Abstract
The Tol proteins are involved in the outer membrane stability of gram-negative bacteria. The C-terminal domain of TolA was mutagenized to identify residues important for its functions. The isolation of suppressor mutants of tolA mutations in the tolB gene confirmed an interaction between TolAIII and the N-terminal domain of TolB.
The Tol system of most gram-negative bacteria comprises five proteins, TolQ, TolR, TolA, TolB, and Pal (31). In Escherichia coli, mutations in any of the tol-pal genes result in hypersensitivity to deleterious agents, release of periplasmic content, formation of outer membrane vesicles at the cell surface, and induction of capsule synthesis which leads to a mucoid phenotype (20). The exact role of the Tol-Pal proteins in maintaining the outer membrane integrity is still largely unknown. The situation is complicated by the fact that even in closely related bacteria, like Salmonella enterica serovar Typhimurium, the Tol system appears to function differently than in E. coli (28). The translocation of filamentous phage DNA and group A colicins requires the TolQRA proteins (16, 19, 34). Some group A colicins also use TolB for this purpose. TolQ is an integral inner membrane protein containing three transmembrane domains (33). TolR and TolA are anchored to the cytoplasmic membrane by a single membrane-spanning segment near the N terminus, leaving most of the protein exposed to the periplasm (23, 27). TolR and TolA have a three-domain structure. In addition to the N-terminal anchoring region, TolA contains a large central domain with a high degree of α-helical structure and a C-terminal domain (TolAIII) which has been crystallized (25). Each of the three domains is separated by a stretch of glycine residues which confers some flexibility to the protein (23). The N-terminal transmembrane domain of colicins and the g3p protein of the filamentous phage interact with TolAIII during translocation or infection (3, 25). TolR also has an N-terminal anchoring domain, a central domain, and a C-terminal domain which has been proposed to form an amphipathic helix interacting with the cytoplasmic membrane (22).
TolQ, TolR, and TolA form a complex in the cytoplasmic membrane (12). Biochemical and genetic studies have shown that these interactions involve the transmembrane domains of the three proteins along with the C-terminal amphipathic helix of TolR (12, 13, 22).
TolB is a periplasmic protein and has been entirely crystallized (1, 6). It contains an N-terminal α+β domain based on a five-stranded mixed β-sheet and a C-terminal six-bladed β-propeller domain. Pal is an outer membrane peptidoglycan-associated lipoprotein. It is anchored to the outer membrane by its N-terminal lipid moiety and strongly interacts with the peptidoglycan layer through its carboxy-terminal region, which contains a particular sequence motif (17, 21). The same region of Pal interacts with the β-propeller domain of TolB (29). The interactions of Pal with TolB and the peptidoglycan appear to be mutually exclusive, since a TolB-Pal complex is not associated with the peptidoglycan (2).
Cell fractionation experiments suggest that the Tol-Pal proteins are preferentially associated with contact regions between the inner and outer membrane (15). Recently, energy-dependent conformational changes in TolA have been characterized (14). They depend on the transmembrane domains of TolA, TolQ, and TolR. Indeed, the transmembrane fragment of TolR and the third transmembrane fragment of TolQ are involved in the pmf-dependent activation of TolA (9). The TolQRA and TolB-Pal complexes are connected by the interaction between Pal and TolA, which requires the proton motive force, TolQ, and TolR (8). In addition, TolAIII has recently been reported to interact with TolB (24). All of these results are consistent with the fact that TolA activation, which requires a functional Tol cytoplasmic membrane complex, drives a signal to Pal via a change of conformation of TolAIII, generating a transient interaction between the TolQRA cytoplasmic and TolB-Pal outer membrane complexes. Interactions between Pal, TolB, and other peptidoglycan-associated proteins have also been demonstrated (7, 10). Three main outer membrane proteins interacting with the peptidoglycan network, namely Lpp, OmpA, and Pal, interact together with TolB and might constitute a structural network to link the peptidoglycan and the outer membrane.
