Abstract
Biofilm formation in Staphylococcus epidermidis is dependent upon the ica operon-encoded polysaccharide intercellular adhesin, which is subject to phase-variable and environmental regulation. The icaR gene, located adjacent to the ica operon, appears to be a member of the tetR family of transcriptional regulators. In the reference strain RP62A, reversible inactivation of the ica operon by IS256 accounts for 25 to 33% of phase variants. In this study, icaA and icaR regulation were compared in RP62A and a biofilm-forming clinical isolate, CSF41498, in which IS256 is absent. Predictably, ica operon expression was detected only in wild-type CSF41498 and RP62A but not in non-IS256-generated phase variants. In contrast, the icaR gene was not expressed in RP62A phase variants but was expressed in CSF41498 variants. An icaR::Emr insertion mutation in CSF41498 resulted in an at least a 5.8-fold increase in ica operon expression but did not significantly alter regulation of the icaR gene itself. Activation of ica operon transcription by ethanol in CSF41498 was icaR dependent. In contrast, a small but significant induction of ica by NaCl and glucose (NaCl-glucose) was observed in the icaR::Emr mutant. In addition, transcription of the icaR gene itself was not significantly affected by NaCl-glucose but was repressed by ethanol. Expression of the ica operon was induced by ethanol or NaCl-glucose in phase variants of CSF41498 (icaR+) but not in RP62A variants (icaR deficient). These data indicate that icaR encodes a repressor of ica operon transcription required for ethanol but not NaCl-glucose activation of ica operon expression and biofilm formation.
Biofilm-forming coagulase-negative staphylococci (CoNS), particularly Staphylococcus epidermidis, are the etiological agents in a significant proportion of biomaterial-related nosocomial infections. Formation of S. epidermidis biofilms is proposed to occur in a two-step manner (20, 24, 26, 39) in which a cellular accumulation process to form the mature biofilm follows a rapid initial attachment to an inert synthetic surface. Initial adherence is mediated by polysaccharide adhesin (PS/A) (24) and/or one of several proteins (including autolysin [19]), and accumulation of cells is due to production of polysaccharide intercellular adhesin (PIA) (20). The PIA is encoded by the ica (intercellular adhesin) operon (20); however, it has been reported recently that this operon also encodes PS/A and that PS/A and PIA are chemically closely related (27). Production of PIA, which represents the key virulence factor of S. epidermidis, is subject to phase-variable regulation (38, 39). Little is known about the molecular basis of this phase variation process; however, recent evidence has indicated that alternating insertion and excision of an insertion sequence element is responsible for between 25 and 33% of ica operon switching in S. epidermidis RP62A under laboratory conditions (39). The insertion element IS257 has also been observed inserting at the ica locus of a clinical S. epidermidis isolate (32).
The role of the S. epidermidis and Staphylococcus aureus ica locus in biofilm infections has been examined by a number of investigators. Ziebuhr et al. (38) reported that 85% of S. epidermidis blood culture isolates contained the ica genes, compared to 6% of saprophytic isolates. A number of studies have indicated the usefulness of targeting the ica locus as a diagnostic marker to distinguish between invasive and contaminating isolates of S. epidermidis (3, 4, 11, 12). An interesting recent report found no significant difference in terms of ica status among CoNS isolated from healthy and sick infants in a neonatal intensive care unit but did note that biofilm-forming capacity was significantly greater in S. epidermidis strains isolated from the blood or skin of neonates with bacteremia (10). These authors concluded that regulation of biofilm expression might play a central role in disease caused by CoNS. Using animal models of catheter infection, Rupp et al. demonstrated the essential requirement for an intact ica operon and production of PS/A in the pathogenesis of S. epidermidis infection (34-36). Finally, the PS/A encoded by the ica operon of S. aureus was successfully used to immunize mice against S. aureus kidney infection (28, 29).
The ica gene cluster, which contains all the genes necessary for production of PIA, was identified using transposon mutagenesis to isolate S. epidermidis mutants defective in biofilm formation (17, 18, 20, 24). The ica locus contains an operon, icaADBC, which contains the structural genes required for PIA synthesis. Expression of the ica operon is regulated under in vivo conditions in S. aureus (29) and by environmental parameters in the laboratory in S. epidermidis. Anaerobic growth (9), the presence of subinhibitory concentrations of certain antibiotics, and environmental stresses (22, 31) all result in elevated expression of the ica operon or PIA synthesis. Transposon mutations in other loci, including rsbU, the first gene in an operon highly homologous to the sigB operon of S. aureus (22), which alter the expression of PIA synthesis have also been identified (22, 25).
Located upstream of the icaADBC operon is the divergently transcribed icaR gene. The role of icaR in biofilm formation has not been determined; however, amino acid sequence alignments suggest that the icaR gene product is a transcriptional regulator (30, 39). In this study we have examined the role of the icaR gene in the regulation of ica operon expression and biofilm formation in a biofilm-forming clinical isolate of S. epidermidis.
MATERIALS AND METHODS
Bacterial strains.
