Abstract
Streptomycetes are filamentous soil bacteria that produce spores through a complex process of morphological differentiation. The ram cluster plays an important part during the development. The ram genes encode a membrane-bound kinase (RamC), a small protein (RamS), components of an ABC transporter (RamAB), and a response regulator (RamR). While the introduction of an extra copy of the ram cluster accelerates development in Streptomyces lividans, ramABR disruption mutants are unable to produce aerial hyphae and spores. The developmental regulation of ram gene transcription was analyzed. Transcription of the ram genes occurred only on solid rich media and not on minimal media. The ramR gene is transcribed from a single promoter during all growth stages, with the highest levels during aerial growth. The ramCSAB genes comprise one operon and are transcribed from one principal promoter, P1, directly upstream of ramC. Transcription of ramCSAB was already observed during vegetative growth, but was strongly upregulated upon initiation of formation of aerial hyphae and was decreased during late stages of development. A large inverted repeat located downstream of ramS terminated the majority of transcripts. The introduction of ramR on a multicopy vector in S. lividans strongly induced P1 activity, while disruption of this regulator eliminated all P1 promoter activity. This shows that ramR is a crucial activator of ramCSAB transcription. Importantly, in bldA, bldB, bldD, or bldH mutants, ramR and ramCSAB are not transcribed, while ram gene transcription was observed in the earliest whi mutant, whiG. This indicates that the transcription of the ram genes marks the transition from vegetative to aerial growth.
Many unicellular bacteria can temporarily adopt a multicellular organization to perform specific tasks. Examples of this include swarming, in which numerous cells aggregate and move collectively over a surface, and myxospore formation, which is preceded by aggregation resulting in the formation of a sacculus (stalk of fruiting body) allowing the subsequent development of resistant cells (myxospores) (31). Different from most prokaryotes, the gram-positive soil bacterium Streptomyces mainly exists as a multicellular organism. Streptomycetes rely on a process of spore formation for propagation and survival. The life cycle of these organisms resembles that of filamentous fungi. After germination of a spore, a network of branched filamentous vegetative hyphae is produced. At late vegetative growth, secondary metabolism is switched on and reproductive growth is initiated. One of the first steps in reproductive growth is the production of hydrophobic aerial hyphae that erect themselves out of the plane of the agar. The initiation of formation of aerial hyphae is believed to be dependent on metabolic status, stress, and a cascade of extracellular signaling molecules (17, 41). Aerial hyphae undergo several transformation steps ultimately leading to the formation of unigenomic spores at the hyphal tips (5, 9). Elemental steps in the process of morphological development have been elucidated by the characterization of developmental mutants. Two classes of developmental mutants are known: those that are unable to produce aerial hyphae (bld mutants) and those that are unable to produce gray mature spores (whi mutants) (3, 4, 11, 15, 24, 27, 29, 41). As the result of genetic analysis of whi and bld mutants, a model of the genetic regulation and interplay of the developmental genes during this process is now emerging (6, 9).
Developmental genes have also been identified because of their ability to stimulate or delay the process of morphological development in mutant or wild-type strains. For example craA, encoding a carbon source-dependent repressor molecule, was identified by the ability to block morphological development of Streptomyces griseus when introduced on a multicopy vector (34). Conversely, Ueda and coworkers identified a cluster of genes (the amf cluster) that restored the development of an A-factor-deficient developmental mutant of S. griseus (HH1) when overexpressed (36). In a similar fashion, the Streptomyces coelicolor ram cluster was identified after it was found to stimulate formation of aerial hyphae of S. lividans when introduced on a low-copy-number vector (22). The amf and the ram cluster appear to be homologous, although the level of DNA sequence identity between both clusters is not as high as one would expect from orthologues. However, the overall architectures of the open reading frames (ORFs) are fully identical, and the proteins encoded are highly similar. Both gene clusters consist of a gene encoding a serine threonine kinase (ramC or amfT), a gene encoding a small protein with unknown function (ramS or amfS), two genes that are translationally coupled encoding components of an ABC transporter (ramA and ramB or amfB and amfA), and a response regulator (ramR or amfR) oriented in the opposite direction. The molecular basis for the accelerated development of Streptomyces lividans complemented with the S. coelicolor ram genes is not known. As observed with the S. coelicolor ram cluster, the S. lividans ram cluster (which is 99.7% identical to the S. coelicolor ram cluster) also accelerated the development of this strain. This indicates that the stimulation of the development in this strain is due to a multicopy effect rather than to complementation of a “defective” S. lividans ram cluster. Transcription analysis revealed that ramAB transcript levels are extremely low in both S. coelicolor and S. lividans. S. lividans transformants carrying an extra copy of the full ram cluster displayed an elevated ramAB transcript level (16). The importance of the ram and amf genes has been demonstrated by the phenotypes observed with disruption mutants. The disruption of amfR in S. griseus results in a fully bld phenotype (35). The disruption of ramABR in S. lividans also resulted in a bld phenotype, while ramR disruption mutants display a delayed development (16).
To further examine the role of the ram genes during morphological differentiation, the transcription of the S. lividans ram genes was analyzed during growth and development. Furthermore, the effect of ramR copy number on ramCSAB transcription was examined. To relate ram gene transcription to a characteristic developmental stage, transcription in various developmental mutants was examined. The results demonstrated the intricate relationship between ram gene expression and the onset of production of aerial hyphae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptomyces strains were routinely grown on the rich solid medium R2YE or the minimal medium SMM (18). For growth in liquid medium, YEME supplemented with 1% glycine and 1% MgSO4 or TSBS (3% [wt/vol] tryptone soya broth with 10% [wt/vol] sucrose) was used (18). The streptomyces strains used in this study are S. lividans 1326, S. coelicolor A3(2) M145, M512 (M145 ΔredD actII orfIV) (10), bldA39 (J1700) (20), bldD53 (J774) (24), and bldH109 (WC109) (3), as well as a whiG::hyg (J2400) strain (9).
