]
- 31.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 33.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S. C., and J. H. Paul. 1998. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64:2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellogg, C. A., J. B. Rose, S. C. Jiang, J. M. Turmond, and J. H. Paul. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 120:89-98. [Google Scholar]
- 36.Kodaira, K. I., K. Nakano, S. Okada, and A. Taketo. 1992. Nucleotide sequence of the genome of the bacteriophage α.3: interrelationship of the genome structure and the gene products with those of the phages, phi.X174, G4 and φK. Biochim. Biophys. Acta 1130:277-288. [DOI] [PubMed] [Google Scholar]
- 37.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S rRNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5:355-359. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Marrec, C., D. Van Sinderen, L. Walsh, E. Stanley, E. Vlegels, S. Moineau, P. Heinze, G. Fitzgerald, and B. Fayard. 1997. Two groups bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luiten, R. G., D. G. Putterman, J. G. Schoenmakers, R. N. Konings, and L. A. Day. 1985. Nucleotide sequence of the genome of Pf3, an IncP-1 plasmid-specific filamentous bacteriophage of Pseudomonas aeroginosa. J. Virol. 56:286-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meacham, C. A., and G. F. Estabrook. 1985. Compatibility methods in systematics. Annu. Rev. Ecol. Syst. 16:431-446. [Google Scholar]
- 44.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. J. Jacobs, R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 45.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.). 1995. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, New York, N.Y.
- 46.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 47.Pace, N. T., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 48.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pajunen, M. I., S. J. Kiljunen, M. E. L. Soderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage φYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul, J. H. 1999. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1:45-50. [PubMed] [Google Scholar]
- 51.Pecenkova, T., and V. Paces. 1999. Molecular phylogeny of φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by gram-positive bacteria. J. Mol. Evol. 48:197-208. [DOI] [PubMed] [Google Scholar]
- 52.Pfister, P., A. Wasserfallen, R. Stettler, and T. Leisinger. 1998. Molecular analysis of Methanobacterium phage PSIM2. Mol. Microbiol. 30:233-244. [DOI] [PubMed] [Google Scholar]
- 53.Read, T. D., C. M. Fraser, R.-C. Hsia, and P. M. Bavoil. 2000. Comparative analysis of Chlamydia bacteriophages reveals variation localized to a putative receptor binding domain. Microbial Comp. Genomics 5:223-231. [DOI] [PubMed] [Google Scholar]
- 54.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with non-marine phages. Limnol. Oceanogr. 42:408-418. [Google Scholar]
- 55.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 56.Sha, Y., U. Melcher, R. E. Davis, and J. Fletcher. 2000. Common elements of Spiroplasma plectroviruses revealed by nucleotide sequence of SVTS2. Virus Genes 20:47-56. [DOI] [PubMed] [Google Scholar]
- 57.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 58.Stassen, A. P. M., E. F. P. M. Schoenmakers, M. Yu, J. G. G. Schoenmakers, and R. N. H. Konings. 1992. Nucleotide sequence of the genome of the filamentous bacteriophage I2-2: module evolution of the filamentous phage genome. J. Mol. Evol. 34:141-152. [DOI] [PubMed] [Google Scholar]
- 59.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H.-W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-Type bacteriophage. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu, A.-H. T., L. L. Voelker, X. Shen, and K. Dybvig. 2001. Complete nucleotide sequence of the Mycoplasma virus P1 genome. Plasmid 45:122-126. [DOI] [PubMed] [Google Scholar]
- 63.Twort, F. W. 1915. An investigation on the nature of the ultra-microscopic viruses. Lancet ii:1241-1243. [DOI] [PMC free article] [PubMed]
- 64.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-783. [Google Scholar]
- 66.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]