In this study we performed an extensive mutagenesis of the C-terminal domain of TolA (TolAIII) to characterize more accurately the residues involved in TolA function. A suppressor analysis of the tolA mutants was then carried out to identify residues of TolB or Pal that were able to interact with TolA.
Mutational analysis of TolAIII.
The TolAIII region is defined by the last 107 residues of TolA, which are separated from the central domain by a stretch of 3 glycine residues (23). Strains and plasmids are described in Table 1. We used a mutagenic PCR technique to isolate mutants in the TolAIII region (5). A 473-bp region including NotI-BamHI sites containing the last 131 residues of tolA was mutagenized by degenerated PCR. The resulting plasmids were used to transform JC9776 [Δ(orf1 tolQRA)::cm]. All of the tolA mutations were constructed in pT71QRA derivatives and then subcloned into the low-copy-number plasmid pLG339 (30) by using an EcoRI-BamHI fragment containing the entire orf1 tolQRA region.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| 1292 | supE hsdS met gal lacY tonA | W. Wood |
| JC7752 | 1292 Δ(tolB pal) | Lab collection |
| JC8056 | 1292 Δ(lacU169) | Lab collection |
| JC9702 | JC8056 degP-lacZ | Lab collection |
| JC9776 | JC8056 Δ(orf1 tolQ tolR tolA)::cm | Lab collection |
| Plasmids | ||
| pT7-5 | Ampr; high copy number | 32 |
| pLG339 | Kanr and Tetr; low copy number | 30 |
| pT71QRA | 3,076-bp EcoRI-HpaI orf1 tolQ tolR tolA region cloned into EcoRI-SmaI sites of PT7-5 | Lab collection |
| pLG1QRA | EcoRI-BamHI orf1 tolQ tolR tolA region of pT71QRA cloned into the same sites of pLG339 | This work |
| pLG1QRΔA | pLG1QRA derivative carrying a 123-bp deletion in tolA, creating a 41-residue in-frame deletion in TolAIII | This work |
| pT7BP2 | 2,893-bp HpaI-EcoRV tolB pal orf2 region cloned into pT7-5 | This work |
| pT7sup | pT7BP2 with the tolB D120N mutation | This work |
The phenotypes of the tolA mutants are presented in Table 2. Most of the mutations led to a total tol phenotype whose consequences were increased sensitivity to cholic acid, release of RNase I into the medium, and tolerance to colicin A. Two TolA substitutions (M374T and L376P) conferred only tolerance to colicin A. These mutants were sensitive to all other colicins so far tested (E1, E2, E3, and K; data not shown). Mutations affecting residues 340, 346, and 348 led to impaired outer membrane stability and an intermediate sensitivity to colicin A.
TABLE 2.
Phenotypes of tolA mutants and suppression by the tolBD120N mutation
| TolB mutationa | TolA mutationb | Release of ribonuclease Ic | Growth on cholic acidd | Relative sensitivity to phage f1e | Relative sensitivity to colicin Af |
|---|---|---|---|---|---|
| None | Wild type | − | + | 1010 | 10−3 |
| None | Y340N | + | − | R | 10−1 |
| None | S346T | + | − | R | 10−2 |
| None | I348N | + | − | R | 10−1 |
| None | F352I | + | − | 1010 | R |
| None | F352C | + | − | 1010 | R |
| None | Y353I | + | − | 1010 | R |
| None | C363R | + | − | 1010 | R |
| None | M374T | − | + | 1010 | R |
| None | L376P | − | + | 1010 | R |
| None | A390V | + | − | R | R |
| None | P421S | + | − | R | R |
| Wild type | Wild type | − | + | 10−3 | |
| D120N | Wild type | − | + | 10−3 | |
| D120N | Y340N | + | − | 10−2 | |
| D120N | S346T | + | − | 10−2 | |
| D120N | I348N | + | − | 10−1 | |
| D120N | F352I | − | + | 10−3 | |
| D120N | F352C | − | + | 10−3 | |
| D120N | Y353I | − | + | 10−3 | |
| D120N | C363R | + | − | R | |
| D120N | M374T | − | + | R | |
| D120N | L376P | − | + | R | |
| D120N | A390V | + | − | R | |
| D120N | P421S | + | − | R |
p417 mutated plasmid carrying the tolB D120N mutation was introduced into strain JC9776 in combination with compatible pLG1QRA derivatives carrying the indicated tolA mutation.
pLG1QRA mutated plasmid carrying the indicated tolA mutation was introduced into strain 9776 (Δorf1 tolQRA). tolA F352C and Y353I mutants were constructed by site-directed mutagenesis and tolA A390V and P421S mutants were induced with nitrosoguanidine. The other mutations were obtained by mutagenic PCR.