Escherichia coli JM109, which was used as a host for pTZ19R clones, was cultivated at 37°C in Luria-Bertani (LB) medium or on LB agar plates (30). The modification-deficient E. coli strain ET12567 was used for the generation of unmethylated DNA, which was used for the transformation of S. coelicolor strains (23). Antibiotics (ampicillin, apramycin, and hygromycin) were purchased from Ducheva (Haarlem, The Netherlands). Thiostrepton was a kind gift of Bristol-Meyers Squibb.
In vitro DNA manipulations.
DNA manipulation and transformation of E. coli were performed according to standard procedures (30). Isolation of Streptomyces plasmid DNA and polyethylene glycol-assisted transformation of Streptomyces protoplasts were performed according to protocols described in reference 18.
PCR was performed in a minicycler (MJ Research, Watertown, Mass.) with Pfu polymerase (Stratagene, La Jolla, Calif.) and the buffer provided by the supplier in the presence of 5% (vol/vol) dimethyl sulfoxide, 25 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate (dNTP), and 10 ng of template. The amplification was performed in 30 cycles consisting of 60 s at 94°C, 60 s at 54°C, and 60s at 72°C.
In vivo promoter-probe experiments.
The low-copy-number plasmid pIJ2587 (approximately 5 to 10 copies per genome) was used to detect promoter activity of the various ram DNA fragments (38). This plasmid contains the promoterless undecylprodigiosin biosynthesis activator gene redD, which allowed promoter activity in cloned fragments to be visualized by the red pigment production induced in the pigmentation-deficient strain S. coelicolor strain M512 (ΔredD actII orfIV) (10). DNA fragments of the upstream regions of ramC, ramA, and ramS (shown in Fig. 3A) as well as ramR were amplified by PCR with the primers listed in Table 1. During amplification, an EcoRI restriction site was created at the 5′ end and a BamHI site was created at the 3′ end, which allowed directional cloning into vector pIJ2587. For construction of pPSt, the unique EcoRI restriction site present upstream of the ramC start codon was used for cloning. Following introduction into S. coelicolor M512, transformants were analyzed by standard plasmid preparation and restriction analysis. All promoter-probing transformants produced normal levels of aerial hyphae and spores. Quantitative analysis of undecylprodigiosin production was performed by harvesting mycelium of the M512 transformants grown on cellophane-overlaid R2YE plates at various moments during growth followed by repeated extraction of 100 mg of mycelium with 500 μl of methanol and measurement of the absorption at 533 nm. From each transformant, undecylprodigiosin was extracted from two independent cultures that both gave comparable results (listed in Table 2).
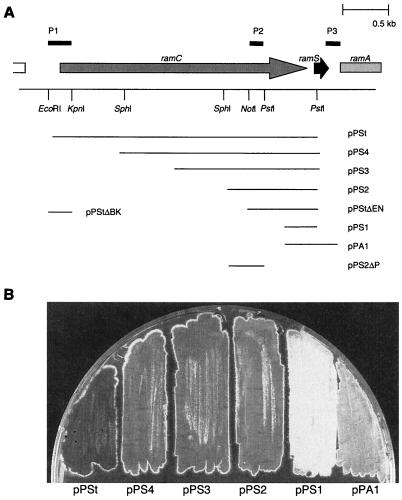
FIG. 3.
Promoter-probe analysis of the ramCSAB operon. (A) The various fragments that were amplified by PCR and cloned in front of the promoterless undecylprodigiosin activator gene redD in vector pIJ2587 are indicated by the horizontal lines. These constructs allowed promoter activity to be monitored visually by the red pigment production elicited in host strain S. coelicolor M512 (M145ΔredD actII orfIV). The lines marked P1, P2, and P3 indicate the regions that showed promoter activity. (B) Bottom view of an R2YE plate with S. coelicolor M512 transformants carrying promoter-probe vectors pPSt, pPS4, pPS3, pPS2, pPS1, and pPA1 after 6 days of growth. pPSt elicited a high level of Red production that became apparent after 30 h of growth. The level of Red production induced by pPS4, pPS3, and pPS2 was markedly lower and became apparent upon the onset of formation of aerial hyphae. pPS1 did not induce any Red production. The level of Red production induced by vector pPA1 was extremely low and was only visible after prolonged incubation. The Red production elicited by clones pPS4, pPS3, and pPS2 was similar to that elicited by deletion variants pPS2ΔP and pPStΔEN. The level of Red production of pPStΔBK was identical to that induced by pPSt.
TABLE 1.
Sequences of the PCR primers used to produce promoter-probe fragmentsa
| Primer | 5′→3′ sequence | Description |
|---|---|---|
| DSR1 | GCC GGA TCC CAT GGC CTC TTC CTT GGG | Reverse primer pPS clones |
| DSF1 | GCC GAA TTC GAC GGG CTC GCC GGA CTC GCC | Forward primer pPS1 |
| CIN2 | GCC GAA TTC GGT TCG TGG CGC CTC CCG ACC | Forward primer pPS2 |
| CIN | GCC GAA TTC GCT GCG CGG TCC GGT CTT CCG | Forward primer pPS3 |
| S1F | CGT GAA TTC GAC GCG CCC GCC GGA CTG CCG | Forward primer pPS4 |
| RCUP | GGC ACG TAC ACG CAG GTG CCG | Forward primer pPSt |
| PRAMA4 | GCC GGA TCC GAG AAC CTC CGT CAG TCG AGC GC | Reverse primer pPA1 |
| PRAMR1 | GCC GGA TCC CCC ACC CCT TGT CAG CGG CGG CC | Forward primer pPR1 |
| PRAMR2 | GCC GAA TTC GGA GCC GAT CGC GCG GTG CC | Reverse primer pPR1 |
Shown are the sequences of PCR primers used for generation of the DNA fragments cloned into vector pIJ2587 and used for promoter-probing experiments. The restriction sites that were introduced for cloning purposes are indicated in boldface.
TABLE 2.