The biofilm-forming strain S. epidermidis CSF41498 was isolated from cerebrospinal fluid and obtained from the Department of Microbiology, Beaumont Hospital, Dublin, Ireland. The well-characterized reference strain S. epidermidis ATCC 35984 (RP62A) is a blood culture isolate originally characterized by Christensen et al. (6, 7) and later by, among others, Ziebuhr et al. (39). The strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strain | ||
| S. epidermidis CSF41498 | Biofilm positive, cerebrospinal fluid isolate | This study |
| S. epidermidis RP62A | Biofilm positive | ATCC 35984a |
| S. epidermidis ICAR1-4 | icaR::Emr | This study |
| S. aureus RN4220 | Restriction-negative, intermediate host for plasmid transfer from E. coli to S. epidermidis | 23 |
| E. coli XL-1 Blue | recA1 endA1 lac [F′ proAB lac1q Tn10(Tetr)] | Stratagene |
| Plasmid | ||
| pEC5 | pBluescript KS+ derivative. Source of ermB gene (Emr). Apr | 5 |
| pBT2 | Temperature-sensitive E. coli-Staphylococcus shuttle vector. Apr (E. coli) Cmr (Staphylococcus) | 5 |
| pSTBlue | PCR cloning vector. Kmr Apr | Novagen |
| pSTicaR | 786-bp PCR product containing the icaR gene amplified from CSF41498 using primers ICAR1 and IPR2 in pSTBlue | This study |
| pGEM3Z | Cloning vector. Apr | Promega |
| pSER1 | 810-bp BamHI-HindIII fragment from pSTicaR containing icaR in pGEM3Z | This study |
| pSER2 | 1,227-bp EcoRI-ClaI ermB fragment from pEC5 cloned into Bpu10I site of icaR gene in pSER1 | This study |
| pSER3 | 2,037-bp BamHI-HindIII fragment containing icaR::Emr in pBT2 | This study |
| pSER4 | 810-bp BamHI-HindIII fragment from pSER1 containing icaR cloned into pBT2 | This study |
ATCC, American Type Culture Collection.
Media and growth conditions.
Escherichia coli strains were grown at 37°C on Luria-Bertani medium supplemented, when required, with ampicillin (100 μg/ml). S. epidermidis and S. aureus strains were routinely grown at 37°C on brain heart infusion (BHI) (Oxoid) medium supplemented when required with the following antibiotics: chloramphenicol (10 μg/ml), erythromycin (10 μg/ml). BHI broth was supplemented with NaCl (4%) and glucose (0.5%) (NaCl-glucose) or ethanol (4%) (EtOH) as required. Bacteria were grown on Congo red agar (CRA) plates, which are composed of BHI agar supplemented with 5% sucrose (Sigma) and 0.8 mg of Congo red/ml (Sigma) to distinguish between wild-type (black, dry colony morphology) and variant (red, smooth colony morphology) phenotypes.
Genetic techniques.
Genomic and plasmid DNA was prepared using Wizard Genomic DNA and Plasmid Purification kits, respectively (Promega, Madison, Wis.). Prior to DNA extraction cells were pretreated with 50 μg of lysostaphin in 100 μl of 50 mM EDTA to facilitate subsequent lysis. Restriction and DNA-modifying enzymes (Roche, New England Biolabs, and MBI Fermentas) were used throughout according to the manufacturer's instructions. All oligonucleotide primers used for PCR, reverse transcription-PCR (RT-PCR), and DNA sequencing were supplied by MWG Biotech (Ebersberg, Germany). Custom automated DNA sequencing was performed by MWG Biotech (Milton Keynes, United Kingdom).
The primers ICAR1 (5′-CTCGAATTTGTTACATACTAG-3′) and ICAC1 (5′-CCATAGCTTGAATAAGGGAC-3′) were used in long-range PCRs to amplify a 4,204-bp fragment comprising the entire ica operon from purified genomic DNA using Expand High-Fidelity Taq DNA polymerase (Roche) under the following conditions: 34 cycles of 94°C for 15 s, 45°C for 30 s, and 68°C for 6 min.
Bacterial transformations.
Plasmid DNA was introduced into E. coli by CaCl 2 and heat shock transformation (37). Transformation of staphylococci was achieved by electroporation using a MicroPulser (Bio-Rad) and the following protocol: 25-ml shaking cultures were grown in BHI broth at 37°C to an optical density at 600 nm (OD600) of approximately 2. The cultures were chilled on ice for 15 min before being washed three times with 50 ml of sterile, ice-cold H2O and being resuspended in 50 μl of sterile H2O. Plasmid DNA (1 to 5 μg in 25 to 50 μl of H2O) was mixed with the cells and subjected to one electroporation pulse (1.8 kV, 2.5 ms). The electroporation mixture was then resuspended in 1 ml of BHI containing 0.5 M sucrose and a subinhibitory concentration of chloramphenicol as recommended by Bruckner (5) and incubated at 30°C for 2 to 3 h. Transformed cells were plated out on BHI agar (0.5 M sucrose) supplemented with chloramphenicol (10 μg/ml) and incubated for 24 to 48 h at 30°C.
Construction of plasmids and icaR::ermB allele replacement.
A 784-bp fragment containing the icaR gene from S. epidermidis CSF41498 was amplified by PCR using the primers ICAR1 (5′-CTCGAATTTGTTACATACTAG-3′) and IPR2 (5′-TTGGATAGAAAAGTAAAAAG-3′), cloned initially into the PCR cloning vector pSTBlue-1 (Novagen) and subsequently into pGEM3Z (Promega) on an 810-bp BamHI-HindIII fragment to create pSER1. DNA sequencing (data not shown) confirmed the absence of mutations in the PCR-amplified icaR gene cloned in pSER1. In order to construct an icaR mutant allele, a 1,227-bp EcoRI-ClaI fragment from pEC5 (5), which contains the ermB gene, was treated with T4 DNA polymerase to generate blunt ends and then cloned into a unique Bpu10I site in the icaR gene. In the resulting plasmid pSER2, the icaR gene is disrupted by the ermB gene inserted 193 bp from the icaR start codon. The orientation of the ermB gene in pSER2 was confirmed to ensure that the ermB promoter would not influence transcription of the ica operon following allele replacement (see Fig. 2B). In order to facilitate delivery of the icaR::ermB allele onto the chromosome of S. epidermidis, a 2,037-bp BamHI-HindIII fragment from pSER2 was subcloned into the temperature-sensitive E. coli-Staphylococcus shuttle vector pBT2 (5), which had also been digested with BamHI and HindIII. The resulting recombinant plasmid, designated pSER3, was first transformed by electroporation into S. aureus RN4220 and finally into S. epidermidis CSF41498, and Cmr (10 μg/ml) colonies were identified on BHI agar (0.5 M sucrose) plates. Plasmid profiles were determined to confirm the presence of intact pSER3 in transformed strains.
