Abstract
Pseudomonas aeruginosa, a γ-proteobacterium, is motile by means of a single polar flagellum and is chemotactic to a variety of organic compounds and phosphate. P. aeruginosa has multiple homologues of Escherichia coli chemotaxis genes that are organized into five gene clusters. Previously, it was demonstrated that genes in cluster I and cluster V are essential for chemotaxis. A third cluster (cluster II) contains a complete set of che genes, as well as two genes, mcpA and mcpB, encoding methyl-accepting chemotaxis proteins. Mutations were constructed in several of the cluster II che genes and in the mcp genes to examine their possible contributions to P. aeruginosa chemotaxis. A cheB2 mutant was partially impaired in chemotaxis in soft-agar swarm plate assays. Providing cheB2 in trans complemented this defect. Further, overexpression of CheB2 restored chemotaxis to a completely nonchemotactic, cluster I, cheB-deficient strain to near wild-type levels. An mcpA mutant was defective in chemotaxis in media that were low in magnesium. The defect could be relieved by the addition of magnesium to the swarm plate medium. An mcpB mutant was defective in chemotaxis when assayed in dilute rich soft-agar swarm medium or in minimal-medium swarm plates containing any 1 of 60 chemoattractants. The mutant phenotype could be complemented by the addition of mcpB in trans. Overexpression of either McpA or McpB in P. aeruginosa or Escherichia coli resulted in impairment of chemotaxis, and these cells had smooth-swimming phenotypes when observed under the microscope. Expression of P. aeruginosa cheA2, cheB2, or cheW2 in E. coli K-12 completely disrupted wild-type chemotaxis, while expression of cheY2 had no effect. These results indicate that che cluster II genes are expressed in P. aeruginosa and are required for an optimal chemotactic response.
Chemotaxis, the directed movement towards chemicals in the environment, is a behavioral response exhibited by most flagellated bacteria. Escherichia coli and Salmonella enterica serovar Typhimurium have served as prototype organisms for studying chemotaxis, and the signal transduction pathway used to effect a chemotactic response in these γ-proteobacteria is a paradigm for “two-component” and histidine kinase phosphosignaling pathways (5, 6, 54, 55). A set of six chemotaxis proteins acts in concert with receptors called methyl-accepting chemotaxis proteins (MCPs). The current model is that MCPs exist as homodimers that are physically associated with a CheW linker protein dimer and a CheA dimer. There is evidence that these dimeric signaling units exist in cells as supermolecular complexes that are arranged as trimers of dimers (30, 51). On binding an amino acid or other attractant, an MCP dimer undergoes a conformational change that initiates sensory signal transduction by altering the activity of CheA, which is a sensor histidine kinase. CheA-P is a phosophodonor for the response regulator protein, CheY. CheY-P is mobile in the cell and interacts with the rotational “switch” protein FliM in the flagellar motors. The flagellar motors of E. coli and S. enterica are in a default counterclockwise rotation status. In this condition, the peritrichous flagella form a bundle that propels the cell in a single direction (smooth swimming). When CheY-P binds to FliM, the flagella rotate clockwise. This causes the flagellar bundle to come apart; each flagellum pushes in a different direction, and the cell “tumbles” and changes direction. The phosphorylation status of CheY thus dictates whether E. coli runs or tumbles. As cells swim up a concentration gradient of attractant, they spend more time smooth swimming than tumbling; this modulation of swimming behavior is manifested as a chemotactic response.
While CheY phosphorylation relies on the activity of the histidine kinase CheA, its dephosphorylation is controlled by CheZ, as well as by an intrinsic dephosphorylation activity. To ensure proper periodic monitoring of the environment, the system is reset by methylation and demethylation of the MCPs. MCP methylation counterbalances the effect of attractant binding and contributes to adaptation by resetting the signaling activity of the receptors, despite the continued presence of stimulus (12). Two proteins regulate the level of methylation of MCPs. CheR, a methyltransferase, adds methyl groups to conserved cytoplasmic glutamate residues. CheB, a methylesterase, which is active when phosphorylated by CheA-P, removes the methyl groups.
The fundamental characteristics of signal reception and transduction that occur during chemotaxis by E. coli are likely conserved among bacteria and archaea. However, with the recent proliferation of genome sequences, we also now realize that there is much more diversity and complexity in chemotactic signaling pathways in prokaryotes than had been previously anticipated. Most motile bacterial species for which genome sequence information is available have multiple homologues of each of the E. coli che genes; most have many more methyl-accepting chemotaxis genes than the five found in the well-studied enteric species (5). Bacillus subtilis has chemotaxis genes (cheC and cheD) not found in enterics (22, 31, 45). The α-proteobacteria Rhodobacter sphaeroides, Sinorhizobium meliloti, and Caulobacter crescentus have several copies of genes encoding products homologous to those of E. coli che genes, but each lacks a cheZ homologue (2, 41).
Pseudomonas aeruginosa is an opportunistic human pathogen and member of the γ-Proteobacteria. It swims in liquid environments by means of a single polar flagellum, and it can also move on solid surfaces by means of swarming (32) and twitching (49). P. aeruginosa is chemotactic to most of the organic compounds that it can grow on, and several repellants have been identified (14, 23, 27, 39, 40, 42). P. aeruginosa has 26 genes that are homologous to E. coli mcp genes. It also has multiple copies of E. coli-like chemotaxis genes arranged in five clusters (Fig. 1) (56). Two che clusters, cluster I and cluster V, which encode homologues of the six che genes found in E. coli, have previously been shown to be essential for chemotaxis by P. aeruginosa (24, 37). Cluster IV has been shown to be involved in twitching motility (8, 25). Here, we investigate the role that cluster II chemotaxis-like proteins may play in P. aeruginosa chemotaxis.