Contribution of P1, P2, and P3 promoters to transcription of the ramCSAB operon
| Time (h) | Red productiona
|
|||||
|---|---|---|---|---|---|---|
| P1 (pPStΔBK) | P2
|
P3 (pPA1) | Negative (M512) | |||
| pPS4 | pPS3 | pPS2 | ||||
| 24 | 2 | 0 | 0 | 0 | 0 | 0 |
| 43 | 47 | 2 | 1 | 2 | 0 | 0 |
| 53 | 100 | 9 | 8 | 7 | 0 | 0 |
| 67 | 116 | 8 | 7 | 7 | 2 | 0 |
To estimate the contributions of the various promoters (P1, P2, and P3) to transcription of the ramCSAB operon, Red production elicited by the promoter-probe constructs in host strain S. coelicolor M512 was determined quantitatively. Undecylprodigiosin was extracted from 100 mg of mycelium, harvested from surface-grown cultures with methanol. The absorption of the methanol fractions was measured at 533 nm. For comparison, a numerical value was calculated as (optical density at 533 nm per mg of cells) × 1,000.
RNA isolation.
Streptomyces mycelium was harvested at various points during growth on R2YE plates that were overlaid with polycarbonate track-etch (PCTE) membranes (0.2-μm pore size) (Osmotics, Livermore, Calif.). Total RNA was isolated by the hot phenol-sodium dodecyl sulfate extraction method as described in reference 18 followed by DNase I treatment. The total RNA concentration for each sample was determined spectrophotometrically. The quality of the isolated RNA was analyzed on a 1.5% denaturing TAE agarose-formaldehyde gel (30).
Nuclease S1 mapping.
For each nuclease S1 protection assay, 0.02 pmol (5 × 104 Cerenkov counts min−1) of labeled probe was coprecipitated with 40 μg of RNA and resolubilized in Na-trichloroacetic acid buffer (26). After denaturation of the RNA-probe mixture at 72°C for 15 min, samples were allowed to cool down overnight to a final temperature of 45°C. Subsequently, samples were diluted in 300 μl of ice-cold S1 buffer (8) containing 100 U of S1 nuclease and chilled on ice. Digestions were performed at 37°C for 45 min, after which the samples were precipitated with 0.1 volume of 3 M sodium acetate (pH 6) and 2 volumes of ethanol. After centrifugation, the pellets were dissolved in standard sequencing gel loading dye, heat denatured, and electrophoresed on denaturing 6% (wt/vol) polyacrylamide sequencing gels. The DNA probe that was used to detect the transcription start upstream of ramC (pcp1) was amplified with primer RCRV (5′-CGT AGA AGT ACG GGT CCG CGT CGC-3′), which was designed based on the sequence approximately 120 bp downstream of the annotated translation start site of RamC, and the M13 reverse sequencing primer (5′-AAC AGC TAT GAC CAT G-3′). As a template we used pTZ19R with an EcoRI-BamHI subclone of the ram cluster encompassing ramC, ramS, ramA, and part of ramB (pBK105). The 254-bp probe contained a 64-bp nonhomologous extension that allowed discrimination of signals originating from reannealing of the probe and full-length protection of the probe by the messenger. Similarly, the DNA probe (pcp2) used to identify the second transcription start site that lies within the ramC ORF was amplified with primer SUP (5′-GTG CGC CAG CCC TCG TCG ACC-3′) and the M13 sequencing primer (5′-GTA AAA CGA CGG CCA GT-3′), with pBK118 as a template. Plasmid pBK118 contains an SphI-BclI fragment of the ram cluster encompassing the region between the 3′ end of ramC and the 5′ end of ramA cloned into pTZ18R. For detection of transcription into ramS, a DNA probe (pcp3) was amplified with pBK156 as a template and primer RSRV (5′-GTG ATG CTC AGG CTG AGG CTG CTG TCG-3′) and the M13 reverse primer. Plasmid pBK156 contains the EcoRI-BamHI insert of pPA1 encompassing parts of ramC, ramS, and ramA cloned into pTZ19R. For the detection of ramR transcription, a 500-bp probe was generated by PCR with primers S1RF (5′-GAC TGC GAG GAC ACG TCC AGC-3′) and RPR2 (5′-GGA GCC GAT CGC GCG GTG CC-3′), with pBK103 containing the BamHI-SphI fragment of ramB and ramR as a template. The ramAB S1 probe (pcp4), which was used to detect the ramAB transcripts, was generated by PCR with pramAB (5′-CGG CCG CCG CGA CCG AGC AC-3′), which was designed based on the sequence approximately 80 bp downstream of the putative translation start site of RamA, and the M13 reverse sequencing primer (5′-AAC AGC TAT GAC CAT G-3′). As a template, we used a PstI-BclI subclone containing parts of ramS and ramA. Probes were labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. To enable the interpretation of the signals, dephosphorylated HaeII-digested pBR322 was labeled with [γ-32P]ATP and run alongside the reaction products. For an accurate determination of the transcription start sites, the S1 samples were accompanied by a sequence ladder on the acrylamide gel. Sequencing was performed with the T7 DNA polymerase sequencing kit (Amersham) according to the manufacturer's recommendations by using PCR probes used for the protection assays as a template. To detect the 3′ end of ramCS transcripts, probe pcp4 was digested at a unique NcoI site located within ramS (Fig. 5B). After isolation of the 270-bp fragment, the 5′ overhang of the NcoI site was filled in with [α-32P]dCTP by using Klenow fragment in the presence of 0.1 mM dATP, dGTP, and dTTP and 1× OnePhorAll buffer (Pharmacia).
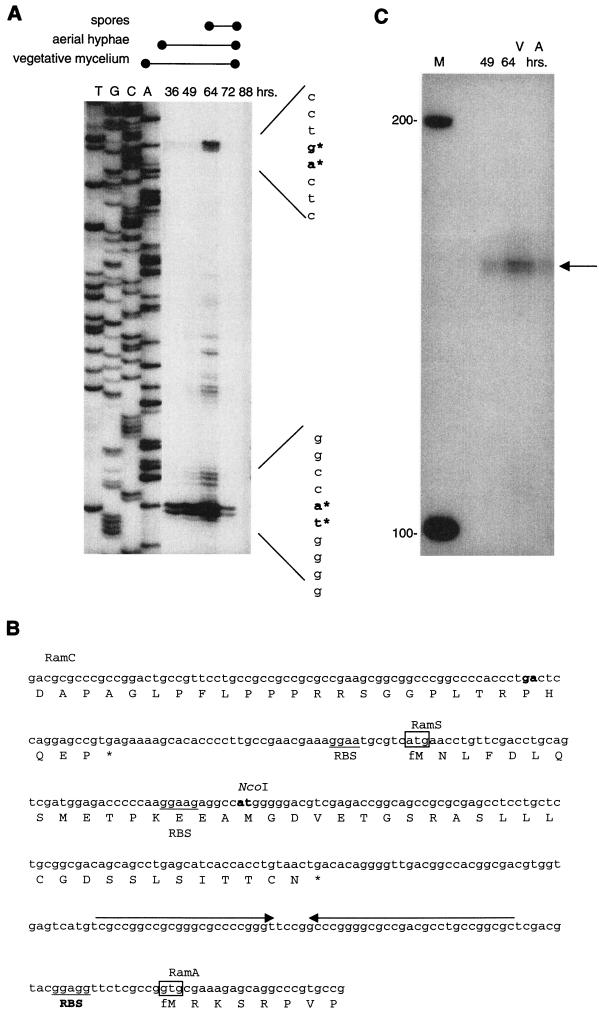
FIG.5.