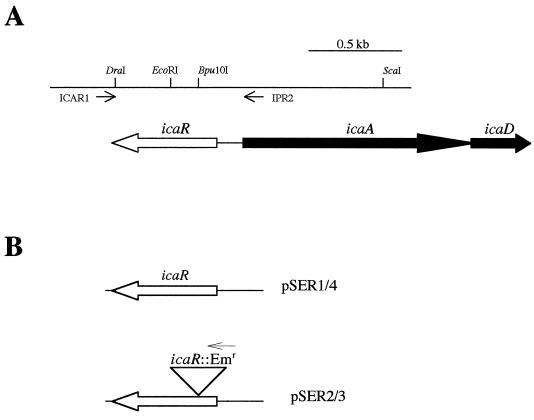
FIG. 2.
(A) Physical and restriction maps and primer binding sites of the S. epidermidis icaR-icaA region. (B) Plasmids used to construct the icaR chromosomal mutation. The position of the icaR::ermB insertion in pSER3 is indicated
Allele replacement of the temperature-sensitive pSER3 in CSF41498 was achieved following two rounds of growth at 42°C for 24 h without antibiotic selection and subsequent selection of erythromycin (10 μg/ml)-resistant colonies on BHI agar plates. Replica plating was then used to identify Emr Cms colonies, and PCR analysis using three different sets of primers to amplify the icaR gene confirmed the presence of the icaR::ermB allele on the chromosome of four independent mutants. Finally, RT-PCR analysis using the KCR1 and KCR2 primers described below (see Fig. 4A) was used to confirm the absence of wild-type icaR transcript in the four independent mutants.
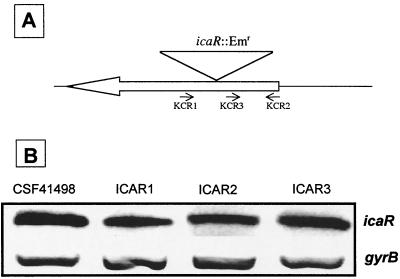
FIG. 4.
(A) Physical maps and icaR primer binding sites used for RT-PCR analysis in CSF41498 and the icaR::Emr mutants. Primers KCR2 and KCR3 were designed to measure icaR transcription in ICAR mutants. (B) Comparative measurement of icaR (primers KCR 2 and KCR3 [panel A]) and gyrB (control) transcription in S. epidermidis CSF41498 and three independent icaR insertion mutants, ICAR1, ICAR2, and ICAR3. RNA was prepared from cultures grown in BHI at 37°C to the early exponential phase of the growth curve (OD600 = 2).
For icaR overexpression and complementation experiments, an 810-bp BamHI-HindIII fragment containing the icaR gene was cloned from pSER1 into pBT2 and the recombinant plasmid was designated pSER4.
RNA purification and RT-PCR.
Bacterial cells were collected and immediately stored in RNAlater (Ambion) to ensure maintenance of RNA integrity prior to purification. Total RNA was subsequently isolated using the GenElute Total RNA purification kit (Sigma) according to the manufacturer's instructions following a 5- to 10-min pretreatment of the cells with 50 μg of lysostaphin in 100 μl of 50 mM EDTA. Purified RNA was eluted and stored in RNAsecure resuspension solution (Ambion), and the integrity of the RNA was confirmed by agarose gel electrophoresis. Residual DNA present in RNA preparations following purification was removed using DNAfree DNase treatment and removal reagents (Ambion).
RT-PCR was performed using the OneStep RT-PCR kit (Qiagen) following the manufacturer's recommended protocol. Master mixes were prepared using primers as follows: for gyrB transcripts, 5′-TTATGGTGCTGGACAGATACA-3′ and 5′-CACCGTGAAGACCGCCAGATA-3′; for icaA transcripts, 5′-AACAAGTTGAAGGCATCTCC-3′ and 5′-GATGCTTGTTTGATTCCCT-3′; for icaR transcripts, KCR1 5′-GGTAAAGTCCGTCAATGGAA-3′ and KCR2 5′-CGCAATAACCTTATTTTCCG-3′. For all three of these RT-PCRs, RT was performed at 55°C for 30 min followed by 23 amplification cycles of 94°C for 20 s, 50°C for 20 s, and 72°C for 20 s.
RT-PCR analysis of icaR expression in the icaR::Emr mutants was carried out using the KCR2 primer together with the primer KCR3 (5′-GCAAAAAATCTATAAAG-3′) (see Fig. 4A). RT for RT-PCRs using these primers was performed at 45°C for 30 min followed by 23 amplification cycles of 94°C for 20 s, 47°C for 20 s, and 72°C for 20 s.
The gyrB gene is constitutively expressed in S. aureus (14) and was used as an internal standard in these experiments. The sequences of the primers used for gyrB RT-PCR were based on the corresponding S. aureus gyr primers (14) but were adjusted for complementarity to the DNA sequence of an S. epidermidis genomic clone with homology to the S. aureus gyrB gene (GenBank accession number AF269920).
Analysis of RT-PCR data.