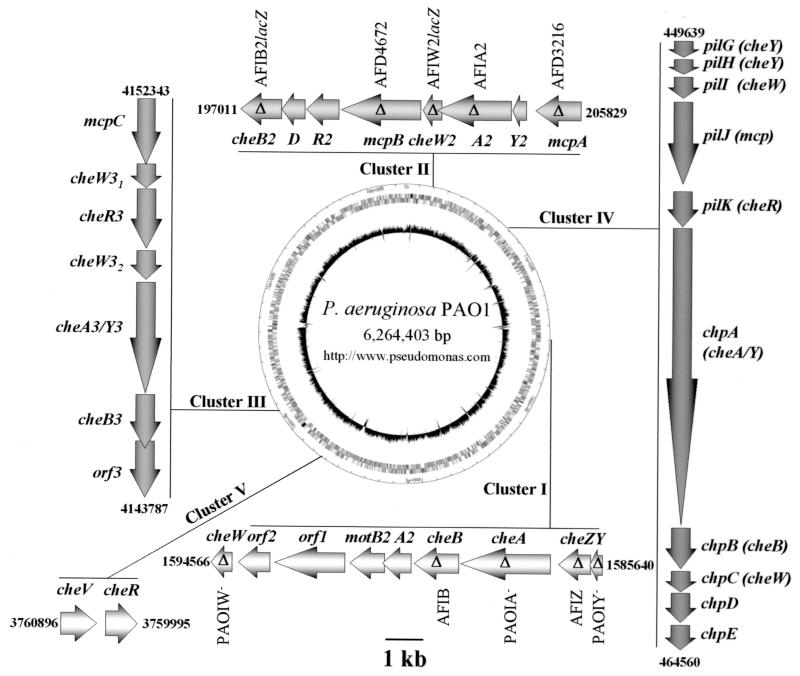
FIG. 1.
Chemotaxis genes in P. aeruginosa. P. aeruginosa has five clusters of chemotaxis-like genes. Clusters I and V have been previously demonstrated to be involved in swimming motility chemotaxis (24, 37). Cluster IV is involved in twitching motility (8, 25). Mutations constructed in this study are indicated by a delta (Δ) within the arrow representing each gene. Names given to each mutant strain are indicated either above or below each mutation (Table 1 provides further information). The P. aeruginosa PAO1 genome map (center) was obtained from the Pseudomonas Genome Project website (http://www.pseudomonas.com), and the positional numbers flanking each cluster of genes are as previously described (Pseudomonas aeruginosa Community Annotation Project [http://www.pseudomonas.com]).
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown on rich medium, Luria-Bertani (LB) medium (46), at 37°C unless otherwise noted. Antibiotics were used at the following concentrations, where appropriate: carbenicillin, 300 μg per ml; chloramphenicol, 100 μg per ml; gentamicin (Gm), 100 μg per ml; and tetracycline (Tc), 100 μg per ml (P. aeruginosa) and ampicillin (Ap), 100 μg per ml; chloramphenicol, 100 μg per ml; gentamicin, 25 μg per ml; kanamycin, 100 μg per ml; and tetracycline, 25 μg per ml (E. coli).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| AFD3216 | mcpA::Gmr derivative of PAO1-Ig; 873 bp deleted from gene | This study |
| AFD4672 | mcpB::Gmr derivative of PAO1-Ig; 234 bp deleted from gene | This study |
| AFIA2 | ΔcheA2 derivative of PAO1-Ig; 1,818 bp deleted from gene | This study |
| AFIB | ΔcheB derivative of PAO1-Ig; 1,065 bp deleted from gene | This study |
| AFIB2lacZ | cheB2::lacZ-Gmr derivative of PAO1-Ig; 1,026 bp deleted from gene | This study |
| AFIW2lacZ | cheW2::lacZ-Gmr derivative of PAO1-Ig; 402 bp deleted from gene | This study |
| AFIZ | ΔcheZ derivative of PAO1-Ig; 738 bp deleted from gene | This study |
| PAO1 | Sequenced wild-type strain, which contains a frame shift mutation in the pilC gene; twitching motility− | 56 |
| PAO1-Ig | Wild-type strain; twitching motility+ | 44 |
| PAOIA− | ΔcheA derivative of PAO1-Ig; 2,226 bp deleted from gene | This study |
| PAOIW− | ΔcheW derivative of PAO1-Ig; 433 bp deleted from gene | This study |
| PAOIY− | ΔcheY derivative of PAO1-Ig; 327 bp deleted from gene | This study |
| E. coli | ||
| CC118 | araD139 Δ(ara leu)7697 ΔlacX74 phoA20 galE galK thi rpsE rpoB argE(Am) recA1 | 28 |
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80lacZΔM15 | Gibco-BRL |
| K-12 | Wild-type strain | 4 |
| S17-1 | thi pro hdsR hdsM+recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | 52 |
| Plasmids | ||
| pAF27 | Apr; mcpB PCR amplified and cloned into EcoRI/HindIII sites of pUC18 | This study |
| pAF27B | Apr/Cbr; mcpB PCR amplified and cloned into EcoRI/HindIII sites of pEX1.8 | This study |
| pAF28 | Apr; SmaI fragment from pUCGm (carrying the gentamicin cassette) cloned into pAF27 cut with AvaI and Klenow treated | This study |
| pAF29 | Tcr; EcoRI/HindIII fragment from pAF28 subcloned into pRK415 | This study |
| pAF33 | Apr; mcpA PCR amplified and cloned into BamHI/HindIII sites of pUC18 | This study |
| pAF35 | Apr; SmaI fragment from pUCGm (carrying the gentamicin cassette) cloned into pAF33 cut with BssHI and Klenow treated | This study |
| pAF37 | Tcr; BamHI/HindIII fragment from pAF35 subcloned into pRK415 | This study |
| pAF56 | Apr; in-frame deletion of cheW2 constructed by PCR and cloned into EcoRI/HindIII sites of pUC19 | This study |
| pAF57 | Apr; SmaI fragment from pUClacZGm containing the lacZ-Gmr cassette cloned into SmaI site of pAF56 | This study |
| pAF58 | Tcr; HindIII/EcoRI fragment from pAF57 cloned into pRK415 | This study |
| pAFB2T | Tcr; in-frame deletion of cheB2 constructed by PCR and cloned into EcoRI/HindIII sites of pEX19Tc | This study |
| pAFB2TlacZGm | Tcr; SmaI fragment from pUClacZGm containing the lacZ-gentamicin cassette cloned into ScaI site of pAFB2T | This study |
| pAFB2TlacZGm2 | Tcr; HindIII/EcoRI fragment from pAFB2TlacZGm cloned into pRK415 | This study |
| pAFBG | Gmr; in-frame deletion of cheB constructed by PCR and cloned into EcoRI/HindIII sites of pEX19Gm | This study |
| pAFZG | Gmr; EcoRI/HindIII fragment from pUCZ subcloned into EcoRI/HindIII sites of pEX19Gm | This study |
| pEX1.8 | Apr/Cbr; high-copy-number cloning and expression vector; contains both ColE1 ori and P. aeruginosa ori; carries lacIq and a Ptac promoter for expression of gene insert | 44 |
| pEX19Gm | Gmr; oriT+sacB+; gene replacement vector with MCS from pUC19 | 19 |
| pEX19Tc | Tcr; oriT+sacB+; gene replacement vector with MCS from pUC19 | 19 |
| pEXA2 | Apr/Cbr; cheA2 PCR amplified from PAO1-Ig chromosome and cloned into EcoRI/HindIII sites of pEX1.8 | This study |
| pEXB2 | Apr/Cbr; cheB2 PCR amplified from PAO1-Ig chromosome and cloned into EcoRI/HindIII sites of pEX1.8 | This study |
| pEXW2 | Apr/Cbr; cheW2 PCR amplified from PAO1-Ig chromosome and cloned into EcoRI/HindIII sites of pEX1.8 | This study |
| pEXY2 | Apr/Cbr; cheY2 PCR amplified from PAO1-Ig chromosome and cloned into EcoRI/HindIII sites of pEX1.8 | This study |
| pHAH128 | Apr/Cbr; mcpA PCR amplified and cloned into EcoRI/HindIII sites of pEX1.8. | This study |
| pRK415 | Tcr; broad-host-range cloning vector | 26 |