Release (+) or absence of release (−) of ribonuclease I activity on plate assay.
Ability (+) or inability (−) to grow on plates containing 2.5% cholic acid.
The titer of a stock of f1 phage was determined on each strain; the phage titer is given as particles per milliliter; R, no phage plaque could be seen at the highest phage concentration.
Serial dilutions of colicin A were spotted on a plate containing the bacteria to be tested. The smallest dilution that enabled the strain to grow is indicated; R, the strain grew in the presence of undiluted colicin A.
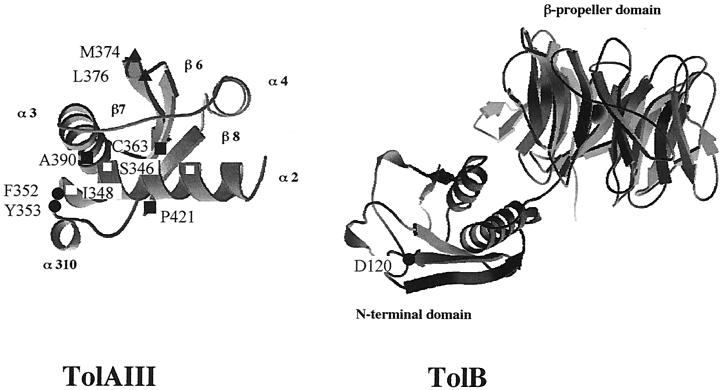
The altered residues were positioned on the three-dimensional structure of the C-terminal domain of TolA (Fig. 1). The localization of the tolA mutants was compared with the positions of the residues involved in the interaction of TolA with g3p (25). Half of the tolA mutants characterized were located in the α2, β6, and β8 domains, which are also involved in the interaction of TolA and g3p. The two substitutions involving residues 352 and 353 were localized in a loop between the α2 and α310 helices, while the two mutations giving only tolerance to colicin A were clearly distinct and localized in a loop between the β6 and β7 domains. All of the mutations leading to a defect in outer membrane stability were localized in a region centered on the α2 helix. At first glance, the result of the mutagenesis allowed us to identify two domains in TolAIII, one involved only in tolerance to colicin A and another one whose alteration gave a more pleiotropic phenotype. This latter domain overlaps the region of interaction of TolA with g3p. The three residues whose alteration led to an intermediate sensitivity to colicin A were localized within the α2 helix.
FIG. 1.
Positions, on the three-dimensional structures of TolAIII and TolB, of the different amino acid substitutions obtained in this study. Shown are mutations leading to tolerance to colicin A (▴), a defect in outer membrane integrity (□), and a defect in outer membrane integrity and tolerance to colicin A suppressed (•) or not suppressed (▪) by the TolB D120N substitution.
Stability of mutated TolA proteins and ability to form TolA-TolQ complexes.
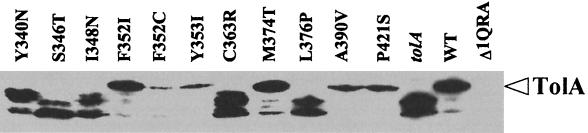
Mutations in the TolA C-terminal domain can affect the function of the tol system in different ways, such as through a modification of a functional domain or by affecting the stability of the protein. Therefore, we examined the pattern of expression of the altered TolA proteins after electrophoresis and Western blot analysis (Fig. 2). In the stationary phase of growth, only five TolA proteins, altered at residues 352, 353, 374, 390, and 421, expressed a stable TolA protein, in most cases at a lower level than that of the parent strain. The other mutations led to a highly unstable TolA protein, as seen by the pattern of protein degradation. This pattern was unchanged in strain JC9772 (degP-lacZ), in which the DegP periplasmic protease, which is involved in the degradation of misfolded proteins, was absent (data not shown).