The intensity of 23S rRNA bands on nondenaturing 1% agarose gels was measured in individual samples prior to RT-PCR to ensure similar RNA loading in RT-PCRs within individual experiments, which are presented below in separate figures. In addition, expression of the constitutively expressed gyrB gene was measured in parallel with measurements of icaA and icaR transcript levels and used to standardize variations in RNA loading between samples in each experiment. All RT-PCRs were optimized to ensure that amplification was terminated in the linear range. For the RT-PCRs described in this study, this was achieved by terminating the PCRs after 23 amplification cycles. Densitometry was performed using the Stratagene Eaglesight software package to compare relative expression levels between samples.
Biofilm assays.
Semiquantitative determinations of biofilm formation in 96-well tissue culture plates (Sigma) were performed based on the method of Christensen et al. (8) as described by Ziebuhr et al. (38) with the following modifications. Bacteria were grown at 37°C in BHI and each strain was tested at least eight times before being washed three times with distilled H2O and dried for 1 h at 56°C as recommended by Gelosia et al. (13) prior to staining with a 0.4% crystal violet solution. The absorbance of the adhered, stained cells was measured at 492 nm using a Multiskan plate reader (Flow Laboratories).
Statistical analysis.
Statistical analysis of data was performed using Microsoft Excel or SPSS software packages.
RESULTS
Non-IS256-mediated regulation of biofilm formation.
Previous data showed that reversible insertion of IS256 into the ica operon could account for between 25 and 33% of phase variants in S. epidermidis RP62A (39) and suggested that unpredictable IS256 insertions at other genetic loci may also contribute to the production of phase variants. In order to investigate non-IS256-mediated ica operon regulation, we performed a PCR screen for the presence of IS256 among a collection of biofilm-forming, ica-positive cerebrospinal fluid isolates of S. epidermidis, using the IS256 primers described by Ziebuhr et al. (39). One isolate, CSF41498, identified as ica positive and IS256 negative (data not shown) was chosen for further analysis for the following reasons: determination of the ica operon nucleotide sequence from CSF41498 and RP62A revealed no DNA sequence variations (data not shown), and in semiquantitative biofilm assays (see Materials and Methods) both were found to have similar biofilm-forming capacities in BHI; the OD492 (± standard deviation) of adherent biofilms stained with crystal violet for CSF41498 was 1.69 ± 0.20, compared to 1.65 ± 0.67 for RP62A.
icaR gene expression in biofilm-positive and biofilm-negative phase variants of CSF41498.
Two phase variants (red, smooth colony morphology) of CSF41498 were isolated on CRA as described previously (39). PCR amplification of the entire ica operon from the two variants generated products of the expected size and confirmed that neither contained an IS256 insertion in the ica operon (data not shown). Plasmid profile analysis of CSF41498 and the two phase variants was performed and revealed that all three strains shared the same plasmid profile (data not shown), thereby confirming their relationship to each other. For comparative analysis two phase variants of RP62A which did not contain IS256 insertions in the ica operon were also isolated.
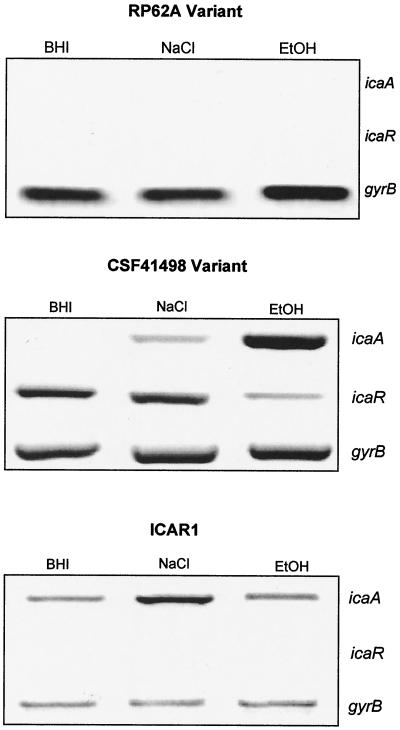
Semiquantitative biofilm analysis demonstrated that the two RP62A phase variants examined were completely biofilm negative and that the two CSF41498 variants displayed only weak biofilm-forming capacity in BHI broth at 37°C (OD492 ± standard deviation, 0.112 ± 0.08).
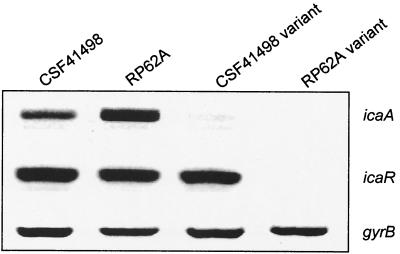
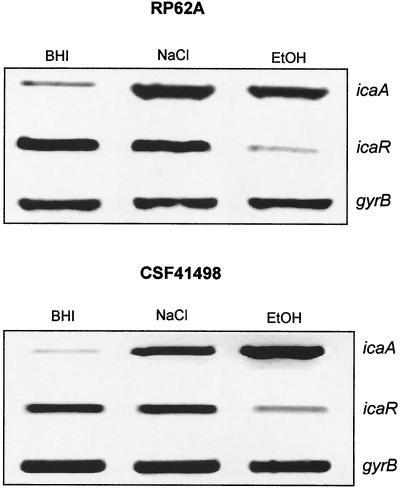
To examine the genetic basis for the variant phenotypes in CSF41498 and RP62A, we conducted a semiquantitative examination of both icaA and icaR gene expression by using RT-PCR. This revealed that icaA transcription levels were two- to threefold higher in the wild-type RP62A during early logarithmic growth than that in CSF41498, although this increased ica operon transcription in RP62A did not correlate with increased biofilm-forming capacity. Predictably, icaA transcription levels were significantly reduced or absent in CSF41498 and RP62A variants when compared to their parental strains (Fig. 1). In RP62A, both icaA and icaR transcription was detected in the wild-type parent, while neither gene was expressed in the phase variants. In contrast, expression of the icaR gene was detected in both wild-type and phase variants of CSF41498 (Fig. 1). Thus, there were two major differences between CSF41498 and RP62A variants: the icaR gene was expressed only in the CS41498 variants and, unlike RP62A variants, the CSF41498 variants retained the capacity to form a weak biofilm. Nucleotide sequence determination of the icaR gene and ica operon promoter region from the CSF41498 variants confirmed the absence of regulatory mutations (data not shown). Detection of icaR gene expression in wild-type and phase variants of CSF41498 suggested that this gene might play a different regulatory role in the two phenotypic variants of this strain. Alignment of the IcaR predicted amino acid sequence using the Domain Architecture Retrieval Tool on the National Center for Biotechnology Information website suggested that this protein is a member of the tetR family of transcriptional regulators.