| pRK600 | Cmr; ori ColE1 RK2-Mob+ RK2-Tra+ | 28 |
| pUC18 | Apr; high-copy-number cloning vector | 61 |
| pUC18Not | Apr; identical to pUC18 except has NotI/polylinker from pUC18/NotI as MCS | 17 |
| pUC19 | Apr; high-copy-number cloning vector | 61 |
| pUCGm | Apr Gmr; pUC19 derivative carrying a Gmr cassette | 48 |
| pUCGmNot | Apr Gmr; 0.8-kb SmaI fragment from pUCGm cloned into HindII site of pUC18Not | This study |
| pUClacZGm | Apr; high-copy-number ori ColE1 vector containing a promoterless lacZ followed by a Gmr gene; 4-kb lacZ-Gmr cassette can be excised as SmaI or BamHI cassette useful for generating lacZ transcriptional fusions | This study |
| pUCZ | Apr; in-frame deletion of cheZ constructed by PCR and cloned into EcoRI/HindIII sites of pUC19 | This study |
| pUTlacZ1 Gm | Gmr; NotI fragment from pUCGmNot containing the Gmr cassette cloned into NotI sites of pUTminiTn5lacZ1, replacing Kmr cassette | This study |
| pUTminiTn5lacZ1 | Kmr; mini-Tn5-based promoter-probe vector | 9 |
Apr, ampicillin resistant; Cbr, carbenicillin resistant; Cmr, chloramphenicol resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; MCS, multiple cloning site.
Chemotaxis assays.
Chemotaxis was examined qualitatively in soft-agar swarm plates. Strains were stab inoculated into the centers of plates that had been solidified with 0.3% agar and that contained one of the following: a defined minimal medium (11) plus 1 mM succinate; diluted LB medium (0.1% [wt/vol] tryptone, 0.05% [wt/vol] yeast extract, and 0.5% [wt/vol] NaCl); or tryptone (0.1% tryptone [wt/vol] and 0.5% NaCl [wt/vol] for P. aeruginosa or 1% tryptone [wt/vol] and 0.5% NaCl [wt/vol] for E. coli). Wild-type cells and the mcpA and mcpB mutants were screened for chemotaxis in minimal medium swarm plates that contained 1 mM concentrations of 60 different growth substrates. The 60 compounds that were tested as potential chemoattractants included amino acids, sugars, organic acids, and several different kinds of aromatic compounds. To test chemotaxis under anaerobic denitrifying conditions, minimal medium-succinate swarm plates were supplemented with 1 mM KNO3 and incubated anaerobically in polycarbonate jars to which GasPak hydrogen plus carbon dioxide generator envelopes (BBL Microbiology Systems, Cockeysville, Md.) had been added. Carbon sources were sterilized separately and added to minimal medium after being autoclaved. Soft-agar plates were incubated at 30°C for 18 to 24 h for P. aeruginosa and at 35°C for 5 to 6 h for E. coli. The assays were repeated at least three times.
Construction of a lacZ-Gmr cassette.
A 0.8-kb SmaI fragment containing a Gmr gene was excised and purified from plasmid pUCGm (48). This fragment was then ligated into HincII-digested pUC18Not (10), giving pUCGmNot. Plasmid pUTminiTn5lacZ1 (9) was digested with NotI, and the Kmr gene was removed. A NotI fragment that contained the Gmr gene from pUCGmNot was ligated into pUTminiTn5lacZ1 digested with NotI, yielding pUTlacZ1Gm. A lacZ-gentamicin fragment was purified out of the latter by digesting it with EcoRI. This fragment was blunt ended by treatment with the Klenow fragment of DNA polymerase I (Roche Molecular Biochemicals, Indianapolis, Ind.) and ligated to pUC18Not that was digested with NotI and blunt ended by treatment with Klenow. Finally, the HindIII and PstI restriction sites within the lacZ and the Gmr genes were removed by partial digestion with HindIII and PstI, Klenow treatment, and then religation. The resulting plasmid, pUClacZGm, is a high-copy-number ori ColE1 vector containing a promoterless lacZ followed by a Gmr gene. This roughly 4-kb fragment can be excised as either an SmaI or BamHI cassette for the generation of lacZ transcriptional fusions.
Construction of cheB2::lacZ-Gmr and cheW2::lacZ-Gmr mutant strains.
An in-frame deletion of cheB2 was created by overlap extension PCR as described previously (18, 20, 21) with the following modifications. A region of DNA from the PAO1-Ig chromosome spanning from approximately 1 kb upstream of cheB2 to the 5′ end of cheB2 was PCR amplified using primers DcheB2 [1] and DcheB2 [2]. A second region, from approximately 1 kb downstream of cheB2 to the 3′ end of cheB2, was PCR amplified using primers DcheB2 [3] (complementary to DcheB2 [2]), and DcheB2 [4]. A mixture of these two DNA fragments (100 ng each) was used as the template in a third PCR amplification including primers DcheB2 [1] and DcheB2 [4]. The product of the third amplification contained a 1,026-bp in-frame deletion in cheB2 including an engineered ScaI site plus approximately 1 kb upstream and 1 kb downstream of cheB2, with engineered HindIII and EcoRI sites on its 5′ and 3′ ends, respectively. This product was digested with EcoRI and HindIII and was ligated into HindIII/EcoRI-digested pEX19Tc, yielding plasmid pAFB2T. The cheB2-lacZ fusion was generated by ligation of a SmaI lacZ-Gmr cassette obtained from plasmid pUClacZGm into ScaI-digested pAFB2T, yielding pAFB2TlacZGm. The HindIII/EcoRI insert from pAFB2TlacZGm was then subcloned into pRK415, giving pAFB2TlacZGm2. This plasmid was then mobilized from E. coli DH5α into PAO1-Ig, using E. coli CC118(pRK600) to provide the transfer functions. The recombinant strain, AFIB2lacZ, was identified by screening for Gmr and Tcs colonies and verified by Southern blot analysis (46). A cheW2::lacZ-Gmr mutant (strain AFIW2lacZ) was constructed using a similar strategy.