FIG. 2.
Stability and expression of proteins containing substitutions in TolA. JC9776 transformed with pLG339 derivatives containing the orf1 tolQRA region was used for this study. For the controls, JC9776 was transformed with pLG339 (Δ1QRA), pLG1QRA (WT), and pLG1QRΔA (tolA). Immunoblots of intact cells were probed with a polyclonal antibody, diluted 1/2,000, and raised against the periplasmic domain of TolA (residues 42 to 421). The position of TolA is indicated.
Cross-linking experiments with formaldehyde were carried out in all mutant strains as previously described (13). The TolA-TolQ and TolA-TolR complexes were still present in strains expressing significant amounts of stable TolA proteins (data not shown). However, as expected, no TolB-Pal complex could be characterized, since most periplasmic proteins, including TolB, were released into the medium.
Isolation and characterization of tolB suppressor mutations of tolA.
Despite the fact that some TolA proteins were unstable, all of the tolA mutants were used for subsequent analysis. Indeed, we made the hypothesis that the instability of TolA might be due to an impaired interaction with another protein and that TolA stability could be restored by a suppressor mutation affecting such a protein. We searched for suppressor mutants in the tolB pal ybgF genes. pT7BP2 was mutagenized with nitrosoguanidine (0.4 mg/ml) as described previously (26).
The resulting plasmids were first used to transform JC8056 and JC7752 (ΔtolB pal), and clones unable to grow in the presence of sodium cholate were identified to calibrate the mutagenesis. In our experimental conditions, we could identify between 3 and 8% of mutants no matter what strain was transformed, indicating that the plasmid-borne mutations were dominant over the wild-type chromosome. A search for suppressor mutations of tolA was carried out with strain JC9776, which does not express the tolQRA genes and has a lower content of TolB, Pal, and YbgF due to the absence of the P1 promoter upstream orf1 and to the polarity of the chloramphenicol cassette, which has been used to replace the orf1 tolQRA region in the deletion. The strain was simultaneously transformed with pLG1QRA and pT7BP2 derivatives. When plasmids containing the corresponding wild-type alleles were used, strain JC9776 was fully complemented. Transformants were selected in the presence of ampicillin and kanamycin and then replicated on the same plates containing sodium cholate. As a consequence of our selection procedure, the two mutants conferring only tolerance to colicin A were not included in our screening. Sixty clones were isolated and further characterized. Of those, seven were able to restore a wild-type phenotype. Plasmids were extracted from the strains, used to transform JC8056, and selected independently for resistance to ampicillin or kanamycin. The tolA and tolB pal ybgF regions of PLG1QRA and pT7BP2, respectively, were sequenced to identify both the suppressor and the tolA mutations. All of the pT7BP2 plasmids carried the same mutation leading to a D120N transition in TolB. This mutation was able to restore a wild-type phenotype to the tolA F352I mutant (Table 2). pT7BP2 carrying the tolB mutation was used to transform strain JC7752. The mutation did not confer any tolB phenotype. This absence of obvious phenotype in suppressor mutants has already been observed in the case of tolR mutants able to restore a wild-type phenotype to tolQ mutants (22). Residue 120 is located in the N-terminal region of TolB. Therefore, the TolB C-terminal region is involved in interactions with Pal and colicins, while the TolB N-terminal region seems to interact with the C-terminal domain of TolA.
Site-directed mutagenesis of tolA and tolB.