Transcript processing and termination (A) High-resolution nuclease S1 protection analysis of the upstream region of ramS with probe pcp3. RNA samples were isolated from S. lividans cultures grown on solid R2YE and harvested at various stages during development (36, 49, 64, 72, and 88 h). The developmental stages (vegetative growth, aerial mycelium, and spore production) are indicated above the panel. Lanes T, G, C, and A represent a sequence ladder generated with oligonucleotide RSRV, which was also used to generate the S1 probe. Since promoter-probe experiments did not indicate any promoter activity in this region, the transcripts detected have most likely been derived from processing of a larger transcript. (B) Nucleotide sequence and the amino acid sequences of the region between the 3′ end of ramC and the 5′ end of ramA. Underlined are the potential ribosome-binding sequences (RBS) upstream of the putative RamS and RamA translation initiation codons (indicated by the box). The horizontal arrows downstream of ramS mark an inverted repeat that may function as transcription terminator. Shown in boldface are the regions corresponding to bands detected by nuclease S1 mapping and primer extension analysis. (C) Mapping of the 3′ end of ramS transcripts with RNA isolated from S. lividans grown on solid R2YE after 49 and 64 h. A band of approximately 175 nt in size was detected, indicating transcription termination corresponding to the position of an inverted repeat downstream of ramS.
RESULTS
ramR is transcribed from a single promoter during all stages of growth on solid media.
The transcription of ramR in S. lividans was analyzed during morphological development by nuclease S1 protection assays. RNA derived from S. lividans grown on PCTE-overlaid R2YE harvested at different time points was hybridized with probe prp1, which encompassed the N-terminal part of ramR and upstream regions (Fig. 1). After processing, the samples were run on a denaturing polyacrylamide gel electrophoresis gel alongside a sequence ladder produced with the oligonucleotide S1RF (see Materials and Methods) (Fig. 2A). A band corresponding to ramR transcripts was detected in all RNA samples, its intensity increasing during vegetative growth to a maximum at the onset of production of aerial hyphae. The 5′ end of the transcript corresponds to nucleotide position −66 relative to the ramR translational start site (Fig. 2B). Promoter-probing experiments revealed promoter activity within this region, indicating that the signals of the S1 protection assays represent de novo transcription (data not shown). Analysis of the DNA sequence for possible promoter consensus sequences revealed the sequence GCACAT around −10, while an apparent −35 sequence is missing. Therefore, PramR does not fall into the class of σ70-like promoters.
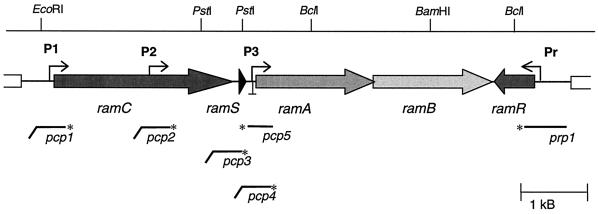
FIG. 1.
Genomic structure of the ram cluster. The arrows indicate the ram genes. ramC encodes a membrane-bound serine/threonine kinase, ramS encodes a small protein with unknown function, ramA and ramB are translationally coupled and encode components of an ABC transporter, and ramR encodes a response regulator. P1, P2, and P3 mark the promoter regions involved in transcription of the ramCSAB genes, while Pr marks the ramR promoter. The symbol ⊥ downstream of ramS marks the position of a transcriptional terminator. Below the map are indicated the DNA probes used in high-resolution nuclease S1 protection assays. The asterisks represent the 32P-labeled end. The horizontal lines indicate homologous regions, while the diagonal parts indicate the nonhomologous parts of the probe (usually derived from cloning vector pTZ19R).
FIG. 2.
Transcription analysis of ramR. (A) High-resolution nuclease S1 protection assay of the 5′ end of ramR transcripts. RNA was isolated from wild-type S. lividans harvested after 36, 49, 64, 72, or 88 h of growth on solid R2YE medium. Developmental stages are indicated above the panel. The transcription of ramR was analyzed with a labeled probe (prp1) directed at ramR and upstream regions (Fig. 1). The lanes C, T, A, and G represent a dideoxide sequence ladder of the same region generated with primer S1RF. The transcription start point is indicated by the asterisk. (B) Nucleotide sequence of the ramR promoter region indicating the ramR transcription start point and the start of the ramR coding sequence.
In vivo promoter probing of the S. coelicolor ramCSAB gene cluster.
Since it has been established that transcription of ramAB is coupled to that of ramS and the region between ramC and ramS is relatively small (37 bp) (Fig. 5B), it seems likely that ramC, ramS, and ramAB are transcriptionally coupled, constituting a single operon (16). To analyze the transcriptional regulation of the putative ramCSAB operon, promoter-probe experiments were carried out. For this purpose, S. coelicolor M512 (M145 ΔredD actII orfIV) was transformed with several derivatives of vector pIJ2587, in which various DNA fragments of the ramCSAB operon were cloned (Fig. 3A). The promoter activity of DNA fragments cloned into pIJ2587 can be assessed visually by the undecylprodigiosin (Red) production elicited in the host strain M512.