Construction of P. aeruginosa cheB, cheZ, cheY, cheW, cheA, and cheA2 in-frame deletion mutants.
An in-frame deletion in cheB was constructed by overlap extension PCR, as described above, using primers DcheB [1]-DcheB [2] and DcheB [3]-DcheB [4]. The product of the third amplification contained a 1,065-bp in-frame deletion in cheB including an engineered ScaI site plus approximately 1 kb upstream and 1 kb downstream of cheB, with engineered HindIII and EcoRI sites on its 5′ and 3′ ends, respectively. This 2,125-bp fragment was cloned as an EcoRI/HindIII cassette into pEX19Gm to give plasmid pAFBG. pAFBG was mobilized from E. coli S17-1 into PAO1-Ig by conjugation. A single recombination event was selected by growth on selective medium containing gentamicin. The double recombinant was selected by growth on LB medium plus 5% sucrose. Correct colonies were identified by screening for Gms and the ability to grow on 5% sucrose. cheB mutations were screened by PCR and further confirmed by Southern blot analysis, yielding the nonpolar, in-frame deletion mutant AFIB. Similar cloning strategies and methods were used to construct in-frame deletion mutations in cheZ, cheY, cheW, cheA, and cheA2 genes in P. aeruginosa strain PAO1-Ig.
Construction of cluster II mcpB::Gmr and mcpA::Gmr mutants.
mcpB was PCR amplified from PAO1-Ig chromosomal DNA using the primers RPA04672F (which includes an engineered EcoRI site) and RPA04672R (which includes an engineered HindIII site). The product was cloned into pUC18 digested with EcoRI/HindIII, and the resulting plasmid was named pAF27. pAF27 was digested with AvaI, removing a 234-bp fragment from the center of mcpB; treated with Klenow and shrimp alkaline phosphatase (Roche Molecular Biochemicals), and ligated to the SmaI fragment containing a Gmr cassette from pUCGm. The resulting plasmid was named pAF28. The EcoRI/HindIII fragment from pAF28 was cloned into pRK415, giving pAF29. This plasmid was transferred from E. coli DH5α to P. aeruginosa PAO1-Ig by conjugation, using E. coli CC118(pRK600) to provide the transfer functions. A double recombinant was selected as described above to give strain AFD4672. The mutation was verified by Southern blotting and PCR. An mcpA::Gmr mutant (AFD3216) was constructed by a similar strategy.
Construction of plasmids for expression of cluster II genes.
The coding sequences of cheA2, cheB2, cheW2, cheY2, mcpA, and mcpB were PCR amplified from PAO1-Ig chromosomal DNA using the appropriate primer pairs. The upstream primers for each PCR contained an additional sequence (CCGAATTCTGATTAACTTTATAAGGAGGAAAAACATATG…) containing an engineered EcoRI site (underlined), an optimized Shine-Delgarno sequence (in boldface) (38), a translational enhancer from gene 10 of phage 7 (in italics) (38), and the start codon of the gene being amplified (double underline). The products from PCR amplifications were digested with EcoRI/HindIII and cloned into the EcoRI/HindIII sites of pEX1.8. These new plasmids were designated pEXA2, pEXB2, pEXW2, pEXY2, pHAH128, and pAF27B and were used to express CheA2, CheB2, CheW2, CheY2, McpA, and McpB, respectively. Each plasmid was then introduced into either P. aeruginosa competent cells (58) or E. coli competent cells (The NEB Transcript, vol. 6, p. 7, New England Biolabs, 1994) and selected for by incorporation of carbenicillin (P. aeruginosa) or ampicillin (E. coli) into the growth medium. Transformants were verified by agarose gel electrophoresis of plasmid DNA isolated by alkaline lysis (3). Proteins were expressed under the control of the Ptac promoter and induced by the addition of 10, 100, or 1,000 μM isopropyl-β-d-thiogalactopyranoside (IPTG) (Research Product International Corp., Mt. Prospect, Ill.) to the growth medium.
RESULTS
Strains with mutations in cluster I che genes are generally nonchemotactic.
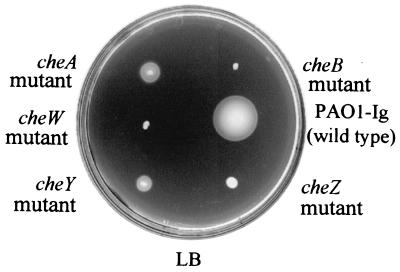
Mutants with in-frame deletions in the cheA, cheB, cheW, cheZ, and cheY genes of cluster I were severely impaired in chemotaxis in LB medium, tryptone, and minimal medium soft-agar swarm plates (Fig. 2). They were also completely defective in chemotaxis when incubated anaerobically in soft-agar swarm plates that included succinate as an attractant and nitrate as a terminal electron acceptor. This is in agreement with the previously described results of Masduki et al. (37) and Kato et al. (24). The cheB and cheZ mutants reversed their swimming directions at high frequency when observed microscopically. The cheA, cheW, and cheY mutants had smooth-swimming phenotypes when observed under the microscope. E. coli cheA, cheW and cheY mutants also smooth swim. E. coli cheB and cheZ mutants have tumbly phenotypes.
FIG. 2.
Cluster I che gene mutants are nonchemotactic. Shown is a dilute LB soft-agar plate with cluster I mutant strains. Each strain was motile but deficient in chemotaxis.
A cheB2 mutant is defective in chemotaxis.