FIG. 1.
Comparative measurement of icaA, icaR, and gyrB (control) transcription in wild-type and phase variants of S. epidermidis CSF41498 and RP62A. RNA was prepared from cultures grown in BHI at 37°C to the early exponential phase of the growth curve (OD600 = 2). Expression of gyrB is constitutive (14) and was used as an internal control in this experiment.
Construction and characterization of an icaR insertion mutation.
To investigate the role of the icaR gene in the regulation of ica operon expression in CSF41498, an IcaR mutant strain was constructed. To achieve this, the icaR gene on the chromosome of CSF41498 was replaced with an icaR::Emr allele in which the icaR gene was interrupted by the ermB gene, which encodes resistance to erythromycin (Fig. 2) (see Materials and Methods). Four independent mutants (ICAR1, ICAR2, ICAR3, and ICAR4) were isolated and characterized. The colony morphology of the ICAR mutant strains grown on CRA at 37°C (black color, smooth regular surface) was different from that of the parental strain (black color, dry irregular surface).
Interestingly, biofilm formation by ICAR1 grown in BHI medium at 37°C for 24 h was unaffected compared to that of CSF41498 (OD492, 1.69 ± 0.20 for CSF41498, compared to 1.57 ± 0.29 for ICAR1), indicating that the icaR gene may not be involved in biofilm formation. Similar results were obtained in biofilm assays with all four ICAR mutants (average OD492 for ICAR1, ICAR2, ICAR3, and ICAR4 was 1.4 ± 0.18).
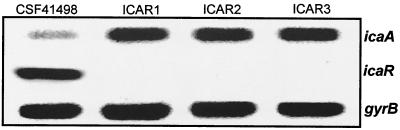
To determine the impact of the icaR::Emr mutation on ica operon expression, transcriptional regulation of the icaA gene was assessed by RT-PCR in all four mutant strains, ICAR1 to -4, and CSF41498 in cultures grown in BHI medium at 37°C. This revealed an at least 5.8-fold increase in icaA transcription in the icaR::Emr mutants above that of the wild-type strain (Fig. 3). These data indicate that the icaR gene encodes a repressor of ica operon expression but that IcaR activity alone does not completely repress ica operon expression. Interestingly, our observation that biofilm formation by ICAR1 was not significantly different from that of wild-type CSF41498 in BHI medium was somewhat at odds with these transcriptional data, which indicated that the overall levels of icaA transcription are significantly higher in the ICAR1 mutant. One possible explanation may be that the biofilm assays performed in this study using BHI medium are not sensitive enough to reflect phenotypic differences associated with increased ica operon expression. This conclusion is supported by our earlier observation that biofilm-forming capacity in RP62A and CSF41498 in BHI medium was similar even though ica operon expression was two- to threefold higher in RP62A (Fig. 1).
FIG. 3.
Comparative measurement of icaA, icaR (KCR1 and KCR2 [see Fig. 4A]), and gyrB (control) transcription in S. epidermidis CSF41498 and three independent icaR insertion mutants, ICAR1, ICAR2, and ICAR3. RNA was prepared from cultures grown in BHI at 37°C to the early exponential phase of the growth curve (OD600 = 2).
The affect of the Emr insertion on the transcription of the icaR gene itself was also examined by RT-PCR. Because the primers used to measure icaR transcription described earlier flank the icaR::Emr insertion, a different RT primer (KCR3) was used for cDNA synthesis in the ICAR mutants (Fig. 4A). Comparative RT-PCR analysis with wild-type CSF41498 revealed that transcription of the icaR gene was not significantly affected in the ICAR mutants (Fig. 4B), which suggested that the icaR gene product does not regulate its own transcription.
Role of the icaR gene in the environmental modulation of ica operon expression.
Our finding that the icaR gene product is a repressor of ica operon transcription prompted us to examine icaA and icaR expression in the wild-type and ICAR mutant strains under known biofilm-promoting growth conditions by using RT-PCR. Biofilm formation by S. epidermidis is subject to environmental regulation: factors such as salt (31) and ethanol (22) can promote biofilm formation. In wild-type CSF41498 we found a strong induction of ica operon expression in both NaCl-glucose (16-fold) and ethanol (26-fold) (Fig. 5). In addition, icaR transcription displayed a 5.6-fold decrease in the presence of ethanol but was not significantly affected by the presence of NaCl-glucose (Fig. 5). Given that the icaR gene product does not appear to regulate its own transcription (Fig. 4), ethanol repression of icaR transcription is unlikely to be the result of altered IcaR activity and may involve an additional transcription factor(s). Similar results were obtained for wild-type RP62A, in which growth in the presence of NaCl-glucose or ethanol resulted in 6.6- and 5.2-fold increases in icaA transcript levels, respectively. Growth in the presence of ethanol caused a 23-fold reduction in icaR transcription, whereas NaCl-glucose had no affect on icaR expression (Fig. 5). Interestingly, in the ICAR1 mutant, growth in the presence of ethanol had no significant effect on ica operon expression, whereas NaCl-glucose resulted in a small but significant 2.4 (± 0.7)-fold (± standard error of the mean) induction of ica operon expression (P = 0.046) (Fig. 6). These findings suggested that ethanol induction of ica operon expression was icaR dependent but that activation of ica operon expression by NaCl-glucose was icaR independent.