No evidence for an interaction between TolA and TolB could be obtained after cross-linking with formaldehyde (data not shown). This could be explained by the fact that no formaldehyde residues are close enough for a cross-link to form. As our suppressor analysis revealed residues of TolA and TolB susceptible to interacting, we tried to construct monocysteine derivatives of tolA and tolB in these regions to perform cysteine cross-linking. Cysteine residues were introduced by site-directed mutagenesis by using a Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.), yielding the variants S350C, F352C, S357C, and Y358C of TolA and D120C, G122C, and G123C of TolB. In addition, a Y353I substitution was also generated in TolA. Only the F352C and Y353I substitutions in TolA led to a total tol phenotype, the other substitutions resulting in a wild-type phenotype (data not shown). These two tolA mutants were tested for their ability to be suppressed by the tolB D120N mutation. Indeed, the tolB suppressor mutation was able to restore a wild-type phenotype to the three tolA mutants altered at residues 352 and 353 (Table 2).
JC9776 transformed with plasmids containing all the combinations of cysteine residues in TolA and TolB was used to characterize potential TolA-TolB complexes by Western blot analysis. For cysteine cross-linking, we used experimental conditions described previously (4). No high-molecular-mass complex could be detected in our experimental conditions, except in the presence of the TolB G123C substitution. In this case, a 90-kDa complex could be detected, but it did not involve TolA or OmpA (data not shown). This complex could reflect TolB dimerization or an interaction between TolB and an unknown protein of similar molecular mass. The stability and expression of the TolA proteins substituted at positions 352 and 353 was not modified in the presence of the tolB D120N mutation (data not shown).
The set of results presented above allowed us to characterize mutants that affected (i) TolA stability, (ii) the interaction between TolA and TolB, and (iii) sensitivity to colicin A. A region potentially important for colicin A activity was identified in a loop between the β6 and β7 sheets, where two mutations giving only tolerance to colicin A were found. The residues involved in or important for the interaction between TolA and TolB were localized in a loop between the α2 and α310 domains. Our results also suggest that the residues of TolAIII involved in the interactions with g3p and colicin A are different. This may explain the difference of structural modification observed when TolAIII interacts with these two proteins (11).
An analysis including 25 TolB proteins from proteobacteria of all subdivisions revealed that an aspartic acid residue was present at this position in 21 cases and was replaced only by asparagine in the 4 other bacteria (Fig. 3). Interestingly, the suppressor mutation we have isolated corresponds to a D120N substitution. This may explain why this substitution does not lead to a tolB phenotype but more probably to a modification allowing the asparagine residue to interact with the altered isoleucine or cysteine residues of TolA in a manner similar to that of the interaction between aspartic acid and tyrosine or phenylalanine present in the wild-type TolB and TolA, respectively.
FIG. 3.
Sequence alignment of the TolB N-terminal region surrounding residue D120 of E. coli. The sequences were extracted from SwissProt or The Institute for Genomic Research and correspond, from top to bottom, to those of Rickettsia prowazekii, Rickettsia conorii, Brucella melitensis, Brucella abortus, Brucella suis, Sinorhizobium meliloti, Mesorhizobium loti, Caulobacter crescentus, Wolbachia sp., Ralstonia solanacearum, Burkholderia mallei, Erwinia chrysanthemi, Yersinia pestis, Escherichia coli, Salmonella enterica serovar Typhimurium, Haemophilus influenzae, Pasteurella multocida, Vibrio cholerae, Pseudomonas putida, Pseudomonas syringae, Pseudomonas aeruginosa, Xylella fastidiosa, Acidithiobacillus ferrooxidans, Geobacter sulfurreducens, and Helicobacter pylori. Sequences were aligned with Clustal W. Strong similarity is indicated by a colon. The position of the D120 residue of E. coli is indicated.
We were unable to find suppressor mutations in the pal gene. Pal may interact with other residues of TolA which could not be identified in our study. As mentioned above, some suppressor mutants devoid of tol phenotype, like those isolated in tolB and tolR, are able to suppress a primary mutation in tolA or tolQ. Therefore, a search for mutants in tolA that are able to complement pal mutants remains to be carried out. Another possibility is that we searched for suppressor mutations by using a pT7-5 derivative which encodes potential suppressor proteins at a higher level than that of the TolA protein it is suppressing and that this may affect the process in some manner.
Conclusion.