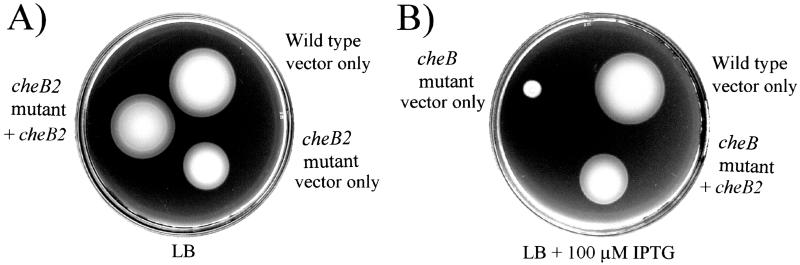
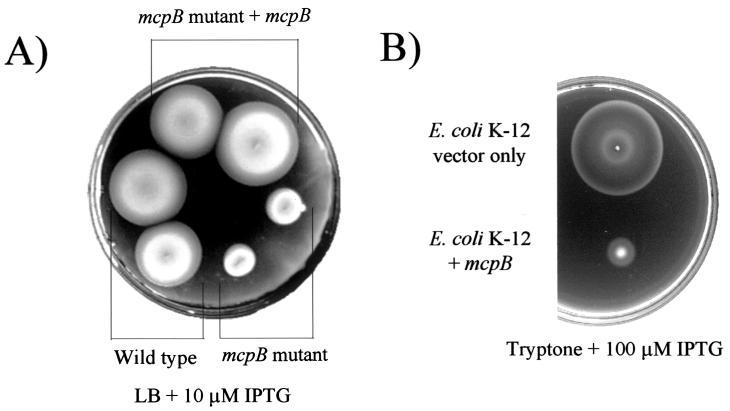
We constructed mutations in cluster II cheB2, cheW2, and cheA2 genes and found that the cheW2 and cheA2 mutants behaved like the wild-type parent in swarm plate assays. The cheB2 mutant, however, formed swarm rings that were smaller than those of the wild type (Fig. 3A). This general defect was observed in minimal swarm media as well as in rich medium. The cheB2 mutant grew at the same rate as its wild-type parent in liquid medium. cheB2 mutant cells did not have an obvious defect in swimming behavior when observed under the microscope. The cheB2 mutant phenotype was complemented by the addition of cheB2 in trans (Fig. 3A).
FIG. 3.
CheB2 is required for optimal chemotaxis by P. aeruginosa, and overexpression of CheB2 partially restores chemotaxis to a nonchemotactic P. aeruginosa cheB mutant. (A) Dilute LB soft-agar plate showing chemotactic rings formed by P. aeruginosa PAO1-Ig (wild type) harboring only vector (pEX1.8), the cheB2 mutant AFIB2lacZ harboring only vector (pEX1.8), and the cheB2 mutant complemented with cheB2 in trans (pEXB2). (B) Dilute LB soft-agar plate showing chemotactic rings formed by P. aeruginosa PAO1-Ig (wild type) harboring only vector (pEX1.8), the cheB mutant AFIB harboring only vector (pEX1.8), and the cheB mutant complemented with cheB2 in trans (pEXB2). Expression of CheB2 from pEXB2 was controlled by the Ptac promoter and was induced by addition of 100 μM IPTG to the medium.
CheB2 restores chemotaxis to the nonchemotactic cheB mutant.
To determine whether cluster I chemotaxis proteins can interact with cluster II proteins, four cluster II proteins, CheA2, CheB2, CheW2, and CheY2, were expressed in strains that contained a defect in the homologous cluster I gene. Of these, only CheB2 could complement a defect in the cluster I paralog, cheB. CheB2 partially restored chemotaxis to the nonchemotactic, cluster I, cheB-deficient strain AFIB (Fig. 3B).
mcpA and mcpB mutants have a general chemotaxis defect.
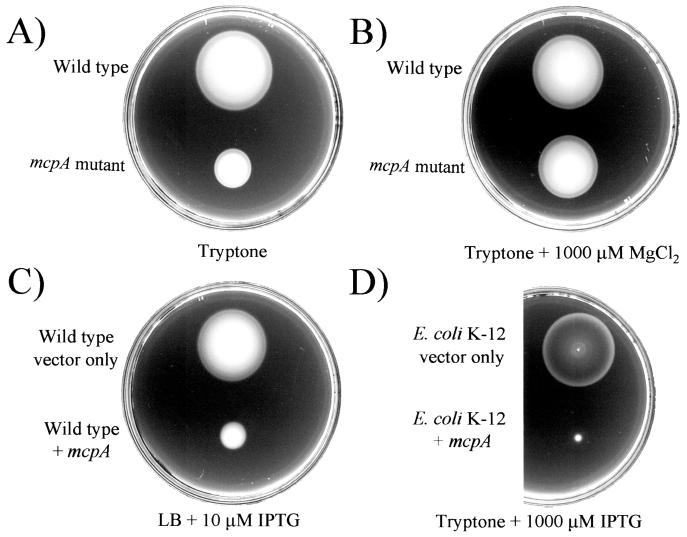
Cluster II contains two genes predicted to encode MCP-like proteins: mcpA and mcpB. Each gene was mutated by replacing part of the gene with a Gmr cassette. The mcpA and mcpB mutants were then screened for chemotaxis to 60 carbon sources in minimal medium using the soft-agar plate assay (data not shown). Carbon sources that were tested as chemoattractants included amino acids, sugars, organic acids, and aromatic compounds. Wild-type cells formed well-defined chemotactic swarm rings in swarm plates that included any of the 60 carbon sources. The mcpA mutant strain, AFD3216, behaved like the wild type in minimal-medium soft-agar plates containing any of the organic chemoattractants that we tested (not shown), but it had a general chemotaxis defect in tryptone soft-agar plates (Fig. 4A). This phenotype was not due to a defect in the growth rate in tryptone media (data not shown). This led us to examine compositional differences between tryptone and our minimal medium. We determined that the presence of magnesium in the minimal medium allowed the mcpA mutant to overcome the mutant phenotype (Fig. 4B). Magnesium, in concentrations as low as 100 μM, added to the tryptone soft-agar plates as either MgCl2 or MgSO4, restored wild-type chemotaxis to the mcpA mutant. Overexpression of McpA in P. aeruginosa led to a nonchemotactic phenotype in soft-agar plates (Fig. 4C). The same phenotype was observed when McpA was overexpressed in E. coli K-12 (Fig. 4D). Individual P. aeruginosa or E. coli cells overexpressing McpA had a smooth-swimming phenotype.
FIG. 4.