The largest fragment, pPSt, encompassing ramC and flanking sequences, strongly stimulated red pigment formation in S. coelicolor M512 on R2YE, indicative of the presence of at least one strong promoter (Fig. 3B). A similar result was obtained with the EcoRI-KpnI fragment of pPSt containing the ramC upstream region and the first 100 bases of ramC, hereafter referred to as P1 (Fig. 3). In both transformants, undecylprodigiosin became apparent after 30 h of growth on R2YE (Table 2). Fragments in which the ramC upstream region as well as portions of ramC were deleted, but the sequence between the NotI and PstI sites within ramC was retained (we hereafter refer to this latter region as P2), all resulted in significantly lower, but still readily detectable, levels of Red production that coincided with production of aerial hyphae (Fig. 3 and Table 2). The region of approximately 250 bp directly upstream of ramS (pPS1) did not elicit any Red production. A construct encompassing 400 bp upstream of ramA (pPA1) did result in low levels of Red production in M512, observable only in older surface-grown cultures (>8 days), suggesting the presence of a weak promoter directly upstream of ramA (P3). To estimate the relative strength of promoter regions P1, P2, and P3, Red production of the various transformants was quantified during growth on solid media. Undecylprodigiosin was extracted from mycelium harvested during growth and quantified by determination of the A533. The values are shown in Table 2. The relative level of Red production induced by P2 was very small compared to that induced by P1, while P3 induced an extremely low level of Red production that was detectable only after 67 h. On rich agar medium (R2YE), promoter activity in regions P1, P2, and P3 was detected with glucose, mannitol, or fructose as the carbon source (1% [wt/vol]). On the contrary, on solid minimal medium with either glucose or mannitol as the carbon source, the ramCSAB promoter-probe constructs did not elicit any Red production. Under these conditions, the abundance of Red production was observed with the control strain harboring pIJ2587 containing the constitutive PermE promoter (data not shown).
In summary, promoter probing shows promoter activity in at least three regions of the ramCSAB operon: one upstream of ramC (P1), one in a region between −700 and −500 relative to ramS (P2), and one located directly upstream of ramA (P3).
The same promoter-probe constructs were introduced into S. lividans 1326. Although this strain produces low levels of undecylprodigiosin, pIJ2587 can be used for the probing of stronger promoters. P1 (pPSt or pPStΔBK) elicited a high level of Red production in S. lividans 1326. The level of Red production in S. lividans transformed with promoter-probe constructs harboring P2 (pPS2, pPS3, or pPS4) was increased in comparison with that in S. lividans carrying the empty vector pIJ2587 (data not shown). These results suggest a regulation that is highly similar to that in S. coelicolor.
Temporal regulation of ramCSAB transcription.
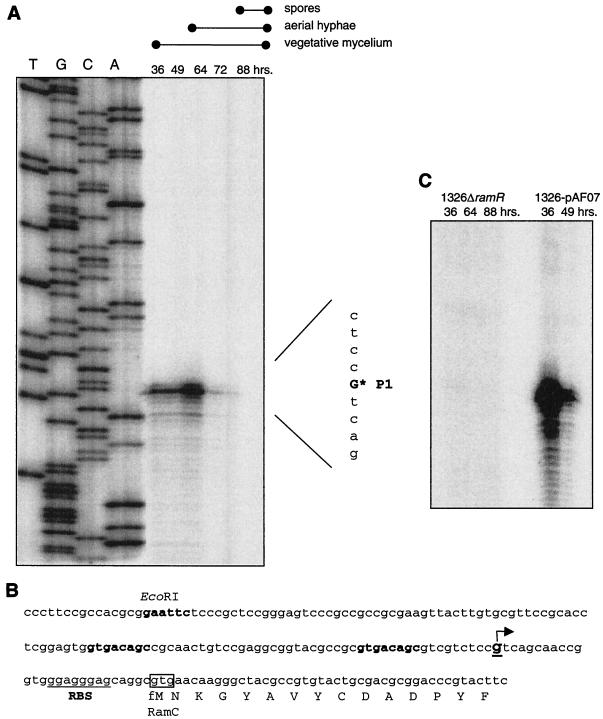
To identify the transcription start in the region upstream of ramC, nuclease S1 mapping was performed with probe pcp1 and RNA derived from S. lividans harvested during growth on R2YE (Fig. 1). One strong band could be discerned, corresponding to a position 30 nucleotides (nt) upstream of the potential ramC translational start site (Fig. 4A). Consistent with promoter-probe data, transcripts were observed already during vegetative growth. Interestingly, transcript levels increased significantly upon the initiation of formation of aerial hyphae and decreased to a low level during aerial growth and spore production. Analysis of the potential ramC promoter regions did not reveal a clear −35 and −10 consensus sequence, but did identify a direct repeat (GTGACAGC-n26-GTGACAGC) potentially overlapping the ramC promoter (P1) (Fig. 4B).
FIG. 4.
Transcription analysis of the ramCSAB operon (A) The transcription start of ramCSAB transcription was analyzed by high-resolution S1 nuclease assay with probe pcp1. This probe encompassed the 5′ end of ramC and upstream regions. The RNA used was isolated at various time points (36, 49, 64, 72, and 88 h) during growth on solid R2YE. The developmental stages are indicated above the panel. The left half of the panel (lanes T, G,C, and A) shows the sequence ladder of the same DNA region that was produced with primer RCRV. The asterisk indicates the transcription start site. (B) Nucleotide sequence of ramC and upstream regions. The arrow indicates the transcription start site that was detected by nuclease S1 mapping. Shown in boldface is a direct repeat overlapping the putative promoter sequence. Underlined is the putative ribosome-binding site (RBS). (C) Transcription of ramC was assessed by nuclease S1 protection assays with pcp1. RNA derived from S. lividans carrying ramR on a high-copy-number vector (1326-pAF07) was isolated after 46 or 49 h of growth corresponding to formation of aerial hyphae. RNA derived from an S. lividans ramR deletion mutant (1326ΔramR) was isolated from cells harvested after 36, 64, or 88 h of growth, during which aerial hyphae (64 h) and spores (88 h) were produced.