FIG. 5.
Comparative measurement of icaA, icaR, and gyrB (control) transcription in S. epidermidis RP62A and CSF41498. RNA was prepared from cultures of each strain grown simultaneously to the mid-exponential phase of the growth curve (OD600 = 4.5) in BHI, NaCl-glucose (BHI supplemented with 4% NaCl and 0.5% glucose) and EtOH (BHI supplemented with 4% ethanol).
FIG. 6.
Comparative measurement of icaA, icaR, and gyrB (control) transcription in phase variants of S. epidermidis RP62A and CSF41498 and the icaR insertion mutant ICAR1. RNA was prepared from cultures grown to the mid-exponential phase of the growth curve (OD600 = 4.5) in BHI, NaCl-glucose (BHI supplemented with 4% NaCl and 0.5% glucose), and EtOH (BHI supplemented with 4% ethanol).
To further assess the role of the icaR gene in the environmental regulation of icaA expression, we tested the ability of ethanol and salt to induce ica operon expression in phase variants of RP62A and CSF41498. These experiments revealed that in the CSF41498 variant in which the icaR gene is expressed, significant increases in icaA expression were detected in the presence of NaCl-glucose and ethanol (Fig. 6), although the effect of the latter was much more potent than the former. Consistent with these data, biofilm assays revealed that ethanol and NaCl-glucose induced biofilm formation in the CSF41498 variants: biofilm-forming capacity (OD492 ± standard deviation) in the standard BHI broth was 0.107 ± 0.06, compared to 0.410 ± 0.22 in the presence of ethanol and 0.21 ± 0.26 in the presence of NaCl-glucose. As in wild-type CSF41498, salt did not influence icaR expression in the CSF41498 variant, while ethanol resulted in a 5.8-fold decrease in icaR transcription (Fig. 6). In contrast, examination of an RP62A variant in which the icaR gene was not expressed revealed that neither ethanol nor salt induced icaA or icaR gene expression (Fig. 6) or biofilm formation (data not shown).
Complementation of ICAR1.
Analysis of the icaR insertion mutants indicated that IcaR acts as a repressor of ica operon expression required for ethanol activation of ica operon expression. In order to complement the ICAR1 mutant and to determine the effect of overexpressing the icaR gene in trans, the icaR gene was amplified by PCR from CSF41498 genomic DNA and ultimately cloned into the E. coli-Staphylococcus shuttle vector pBT2 as described in Materials and Methods to generate pSER4. Electroporation was used to transform pSER4 and the control vector pBT2 into CSF41498. Because pBT2 contains a temperature-sensitive origin of replication, strains containing pBT2 and derivatives were cultured at 30°C with chloramphenicol (10 μg/ml) selection.
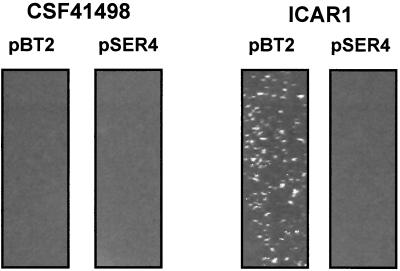
Broth cultures of ICAR1 bearing pBT2 and pSER4 displayed a striking phenotypic difference. Cultures of ICAR1(pBT2) formed macroscopically visible large cell aggregates or clusters, whereas ICAR1(pSER4) cultures displayed little cell clumping and were similar to wild-type CSF41498(pBT2) (Fig. 7). Growth of CSF41498 and ICAR1 plasmid-bearing strains in the presence of ethanol induced cell cluster formation in all cultures. Enhanced cell cluster formation or intercellular adhesion is consistent with elevated expression of the ica operon in the ICAR1 mutant and in CSF41498 grown in the presence of ethanol. Moreover, this was the first indication that the introduction of the icaR gene in trans was sufficient to complement the icaR::Emr phenotype. It may also be significant that the ICAR1 cell clumping phenotype was more pronounced in cultures of this strain containing pBT2, which were grown at 30°C with chloramphenicol selection.
FIG. 7.
Cell cluster formation in overnight cultures of S. epidermidis CSF41498 (left) and ICAR1 (right) bearing plasmids pBT2 (control) (lanes 1 and 3) and pSER4 (icaR gene) (lanes 2 and 4) grown at 30°C in BHI medium supplemented with chloramphenicol (10 μg/ml).