As in the case of the Ton system (18), the Tol system appears to function via an energy-dependent conformational change of TolA in the periplasm, leading to an interaction between inner and outer membrane proteins on the periplasmic side. This kind of signalization may contribute to the role of the Tol system in maintaining outer membrane stability. The TolAIII domain appears to be crucial in this process by interacting with TolB and Pal. As many alterations in TolAIII lead to a highly unstable protein, the structural constraints of the TolAIII domain are probably fundamental for its role.
Acknowledgments
Work in our laboratory was supported by the Life Science Department of the CNRS, the University of Lyon, and a joint BQR between the University and the INSA of Lyon. J.F.D. has an MENRT fellowship.
REFERENCES
- 1.Abergel, C., E. Bouveret, J. M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Benedetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Struct. Fold Des. 7:1291-1300. [DOI] [PubMed] [Google Scholar]
- 2.Bouveret, E., H. Benedetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouveret, E., A. Rigal, C. Lazdunski, and H. Benedetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 4.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadwell, R. C., and G. F. Joyce. 1995. Mutagenic PCR, p. 583-589. In C. W. Dieffenbach and G. S. Dveskler (ed.), PCR primer, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Struct. Fold Des. 8:57-66. [DOI] [PubMed] [Google Scholar]
- 7.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubes. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38:904-915. [DOI] [PubMed] [Google Scholar]
- 9.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 10.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29:359-367. [DOI] [PubMed] [Google Scholar]
- 11.Deprez, C., L. Blanchard, F. Guerlesquin, M. Gavioli, J. P. Simorre, C. Lazdunski, D. Marion, and R. Lloubes. 2002. Macromolecular import into Escherichia coli: the TolA C-terminal domain changes conformation when interacting with the colicin A toxin. Biochemistry 41:2589-2598. [DOI] [PubMed] [Google Scholar]
- 12.Derouiche, R., H. Benedetti, J. C. Lazzaroni, C. Lazdunski, and R. Lloubes. 1995. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270:11078-11084. [DOI] [PubMed] [Google Scholar]
- 13.Germon, P., T. Clavel, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 180:6433-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germon, P., M. C. Ray, A. Vianney, and J. C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guihard, G., P. Boulanger, H. Benedetti, R. Lloubes, M. Besnard, and L. Letellier. 1994. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J. Biol. Chem. 269:5874-5880. [PubMed] [Google Scholar]
- 16.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569-1580. [DOI] [PubMed] [Google Scholar]
- 17.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha helical motif in the C-terminal region of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 19.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubes, and H. Benedetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 21.Lazzaroni, J. C., and R. Portalier. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6:735-742. [DOI] [PubMed] [Google Scholar]
- 22.Lazzaroni, J. C., A. Vianney, J. L. Popot, H. Benedetti, F. Samatey, C. Lazdunski, R. Portalier, and V. Geli. 1995. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J. Mol. Biol. 246:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Levengood, S. K., W. F. Beyer, and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88:5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lloubes, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523-529. [DOI] [PubMed] [Google Scholar]
- 25.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Struct. Fold Des. 7:711-722. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course of bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Muller, M. M., A. Vianney, J. C. Lazzaroni, R. E. Webster, and R. Portalier. 1993. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J. Bacteriol. 175:6059-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prouty, A. M., J. C. Van Velkinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray, M. C., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 2000. Identification by genetic suppression of Escherichia coli TolB residues important for TolB-Pal interaction. J. Bacteriol. 182:821-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoker, N. G., N. F. Fairweather, and B. G. Spratt. 1982. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18:335-341. [DOI] [PubMed] [Google Scholar]
- 31.Sturgis, J. N. 2001. Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 3:113-122. [PubMed] [Google Scholar]
- 32.Tabor, I. 1990. Expression using the T7 RNA polymerase:promoter system, p. 16.2.1-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 33.Vianney, A., T. M. Lewin, W. F. Beyer, J. C. Lazzaroni, R. Portalier, and R. E. Webster. 1994. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 176:822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster, R. E. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]