To detect transcription into ramS, nuclease S1 protection assays were carried out with probe pcp3 (Fig. 1). This probe overlaps the 3′ end of ramC and the start of ramS and has a 64-bp nonhomologous tail at the 5′ end, allowing discrimination between probe reannealing and full-length protection by mRNA. A strong band of approximately 300 nt was observed. The size of this band corresponds to full-length protection of the homologous part of the probe, confirming that the transcription of ramS is coupled to that of ramC (data not shown). Two additional bands with high intensity could be discerned (Fig. 5A), the first corresponding to a position 16 bp upstream of the ramC translational stop and the second corresponding to one of the potential ramS translational start sites (Fig. 5B). These results were also observed in experiments with different RNA isolates and confirmed by primer extension. Furthermore, such a high level of ramS transcripts was confirmed by Northern hybridization (data not shown). Since promoter-probe experiments could not detect any promoter activity in this area, we believe that these bands are due to processing of a larger transcript rather than de novo transcription.
Promoter-probe experiments revealed a weak internal promoter in the region between −700 and −500 relative to the ramS translational start site (Fig. 3). In an attempt to identify transcripts originating from this promoter region, nuclease S1 protection assays were performed with probe pcp2 (Fig. 1). In addition to the band derived from full-length protection of the homologous part of the probe by the messenger, many relatively weak bands were detected (data not shown), most of which probably reflect processed ramC transcripts. This made it impossible to discriminate between de novo transcription from P2 and processed P1-derived transcripts. Since the same RNA was used for mapping of other genes, including ramR (Fig. 2A), which revealed no bands other than the full-length transcript, this is most likely due to specific turnover of ramC transcripts.
Identification of a transcription terminator downstream of ramS.
Transcripts originating from P1 were readily detectable by S1 nuclease mapping with probe pcp1, pcp2, or pcp3. Conversely, by using probe pcp4 to detect ramA transcripts, signals could only be detected after prolonged exposure of the phosphorimager screen. We took care to label the ramC and ramA probes (pcp1 and pcp4, respectively) to very similar intensities. The relatively low level of ramA transcripts that were detected in S. lividans confirms our results published earlier (16). Sequence analysis revealed the presence of a large inverted repeat located directly downstream of the ramS ORF (Fig. 5B). The inverted repeat can potentially form a stable stem-loop structure in the nascent mRNA strand. This prompted us to map the 3′ end of ramCS transcripts by nuclease S1 assays with probe pcp5 (Fig. 1). A band of approximately 175 nt was detected (Fig. 5C), corresponding to a position at the 3′ end of the inverted repeat. This indicates that transcription is terminated at this position and strongly suggests a role for the stem-loop structure as a termination signal (Rho independent).
The effect of the ramR copy number on transcription of the ram operon.
The effect of an increased copy number of ramR on ramCSAB transcription was examined. P1 activity was analyzed in RNA derived from surface-grown cultures of S. lividans carrying ramR on the high-copy-number vector pUWL219 (pAF07) and the S. lividans ramR::hyg disruption mutant. The disruption of ramR caused a delay in the onset of formation of aerial hyphae of approximately 24 h compared to the wild type, S. lividans 1326. The aerial hyphae produced by this mutant strain subsequently formed mature and viable spores (16). The introduction of ramR on a high-copy-number plasmid (pAF07) into S. lividans resulted in an early onset of formation of aerial hyphae (Fig. 6). Analysis of P1 activity (ramCSAB) was performed by S1 nuclease protection assays with probe pcp1. The level of transcription from P1 was extremely high in RNA samples obtained from S. lividans carrying ramR on a high-copy-number vector (Fig. 4C). In accordance with P1 activity in S. lividans 1326, transcript levels were found to decrease at later stages of formation of aerial hyphae. Conversely, the normally abundant P1-derived transcripts were not detected in the ramR disruption mutant, strongly suggesting that RamR is essential for ramCSAB transcription initiation.
FIG. 6.
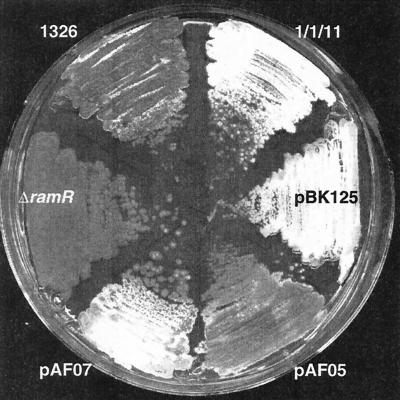
Morphological effects induced by ram gene copy number. While S. lividans (strain 1326) initiated production of aerial hyphae after 3 days of growth on R2YE plates, abundant aerial hyphae were formed by S. lividans carrying the full ram cluster on the low-copy-number plasmid pIJ698 (1/1/11) or integrated as an extra copy in the genome at the ΦC31 integration site (pBK125). In addition, S. lividans transformants carrying ramR in the high-copy vector pUWL219 (pAF07) also displayed an accelerated onset of development. The development of strains carrying ramR as an extra copy integrated at the ΦC31 integration site in the genome (pAF05) was not accelerated. The development of S. lividans carrying an insertional mutation in ramR was delayed in comparison with that of wild-type strains.
The ram genes are not expressed in bld mutants.
To relate ramR and ramCSAB transcription to a characteristic stage during development, transcription of the ram genes was analyzed in the S. coelicolor developmental mutants (bldA, bldB, bldH, bldD, and whiG) as well as in wild-type S. coelicolor strain M145. For this purpose, S. coelicolor, the bld strains, and whiG were grown on PCTE membrane-overlaid R2YE medium and harvested after 40 and 64 h, when S. coelicolor and whiG produced aerial hyphae. RNA was isolated and analyzed by nuclease S1 mapping, with prp1 for the detection of ramR transcripts and pcp1 for ramCSAB transcription. Although the S. coelicolor ram genes are highly similar to the S. lividans ram cluster, S. coelicolor-specific variants of probes pcp1 and prp1 were generated by performing the PCRs on the corresponding clones of the S. coelicolor ram cluster. In full analogy to S. lividans, transcription of ramR and ramCSAB in S. coelicolor M145 was readily detectable. Surprisingly, we were unable to detect any transcription from either Pr (ramR) or P1 (ramCSAB) in any of the bld mutants, while the transcription was detected in the earliest whi mutant, whiG. These findings show the intricate relationship between ram gene expression and initiation of formation of aerial hyphae.