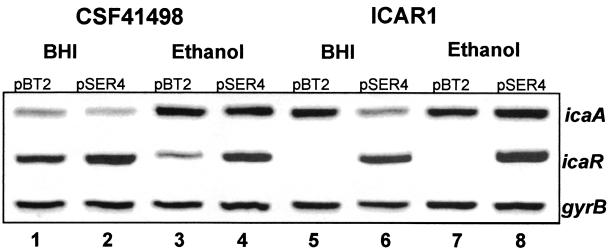
When the plasmid-bearing CSF41498 and ICAR1 strains were grown at 30°C on CRA containing chloramphenicol (10 μg/ml), strains carrying pSER4 were visibly red compared to the darker color of the control strains containing pBT2. This observation was consistent with the proposed negative regulatory role of the icaR gene product. However, we also observed that strains carrying pBT2 grown at 30°C on CRA supplemented with chloramphenicol (10 μg/ml) were not as dark as their plasmid-free parental strains, a phenotypic difference which appeared to be primarily due to the presence of the antibiotic rather than the altered temperature. RT-PCR was used to measure the effect of pBT2 and pSER4 on icaA and icaR transcript levels in plasmid-bearing CSF41498 cultures grown to an OD600 of 1.0 in BHI medium. These experiments confirmed that levels of icaR transcript were elevated in the CSF41498 strain containing pSER4 but not in the strain bearing pBT2 (Fig. 8). Associated with this increased icaR expression was an approximately twofold decrease in icaA expression (Fig. 8). These findings are consistent with our earlier data indicating that icaR encodes a repressor of ica operon expression.
FIG. 8.
Comparative measurement of icaA, icaR, and gyrB (control) transcription in S. epidermidis CSF41498 and ICAR1 strains complemented with plasmids pBT2 (control) and pSER4 (icaR). RNA was prepared from cultures grown to an OD600 of 1 at 30°C in BHI and BHI EtOH (4% ethanol), with chloramphenicol (10 μg/ml) selection. Lane 1, CSF41498(pBT2) in BHI; lane 2, CSF41498(pSER4) in BHI; lane 3, CSF41498(pBT2) in BHI-EtOH; lane 4, CSF41498(pSER4) in BHI-EtOH; lane 5, ICAR1(pBT2) in BHI; lane 6, ICAR1(pSER4) in BHI; lane 7, ICAR1(pBT2) in BHI-EtOH; lane 8, ICAR1(pSER4) in BHI-EtOH.
In order to determine if the effect of icaR::Emr mutation on ica operon transcription could be complemented to wild type, RT-PCR was used to measure icaA and icaR transcript levels in RNA extracted from plasmid-bearing CSF41498 and ICAR1 cultures. In these experiments, complementation was defined as the restoration of icaA transcript levels to those of CSF41498 in complemented strains grown in the presence and absence of ethanol. These experiments revealed that icaA expression levels in ICAR1 were restored to wild-type levels observed in CSF41498(pBT2) (Fig. 8). In addition, ethanol failed to induce icaA expression in ICAR1(pBT2), but an approximately 2.6-fold icaA induction was measured in ICAR1(pSER 4) grown in ethanol (Fig. 8). These results corroborate our earlier findings that identified the icaR gene product as a negative transcriptional regulator which contributes to the environmental modulation of ica operon expression and biofilm formation.
DISCUSSION
The persistent and recurrent characteristics of S. epidermidis biofilm infections highlight the need to develop new therapeutic strategies targeted at this organism. In this context, understanding the regulation of ica operon expression and biofilm formation is of primary importance. This study provides the first experimental evidence to demonstrate that expression of the ica operon in a biofilm-forming clinical isolate of S. epidermidis, CSF41498, is regulated by a transcriptional repressor encoded by the divergently transcribed icaR gene. The function of icaR was examined in CSF41498 for a number of reasons. The insertion sequence element IS256, which contributes directly (39) and possibly indirectly to ica operon regulation, is absent in this strain. In addition, the biofilm-forming capacity of CSF41498 is similar to that of the well-characterized reference strain RP62A, and the nucleotide sequence of the ica operons from both strains is identical. Finally, when phase variants of CSF41498 were compared with non-IS256-generated RP62A variants, icaR gene expression was detected only in the CSF41498 variants (Fig. 1), indicating that there may be a functional role for this gene in both phenotypic variants of this strain.
Differential regulation of icaR gene expression suggests that ica operon regulation in RP62A and CSF41498 phase variants may be fundamentally different. In CSF41498 variants, it appears that IcaR repression of ica operon transcription contributes to impaired biofilm-forming capacity. However, because icaR transcript levels are similar in wild-type and phase variants of CSF41498, an additional factor(s) may act in concert with IcaR to repress ica operon transcription in these phase variants. In RP62A variants, the absence of icaR expression indicates that an alternative regulatory mechanism results in ica operon repression.
In CSF41498, a chromosomal icaR::Emr mutation led to increased ica operon expression, indicating that the icaR gene product is a transcriptional repressor. Consistent with this finding, overexpression of icaR on a multicopy plasmid resulted in repression of ica operon transcription. Predicted IcaR amino acid sequence alignments suggest that IcaR is a member of the tetR family of transcriptional regulators. This family of proteins, which includes the qacR repressor of the QacA multidrug efflux pump system in S. aureus, are characterized by conserved helix-turn-helix DNA-binding domains at the amino-terminal ends and divergent carboxy-terminal domains which may be involved in interactions with compounds that modulate their regulatory activity (2, 15, 33). Thus, QacR binding to a range of inducing compounds interferes with its ability to repress qacA transcription (15). It is tempting to speculate that IcaR activity may also be modulated by interaction with inducing compounds.
Although regulation of the ica operon was altered in the icaR::Emr mutants, IcaR did not appear to autoregulate expression of its own gene. Most other members of the tetR family of regulatory proteins that are divergently transcribed from the structural gene that they regulate appear to regulate the expression of their own genes, including TetR (21), Pseudomonas putida CamR (1), and Streptomyces glaucescens TcmR (16). It therefore appears somewhat anomalous that icaR transcription was not altered in the icaR::Emr mutant. However, it is interesting that in S. aureus the QacR protein does not autoregulate transcription of its own gene either (15), suggesting that this may be a shared regulatory feature of tetR family proteins in staphylococci.