McpA is required for a wild-type chemotactic response by P. aeruginosa, and overexpression of McpA inhibits the chemotactic response. (A) Tryptone soft-agar plate showing chemotactic rings formed by P. aeruginosa PAO1-Ig (wild type) and an mcpA mutant (AFD3216). (B) Tryptone soft-agar plate with 1,000 μM MgCl2, comparing the chemotactic responses of wild-type cells and mcpA mutant cells. Addition of Mg2+ to the medium restores wild-type chemotaxis to the mcpA mutant. (C) Dilute LB soft-agar plate showing chemotactic rings formed by P. aeruginosa PAO1-Ig (wild type) harboring only vector (pEX1.8) and P. aeruginosa PAO1-Ig (wild type) overexpressing McpA (pHAH128). Expression of McpA from pHAH128 was induced by the addition of 10 μM IPTG to the medium. (D) Tryptone soft-agar plate showing chemotactic rings formed by E. coli K-12 harboring only vector (pEX1.8) and E. coli K-12 expressing McpA (pHAH128). Expression of McpA from pHAH128 was controlled by the Ptac promoter and was induced by the addition of 1,000 μM IPTG to the medium.
The mcpB mutant strain, AFD4672, grew at wild-type rates (data not shown) but had a general chemotaxis defect on LB medium, tryptone, and minimal-medium soft-agar swarm plates containing any of 60 different attractants. This defect was observed when plates were incubated either aerobically or anaerobically under denitrifying conditions (Fig. 5A). The defect was complemented when mcpB was expressed in trans (Fig. 5A). Overexpression of McpB in P. aeruginosa led to a nonchemotactic phenotype on soft-agar plates (not shown). The same phenotype was observed when McpB was overexpressed in E. coli (Fig. 5B). Individual P. aeruginosa cells overexpressing McpB had a constantly smooth-swimming phenotype when observed microscopically. The same was observed when McpB was overexpressed in E. coli.
FIG. 5.
McpB is required for optimal chemotaxis by P. aeruginosa, and overexpression of McpB inhibits the chemotactic response. (A) Dilute LB soft-agar plate showing chemotactic rings formed by P. aeruginosa PAO1-Ig (wild type) harboring only vector (pEX1.8), an mcpB mutant (AFD4672) harboring only vector (pEX1.8), and the mcpB mutant complemented with mcpB in trans (pAF27B). Expression of McpB from pAF27B was induced by the addition of 10 μM IPTG to the medium. (B) Tryptone soft-agar plate showing chemotactic rings formed by E. coli K-12 harboring only vector (pEX1.8) and E. coli K-12 expressing McpB (pAF27B). Expression of McpB from pAF27B was induced by the addition of 100 μM IPTG to the medium.
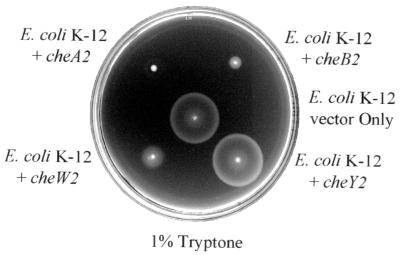
Cluster II proteins CheA2, CheB2, and CheW2 disrupt chemotaxis when expressed in E. coli K-12, but not when expressed in P. aeruginosa PAO1-Ig.
CheA2, CheB2, and CheW2 were expressed in E. coli K-12 from a high-copy-number ori ColE1 plasmid (pEX1.8). Each of these proteins disrupted normal chemotaxis in 1% (wt/vol) tryptone soft-agar plates (Fig. 6). Overexpression of CheY2 had no effect on E. coli chemotaxis. Each strain was motile when a sample was removed from the plate and observed microscopically. Thus, the effect was not due to a disruption of motility. Overexpression of CheA2, CheB2, CheW2, and CheY2 in wild-type P. aeruginosa cells had no effect on chemotaxis as assayed in soft-agar swarm plates. Expression was induced by IPTG concentrations up to1,000 μM (data not shown).
FIG. 6.
Effect of expression of cluster II Che proteins on E. coli K-12 chemotaxis. Shown is a tryptone soft-agar plate with chemotactic rings formed by E. coli K-12 harboring only vector (pEX1.8) or expressing CheA2 (pEXA2), CheB2 (pEXB2), CheW2 (pEXW2), or CheY2 (pEXY2). No IPTG was added to the medium. CheY2 failed to inhibit E. coli chemotaxis even when expression was induced with 100 μM IPTG.
DISCUSSION
The P. aeruginosa cluster I CheA, CheB, CheW, CheY, and CheZ proteins have 32, 36, 30, 58, and 32% amino acid identity with the corresponding E. coli Che proteins. CheR proteins from P. aeruginosa (encoded in cluster V) and E. coli are 30% identical. Given that the cluster I che gene mutants are completely defective in chemotaxis, it was surprising to find, on inspection of the P. aeruginosa genome sequence, an additional cluster of che genes (cluster II) whose predicted protein products, with the exception of cheY2, had even higher overall sequence identities to the orthologous E. coli chemotaxis proteins (Table 2). Key amino acid residues shown to be important for E. coli chemotaxis protein function are conserved in both the cluster I and the cluster II Che proteins from P. aeruginosa.
TABLE 2.
Percent identities of P. aeruginosa cluster II proteins to chemotaxis proteins from clusters in other organisms
| P. aeruginosa cluster II protein | % identity
|
||||
|---|---|---|---|---|---|
| P. aeruginosa cluster I paralog | E. coli ortholog | P. syringae pv. tomato cluster II ortholog | S. oneidensis MR-1 cluster II ortholog | V. cholerae cluster II ortholog | |
| McpA | NAa | NA | 25 | 30 | 29 |
| CheY2 | 33 | 36 | 49 | 60 | 50 |
| CheA2 | 33 | 42 | 37 | 43 | 40 |
| CheW2 | 30 | 63 | 35 | 59 | 37b |
| 33c | |||||
| McpB | NA | NA | 29 | 49 | 37 |
| CheR2 | 31 | 51 | 39 | 49 | 39 |
| CheD | NA | NA | 29 | 52 | 33 |
| CheB2 | 39 | 60 | 47 | 53 | NAd |
NA, not applicable; this cluster does not contain a paralog or ortholog. Identities have been rounded to the nearest whole percentage and were calculated with the ClustalW Multiple Alignment algorithm within the BioEdit sequence alignment editor (13).
First cheW2 in this cluster.
Second cheW2 in this cluster.
V. cholerae has a frame shift mutation in the cheB2 open reading frame (16).
Our data suggest that P. aeruginosa cluster II che genes participate in chemotaxis. A cheB2 mutant is impaired in chemotaxis. Also, CheB2, when overexpressed, can complement a P. aeruginosa cheB mutant and restore it to a wild-type pattern of motile behavior and chemotactic response. Although the cheA2 and cheW2 mutants did not have a discernible chemotaxis defect, CheA2 and CheW2 proteins that were expressed in an E. coli K-12 background disrupted E. coli chemotaxis. This suggests that CheA2 and CheW2 can compete with endogenous E. coli chemotaxis proteins and interfere with the normal chemotactic response. There are a number of ways in which cluster II Che proteins might function in chemotaxis. They may contribute to an optimal chemotactic response by coexisting and participating in a major signal transduction pathway that is dominated by cluster I proteins. Alternatively, cluster II Che proteins may form complexes that are physically and functionally distinct from cluster I signaling complexes. The observation that overexpression of cluster II proteins interferes with E. coli, but not P. aeruginosa, chemotaxis favors the second possibility.