DISCUSSION
The initiation of morphological development is a poorly understood process that depends on a complex interplay of regulatory circuits, metabolism, cell signaling, and stress (6, 17). The ram gene cluster plays an important role in the initiation of differentiation of S. lividans and S. coelicolor. This cluster consists of genes encoding a sensor kinase (RamC), a small protein (RamS), the components of an ABC transporter (RamAB), and a response regulator (RamR) (16, 22). Homologous gene clusters have been identified in S. griseus (amf) and Streptomyces avermitilis (36, 37). Involvement of the ram genes in development has been demonstrated by both the accelerated development observed after this gene cluster was introduced as a single extra copy or on a multicopy plasmid into S. lividans and the negative effects on differentiation when disrupted. A functional correlation between the ram gene products is indicated by the similarity of the gene organizations of the S. griseus amf cluster and S. lividans ram, although the level of DNA sequence identity is relatively low. Transcription analysis showed that ramC, ramS, ramA, and ramB form one operon. By promoter probing, three promoter regions involved in transcription of this operon were identified. A strong promoter (P1) was identified directly upstream of ramC. Two promoters that have a very limited contribution to transcription of this operon were identified in a DNA segment situated between 500 and 700 bp upstream of ramS (P2) and directly upstream of ramA (P3), respectively.
Key steps during morphological development on rich media have been found to be less important for development on minimal media. This was shown by the conditional phenotype of the bld mutants (3, 24). A conditional bld phenotype was also observed for S. lividans ramABR mutants, which produced abundant aerial hyphae and spores when grown on minimal agar medium containing mannitol as a carbon source (16). Apparently, under these conditions, an alternative developmental pathway is used that is independent of the ram or bld genes (16, 17, 28). This was supported by promoter-probing data that showed that ramR and ramCSAB promoter activity occurs only on rich media and not on minimal medium containing either mannitol or glucose. On both types of media, sporulation was normal.
High-resolution nuclease S1 analysis of the upstream regions of ramC identified a single transcription start site 27 bp upstream of the RamC translational start (P1). Transcripts were detected during vegetative growth, but the transcript level was strongly upregulated upon the formation of aerial hyphae. In samples obtained during later stages of development of aerial hyphae and spores, transcript levels decreased to a low level. A clear −35 and −10 consensus sequence could not be identified upstream of the transcription start. Interestingly, a direct repeat (GTGACAGC-n26-GTGACAGC) was identified that possibly overlaps the putative −35 and −10 sequence. While the importance of this repeat for transcription regulation is unclear, it may well constitute a transcription factor-binding site. Similar repeat sequences were identified on the S. coelicolor genome at approximately 180 bp upstream of a putative ECF sigma factor gene (CAC33073) and approximately 20 bp upstream of a gene encoding a signal peptidase (CAC36379) (www.sanger.ac.uk/Projects/S_coelicolor/). However, whether this implies a regulatory mechanism that is similar to that of the ramCSAB operon remains to be established.
A relatively large number of transcripts originating from P1 are terminated downstream of ramS, explaining the low level of transcription that has been detected for ramA (16). Transcripts terminate at a position corresponding to the 3′ end of a large inverted repeat downstream of ramS. This region may form a stable stem-loop structure in the nascent messenger. Similar Rho-independent transcription-terminator mechanisms have been shown to provide an extra level of regulation for cotranscribed genes (39, 42). In addition, a high level of short ramS transcripts was found to accumulate during growth. This was shown by S1 nuclease mapping and Northern hybridization with RNA derived from several independent isolations. The stem-loop structure may be an important cis-acting structure element providing stability of the ramS mRNA by inhibiting 3′-5′ exonuclease activity (2, 12, 19). This mechanism allows the differential expression of the ram genes within the polycistronic operon and may be important for the function of the ram gene products.
The transcription of ramR was shown to originate from a single start site 66 nt upstream of the apparent translational start codon. Transcription occurs during all growth stages and reaches a maximum during formation of aerial hyphae. The ramR promoter does not fall into the class of σ70-like promoters and is thus likely to be dependent on a different RNA polymerase sigma factor for transcription (33). In addition, ramR transcription was shown not to be dependent on the developmental sigma factor σWhiG, since normal transcript levels were detected in whiG mutants.
RamR has often been suggested to play a role in the regulation of ramCSAB (16, 22, 37). The dependence of the ramCSAB genes on ramR has been demonstrated by complementation experiments. While the introduction of the full ram cluster as a single extra copy into S. lividans resulted in an accelerated development, the introduction of the ramCSAB genes does not bring about this effect. On the other hand, the introduction of ramR results in an accelerated development only when placed on a multicopy vector and not as a single copy integrated in the genome (16, 22). The disruption of ramR eliminated virtually all P1 promoter activity, while the introduction of ramR on a multicopy vector resulted in a strong increase in ramCSAB transcript levels. This strongly suggests that RamR is involved in ramCSAB transcription. RamR may stimulate ramCSAB transcription indirectly, perhaps by stimulating other developmental genes that resort to a higher ramCSAB transcript level. Alternatively, RamR may act directly as a positive regulator of the ramCSAB operon, which would explain the conserved genetic architecture in S. lividans, S. coelicolor, and S. griseus. Perhaps a low, undetectable level of ramCSAB transcription from P1 in ramR mutants is sufficient to initiate development, albeit with a delay. Alternatively, promoter activity of the P2 and/or P3 promoter allows ramR mutants to initiate development.