Recent reports have provided evidence that environmental stresses such as high osmolarity (NaCl) or ethanol concentrations can increase ica operon expression and promote biofilm formation in S. epidermidis (23, 32). In this study we observed that biofilm formation was induced by ethanol in phase variants of CSF41498 which were icaR+, but not in RP62A variants which were icaR deficient. To investigate the role of the icaR gene in the environmental modulation of biofilm formation, we examined the effect of ethanol and NaCl-glucose on the regulation of ica operon expression in RP62A, CSF41498, and the ICAR1 mutant. These data revealed that ethanol and NaCl-glucose strongly induced ica operon expression in wild-type CSF41498 and phase variants (both icaR+). Similarly, ethanol and NaCl-glucose induced ica operon expression in the wild-type RP62A (icaR+) but not in a phase variant (icaR deficient). In contrast, there was no significant induction of ica expression by ethanol in the icaR insertion mutant, and a small but significant induction was observed in the presence of NaCl-glucose. In both CSF41498 and RP62A, ethanol induction of icaA gene expression was associated with decreased icaR transcription. In contrast, NaCl-glucose did not significantly affect icaR regulation. Apparently, in CSF41498 induction of ica operon expression in the presence of ethanol is icaR dependent and ethanol directly or indirectly represses icaR transcription. In contrast, NaCl-glucose, which also induces ica operon expression, does not significantly alter icaR regulation and appears to activate the ica operon through a separate pathway which does not involve icaR. Recently, Knobloch et al. proposed that separate pathways can activate ica operon expression in response to different environmental stresses (22). This proposal was based on evidence that a biofilm-negative rsbU transposon mutant of S. epidermidis 1457 (rsbU is a positive regulator of the alternative sigma factor SigB) was enabled to form a biofilm again by ethanol only and not by NaCl (22). Our evidence that NaCl-glucose-induced ica operon expression is icaR independent but that activation of ica operon transcription by ethanol does require the icaR gene may also suggest the existence of separate ica operon-inducing regulatory pathways. Moreover, these findings may also suggest that icaR-dependent ethanol activation of the ica operon does not involve SigB but that a separate pathway involving the alternative sigma factor can independently induce ica expression. Finally, the possible existence of separate pathways is further supported by our observation that, in CSF41498, ethanol is a more potent activator of ica expression than NaCl-glucose.
The evidence presented here suggests that the icaR gene product is a transcriptional repressor which plays an adaptive role in S. epidermidis biofilm formation by modulating the regulation of ica expression in response to specific environmental conditions.
Acknowledgments
This work was funded by a grant from the Research Committee of the Royal College of Surgeons in Ireland to J.O'G. We are grateful to Pfizer (Ireland) for generously supporting the establishment of the RCSI Microbiology Laboratory at the RCSI Education and Research Centre.
Plasmids pBT2 and pEC5 were a kind gift from R. Bruckner. Grateful thanks go to C. J. Dorman for critical reading of the manuscript. We thank Peadar Clarke for experimental advice and assistance throughout this study.
REFERENCES
- 1.Aramaki, H., Y. Sagara, H. Kabata, N. Shimamoto, and T. Horiuchi. 1995. Purification and characterization of a cam repressor (CamR) for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J. Bacteriol. 177:3120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aramaki, H., N. Yagi, and M. Suzuki. 1995. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 8:1259-1266. [DOI] [PubMed] [Google Scholar]
- 3.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arciola, C. R., S. Collamati, E. Donati, and L. Montanaro. 2001. A rapid PCR method for the detection of slime-producing strains of Staphylococcus epidermidis and S. aureus in periprosthesis infections. Diagn. Mol. Pathol. 10:130-137. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, G. D., A. L. Bisno, J. T. Parisi, B. McLaughlin, M. G. Hester, and R. W. Luther. 1982. Nosocomial septicemia due to multiply antibiotic-resistant Staphylococcus epidermidis. Ann. Intern. Med. 96:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Silva, G. D., M. Kantzanou, A. Justice, R. C. Massey, A. R. Wilkinson, N. P. Day, and S. J. Peacock. 2002. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 13.Gelosia, A., L. Baldassarri, M. Deighton, and T. Van Nguyen. 2001. Phenotypic and genotypic markers of Staphylococcus epidermidis virulence. Clin. Microbiol. Infect. 7:193-199. [DOI] [PubMed] [Google Scholar]
- 14.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 16.Guilfoile, P. G., and C. R. Hutchinson. 1992. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J. Bacteriol. 174:3659-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann, C., and F. Gotz. 1998. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl. Bakteriol. 287:69-83. [DOI] [PubMed] [Google Scholar]
- 19.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 21.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 22.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. Knobloch, H. A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenney, D., K. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 2000. Vaccine potential of poly-1-6 beta-d-N-succinylglucosamine, an immunoprotective surface polysaccharide of Staphylococcus aureus and Staphylococcus epidermidis. J. Biotechnol. 83:37-44. [DOI] [PubMed] [Google Scholar]
- 29.McKenney, D., K. L. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523-1527. [DOI] [PubMed] [Google Scholar]
- 30.O'Gara, J. P., and H. Humphreys. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50:582-587. [DOI] [PubMed] [Google Scholar]
- 31.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohde, H., J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2001. Correlation of biofilm expression types of Staphylococcus epidermidis with polysaccharide intercellular adhesin synthesis: evidence for involvement of icaADBC genotype-independent factors. Med. Microbiol. Immunol. 190:105-112. [DOI] [PubMed] [Google Scholar]
- 33.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4:2051-2062. [DOI] [PubMed] [Google Scholar]
- 34.Rupp, M. E., P. D. Fey, C. Heilmann, and F. Gotz. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183:1038-1042. [DOI] [PubMed] [Google Scholar]
- 35.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]