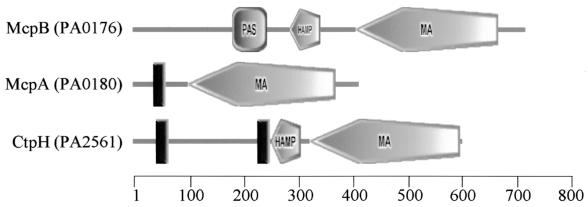
A distinct cluster II chemotaxis-signaling complex may be important for sensing some general parameter of cellular physiology through associated McpA and McpB proteins. Such a role is consistent with the observation that mcpA and mcpB mutants have general chemotaxis defects and unusual molecular architectures. Most mcp mutants that have been described are defective in chemotaxis to a subset of the chemoattractants that a particular bacterial species can detect. For example, mutants in the P. aeruginosa mcp genes ctpH and ctpL that are specifically nonchemotactic to inorganic phosphate have been described (59). Similarly, the mcp mutant pctA is defective in chemotaxis to l-serine but attracted to other amino acids (33). Most of the 26 predicted P. aeruginosa MCPs have a typical E. coli-like MCP architecture that consists of two transmembrane regions in the N-terminal half of the protein and a domain, called the highly conserved domain, that is involved in sensory signaling (Fig. 7). Analysis of the amino acid sequence of McpA using the Simple Modular Architecture Research Tool (SMART) (35, 47) indicates that McpA has a highly conserved domain (34) but only one predicted transmembrane domain (Fig. 7). McpB (35, 47) has a highly conserved domain (34), a PAS (for Per, ARNT, Sim) (57) domain, and no predicted transmembrane domains (Fig. 7). PAS domains are typically involved in sensing redox potential, oxygen, or light (57). McpA and McpB are the only P. aeruginosa MCPs that have C-terminal pentapeptides (EVELF in the case of McpA and GWEEF in the case of McpB) related to that found on the high-abundance receptors of E. coli (NWETF) (43, 60). This reinforces the concept that McpA and McpB may play a more general and central role in chemotactic signal transduction than do typical MCPs.
FIG. 7.
Domain architectures of 3 of the 26 P. aeruginosa MCPs. The structures were created with the SMART server (http://smart.embl-heidelberg.de/) (35, 47). PA numbers are indicated in parentheses and are according to the Pseudomonas aeruginosa Community Annotation Project (http://www.pseudomonas.com). The roles of McpA and McpB in P. aeruginosa chemotaxis are discussed in the text. CtpH is an MCP for inorganic phosphate in P. aeruginosa and has been previously described (59). CtpH represents the common structural motif of an MCP and is similar to E. coli MCPs. Domain representations are as follows: rectangle, transmembrane domain; square, PAS domain; elongated pentagon, highly conserved domain of MCP (MA, methyl accepting); small pentagon, HAMP domain (1). The scale represents amino acid positions.
Cluster II has previously been referred to as the α-subgroup-like cluster of che genes (53). However, genetic organization and physiological observations suggest that α-proteobacteria and P. aeruginosa have different chemotaxis systems. None of the multiple clusters of chemotaxis genes identified in the genomes of the sequenced α-proteobacteria R. sphaeroides, S. meliloti, and C. crescentus resembles P. aeruginosa cluster II in gene organization (7, 15, 36). Furthermore, the proteins predicted to be encoded by cluster II genes, with the exception of CheY2, have the highest degree of sequence identity to E. coli rather than to α-proteobacteria orthologs (Table 2) (56). The best-described chemotaxis system in a bacterium belonging to the α subgroup of proteobacteria is that of R. sphaeroides (50). A model for R. sphaeroides chemotaxis has been proposed in which two chemotaxis-signaling complexes, one encoded by operon 1 and a second encoded by operon 2, contribute to an optimal chemotactic response (50). In the absence of operon 2, a repellent response to the attractant propionate was seen (50). An operon 1 deletion mutant had no obvious chemotaxis phenotype (50). Significantly, motor bias phenotypes (in the case of R. sphaeroides, either smooth swimming or stopped) were seen only in mutants that had defects in both operon 1 and operon 2 (50). P. aeruginosa differs from R. sphaeroides and other α-proteobacteria in that a single set of chemotaxis genes clearly dominates the chemotactic response, as evidenced by the observation that cluster I mutants have profound effects on the rotational bias of the flagellar motor. This is likely to facilitate studies of the contribution of cluster II genes to the ability of P. aeruginosa to sense and respond to its environment.
One cannot, at this point, exclude the possibility that the major output from a cluster II signaling complex may be something other than a chemotactic response. Such a hypothetical output would likely be processed through CheY2. Overexpression and complementation data offer no support for the idea that CheY2 interacts with the flagellar motor. Moreover, CheY2 did not complement a P. aeruginosa cheZ mutant. These data suggest that CheY2 may not act as a phosphate sink, as has been reported for R. sphaeroides and S. meliloti CheY proteins (53).