Like most response regulators, RamR most likely requires phosphorylation by a sensor kinase for activity (32, 40). The ramCSAB transcription profile showing a sudden increase upon the initiation of formation of aerial hyphae may reflect the triggering of RamR activity by phosphorylation. A question that remains is what factors are responsible for activation of RamR. Unlike 62 of the total of 87 response regulator genes identified on the S. coelicolor genome (www.sanger.ac.uk/Projects/S_coelicolor), ramR is not localized adjacent to the cognate sensor kinase genes. There are examples of response regulator genes that do not lie adjacent to a histidine kinase gene that are also not activated by phosphorylation of the conserved aspartate residue (1, 13, 25). However, the integrity of the phosphorylation pocket and the conserved aspartate residue was shown to be crucial for AmfR activity in S. griseus and suggests that RamR also relies on phosphorylation for activation (36). Perhaps RamR is activated by one of the “lone” kinases encoded by genes that do not lie adjacent to an apparent response regulator gene. Alternatively, RamR may share a histidine kinase with another response regulator, as has been observed with the motility regulators CheB and CheY in E. coli and osmoregulators Ssk1p and Skn7p in S. cerevisiae (14, 21). An interesting candidate is a histidine kinase located on S. coelicolor cosmid 9E12, which is cotranslated with a ramR homologue and lies adjacent to orthologues of genes that in S. griseus are cotranscribed with amfR. The similarities in gene architecture between the S. griseus amf cluster, the ram cluster, and the genes downstream of the S. coelicolor ramR homologue may indicate a functional relationship, although this needs to be examined.
The transcription of ramR and the ramCSAB operon was studied in developmental mutants to relate the expression of these genes to a characteristic developmental stage. To our surprise, we were unable to detect any ramR or ramCSAB transcription in bldD, bldA, bldB, or bldH mutants. Conversely, transcription of ramR and ramCSAB was readily detected in samples obtained from the earliest whi mutant, whiG. The bldD gene product is known to repress the transcription of key developmental genes, such as whiG and bldN, (7). Mutation of the bldD gene leads to ectopic activation of sporulation genes, which could drastically disturb normal physiology and development. ramR and ramCSAB are the first developmental genes characterized as silent in bldD. Considering the absence of ram gene transcription in all of the bld mutants tested and its presence in the whiG mutant, we suspect that ram transcription correlates temporally to the initiation of formation of aerial hyphae. The transcriptional coupling of ramC, ramS, and ramAB strongly suggests a functional relationship between the gene products. Recently, Ueda and coworkers demonstrated that the C-terminal part of AmfS partially restores the development of an amfS mutant when supplied exogenously. This led to the proposal of a model in which AmfA and -B are involved in the export and processing of AmfS, which functions as an extracellular signaling molecule (37). Additional experiments will be required to reveal the function of the ram gene products and the relationship to formation of aerial hyphae. The ram gene expression may be a final checkpoint prior to aerial growth and thus bridge the gap between the bld and whi mutants. Further insight into the function of the ram gene cluster will therefore improve our understanding of the mechanisms that regulate the switch from vegetative to aerial growth in the Streptomyces life cycle.
Acknowledgments
We thank Anders Fuglsang for technical assistance and Keith Chater for providing the Streptomyces developmental mutant strains.
REFERENCES
- 1.Ainsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol 34:607-619. [DOI] [PubMed] [Google Scholar]
- 2.Belasco, J. G., and C. F. Higgins. 1988. Mechanisms of mRNA decay in bacteria: a perspective. Gene 72:15-23. [DOI] [PubMed] [Google Scholar]
- 3.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chater, K. F. 1972. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 72:9-28. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 7.Elliot, M. A., M. J. Bibb, M. J. Buttner, and B. K. Leskiw. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269. [DOI] [PubMed] [Google Scholar]
- 8.Favaloro, J., R. Treisman, and R. Kamen. 1980. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 65:718-749. [DOI] [PubMed] [Google Scholar]
- 9.Flardh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 10.Floriano, B., and M. Bibb. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385-396. [DOI] [PubMed] [Google Scholar]
- 11.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 183:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie, E. P., C. S. Flaxman, J. White, D. A. Hodgson, M. J. Bibb, and K. F. Chater. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144:727-738. [DOI] [PubMed] [Google Scholar]
- 14.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood, D. A., H. Wildermuth, and H. M. Palmer. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J. Gen. Microbiol 61:397-408. [DOI] [PubMed] [Google Scholar]
- 16.Keijser, B. J., G. P. van Wezel, G. W. Canters, T. Kieser, and E. Vijgenboom. 2000. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2:565-574. [PubMed] [Google Scholar]
- 17.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., M. Bibb, M. Buttner, and K. Chater. 2001. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 19.Klug, G. 1993. The role of mRNA degradation in the regulated expression of bacterial photosynthesis genes. Mol. Microbiol. 9:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 21.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, H., and K. Kendall. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 24.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 25.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 26.Murray, M. G. 1986. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal. Biochem 158:165-170. [DOI] [PubMed] [Google Scholar]
- 27.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 28.Pope, M. K., B. Green, and J. Westpheling. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180:1556-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryding, N. J., M. J. Bibb, V. Molle, K. C. Findlay, K. F. Chater, and M. J. Buttner. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 32.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 33.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda, K., K. Endo, H. Takano, M. Nishimoto, Y. Kido, Y. Tomaru, K. Matsuda, and T. Beppu. 2000. Carbon-source-dependent transcriptional control involved in the initiation of cellular differentiation in Streptomyces griseus. Antonie Leeuwenhoek 78:263-268. [DOI] [PubMed] [Google Scholar]
- 35.Ueda, K., C.-W. Hsheh, T. Tosaki, H. Shinkawa, T. Beppu, and S. Horinouchi. 1998. Characterization of an A-factor-responsive repressor for amfR essential for onset of aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 180:5085-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda, K., K. Miyake, S. Horinouchi, and T. Beppu. 1993. A gene cluster involved in aerial mycelium formation in Streptomyces griseus encodes proteins similar to the response regulators of two-component regulatory systems and membrane translocators. J. Bacteriol. 175:2006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda, K., K.-I. Oinuma, G. Ikeda, K. Hosono, Y. Ohnishi, S. Horinouchi, and T. Beppu. 2002. AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J. Bacteriol. 184:1488-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wezel, G. P., J. White, G. Hoogvliet, and M. J. Bibb. 2000. Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Mol. Microbiol. Biotechnol. 2:551-556. [PubMed] [Google Scholar]
- 39.von Hippel, P. H., and T. D. Yager. 1992. The elongation-termination decision in transcription. Science 255:809-812. [DOI] [PubMed] [Google Scholar]
- 40.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 41.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, K. S., and P. H. von Hippel. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci USA 92:8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]