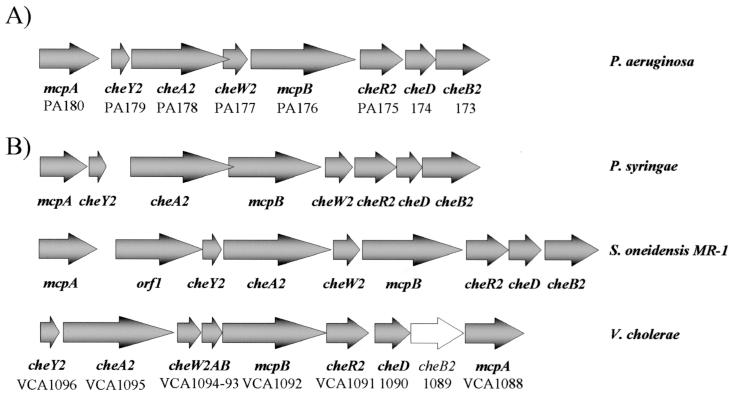
The γ-proteobacteria Pseudomonas syringae pv. tomato, Shewanella oneidensis MR-1, and Vibrio cholerae each have a set of cluster II-like chemotaxis genes (Table 2 and Fig. 8). Pseudomonas putida (The Institute for Genomic Research [TIGR] unfinished genomes BLAST search [http://tigrblast.tigr.org/ufmg/]) and Pseudomonas fluorescens (Pseudomonas fluorescens Genome Project [http://www.jgi.doe.gov/JGI_microbial/html/pseudomonas/pseudo_homepage.html]), species that are closely related to P. aeruginosa, do not have cluster II chemotaxis genes. They do, however, have sets of cluster I, III, IV, and V chemotaxis genes with high amino acid sequence identity (on the order of 70 to 80% identity) to the orthologous P. aeruginosa che genes. The function of a cluster II signal transduction pathway could be associated with physiological capabilities that cluster II bacteria have in common. However, it is not immediately clear what these commonalities may be. V. cholerae, S. oneidensis, and P. aeruginosa are able to grow anaerobically by denitrification. Yet P. syringae is an obligate aerobe and P. fluorescens, which lacks cluster II genes, can denitrify. P. syringae, V. cholerae, and P. aeruginosa are each either an animal or plant pathogen. S. oneidensis strain MR-1 (formerly Shewanella putrefaciens MR-1) is not known to be pathogenic, but some Shewanella species are opportunistic human pathogens (29). This suggests that it might be worth exploring a possible connection between the activity of a cluster II signal transduction pathway and environmental stress conditions of the type that pathogens are known to experience.
FIG. 8.
Cluster II-like gene arrangements in γ-proteobacteria. (A) P. aeruginosa cluster II. PA numbers are according to the Pseudomonas aeruginosa Community Annotation Project (http://www.pseudomonas.com). (B) Cluster II-like gene arrangements in other γ-proteobacteria. P. syringae cluster II and S. oneidensis MR-1 cluster II were accessed through the TIGR Microbial Genome Database (http://www.tigr.org/tdbl). V. cholerae VCA numbers are according to the TIGR Complete Microbial Resource (http://www.tigr.org/tigr.scripts/CMR2/CMRHomePage.spl). V. cholerae cheB2 may not be functional due to a frame shift mutation (16).
Acknowledgments
This work was supported by Public Health Service grant GM56665 from the National Institute of General Medical Sciences. A.F. was supported by a postdoctoral fellowship from the Fundación Ramón Areces (Spain). A.C.H. was supported by a National Science Foundation Research Training Grant (DBI9602247) and by a fellowship from the University of Iowa Center for Biocatalysis and Bioprocessing.
We thank Steve Lory and Barbara Iglewski for providing P. aeruginosa strains. We also gratefully acknowledge Rebecca Parales for critical review of the manuscript.
REFERENCES
- 1.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signaling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 2.Armitage, J. P., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl, L. M. Albright, D. M. Coen, and A. Varki (ed.). 1998. Current protocols in molecular biology, vol. 1. John Wiley and Sons Inc., New York, N.Y.
- 4.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, 7th ed., p. 807-876. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 5.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 6.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darzins, A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11:137-153. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived mini-transposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 11.Ditty, J. L., A. C. Grimm, and C. S. Harwood. 1998. Identification of a chemotaxis gene region from Pseudomonas putida. FEMS Microbiol. Lett. 159:267-273. [DOI] [PubMed] [Google Scholar]
- 12.Djordjevic, S., and A. M. Stock. 1997. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5:545-558. [DOI] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hamilton, P. B., and G. Sheeley. 1971. Chemotactic response to amino acids by Pseudomonas aeruginosa in a semisolid nitrate medium. J. Bacteriol. 108:596-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauwaerts, D., G. Alexandre, S. K. Das, J. Vanderleyden, and I. B. Zhulin. 2002. A major chemotaxis gene cluster in Azospirillum brasilense and relationships between chemotaxis operons in alpha-proteobacteria. FEMS Microbiol. Lett. 208:61-67. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 20.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 22.Karatan, E., M. M. Saulmon, M. W. Bunn, and G. W. Ordal. 2001. Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis. J. Biol. Chem. 276:43618-43626. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J., A. Ito, T. Nikata, and H. Ohtake. 1992. Phosphate taxis in Pseudomonas aeruginosa. J. Bacteriol. 174:5149-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, J., T. Nakamura, A. Kuroda, and H. Ohtake. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63:155-161. [DOI] [PubMed] [Google Scholar]
- 25.Kearns, D. B., J. Robinson, and L. J. Shimkets. 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J. Bacteriol. 183:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 27.Kelly-Wintenberg, K., and T. C. Montie. 1994. Chemotaxis to oligopeptides by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 29.Khashe, S., and J. M. Janda. 1998. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens. J. Clin. Microbiol. 36:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 31.Kirby, J. R., C. J. Kristich, M. M. Saulmon, M. A. Zimmer, L. F. Garrity, I. B. Zhulin, and G. W. Ordal. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42:573-585. [DOI] [PubMed] [Google Scholar]
- 32.Köhler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda, A., T. Kumano, K. Taguchi, T. Nikata, J. Kato, and H. Ohtake. 1995. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J. Bacteriol. 177:7019-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 35.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 37.Masduki, A., J. Nakamura, T. Ohga, R. Umezaki, J. Kato, and H. Ohtake. 1995. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J. Bacteriol. 177:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 39.Moench, T. T., and W. A. Konetzka. 1978. Chemotaxis in Pseudomonas aeruginosa. J. Bacteriol. 133:427-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulton, R. C., and T. C. Montie. 1979. Chemotaxis by Pseudomonas aeruginosa. J. Bacteriol. 137:274-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohga, T., A. Masduki, J. Kato, and H. Ohtake. 1993. Chemotaxis away from thiocyanic and isothiocyanic esters in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 113:63-66. [DOI] [PubMed] [Google Scholar]
- 43.Okumura, H., S. Nishiyama, A. Sasaki, M. Homma, and I. Kawagishi. 1998. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J. Bacteriol. 180:1862-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosario, M. M., and G. W. Ordal. 1996. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol. Microbiol. 21:511-518. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 49.Semmler, A. B., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 50.Shah, D. S., S. L. Porter, A. C. Martin, P. A. Hamblin, and J. P. Armitage. 2000. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J. 19:4601-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu, T. S., N. Le Novere, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell. Biol. 2:792-796. [DOI] [PubMed] [Google Scholar]
- 52.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1:784-789. [Google Scholar]
- 53.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327-2335. [DOI] [PubMed] [Google Scholar]
- 54.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 55.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 56.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, H., J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 182:3400-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 61.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]