Abstract
An individual Borrelia burgdorferi bacterium can encode as many as 13 different Erp (OspE/F-related) proteins from mono-and bicistronic loci that are carried on up to 10 separate plasmids. We demonstrate through multilabel immunofluorescence analyses that individual bacteria simultaneously coexpress their entire Erp protein repertoire. While it has been proposed that B. burgdorferi controls expression of Erp and other plasmid-encoded proteins through changes in DNA topology, we observed regulated Erp expression in the absence of detectable differences in DNA supercoiling. Likewise, inhibition of DNA gyrase had no detectable effect on Erp expression. Furthermore, expression of loci physically adjacent to erp loci was observed to be independently regulated. It is concluded that Erp expression is regulated by a mechanism(s) directed at erp loci and not by a global, plasmid-wide mechanism.
The Lyme disease etiological agent, Borrelia burgdorferi, is spread to humans and other vertebrates through the bites of infected Ixodes ticks (11). To effectively survive in nature, B. burgdorferi must sense its environment and synthesize proteins appropriate for the interactions within the different tick and mammalian tissues that it encounters. Indeed, some B. burgdorferi proteins have been observed to be differentially synthesized during transmission and infection (19, 40, 41). The importance of precise gene regulation in the B. burgdorferi infection cycle was underscored by a recent study, which found that mutants defective in genetic regulation were unable to disseminate in the mammalian host (5).
Among the B. burgdorferi proteins synthesized during mammalian infection are members of the Erp (OspE/F-related) protein family (3, 32, 42, 46, 48). A single bacterium can carry multiple erp loci, located on as many as 10 different, but homologous, plasmids (14, 15, 47). Recent studies strongly suggested that those plasmids, known as cp32s, are prophages (23, 24). Erp proteins are located on the bacterial outer surface, and those encoded by an individual bacterium may share anywhere between 16 and 100% identical amino acid sequences among themselves (2, 15, 25, 42, 43, 47). Recently, it was shown that Erp proteins bind complement inhibitory factor H (30, 44), suggesting that they contribute to the ability of B. burgdorferi to infect mammals by blocking host complement-mediated killing.
Previous studies have shown that B. burgdorferi regulates synthesis of Erp proteins (1, 6, 29, 43, 45, 48). Among the factors that influence erp transcription levels is temperature, with significantly higher levels of erp mRNA and Erp proteins being produced by bacteria grown at 34°C than by those grown at 23°C. These temperatures are hypothesized to model those experienced by the bacteria during mammalian and tick infection, respectively. All erp loci contain highly similar promoter regions, suggesting that they are all transcriptionally coregulated (47). Furthermore, a DNA-binding protein was recently observed to bind specifically to all tested erp promoter DNAs (6). For those reasons, it has been hypothesized that all Erp proteins are coexpressed and that circumstances affecting transcription of one erp locus will have similar effects on other erp genes (42, 47). However, some studies have suggested that a bacterium can independently regulate expression of erp genes, transcribing some genes while silencing others (1, 3, 18, 48). It has also been suggested that erp gene regulation may largely be due to temperature-related differences in plasmid supercoiling (39). Several other pathogenic bacteria thermoregulate gene expression of proteins coincidental with changes in DNA supercoiling (20, 36, 37, 49).
To test these hypotheses, we investigated the abilities of B. burgdorferi to express individual Erp proteins. We also investigated DNA supercoiling of B. burgdorferi plasmids after they had been grown at different culture temperatures. Furthermore, we analyzed whether the enzyme responsible for controlling negative DNA supercoiling, DNA gyrase, influenced the regulation of erp gene expression. Additionally, if regulation of Erp proteins was due to a global mechanism such as plasmid supercoiling, we would expect similar regulation of other cp32-encoded proteins. With this in mind, we also analyzed the regulation of three additional genes located adjacent to each erp locus.
MATERIALS AND METHODS
Bacteria strain and culture conditions.
B. burgdorferi B31 was originally isolated from an infected Ixodes scapularis tick collected on Shelter Island, N.Y., and was cloned by limiting dilution (7, 11). The B31 culture used in this study is infectious to both mice and ticks and contains cp32-1 (erpAB), cp32-3 (erpG), cp32-4 (erpHY), cp32-5 (erpIJ), cp32-6 (erpK), cp32-7 (erpLM), cp32-8 (erpNO), cp32-9 (erpPQ), and lp56 (erpX) (14, 42). The ErpA, ErpI, and ErpN proteins are identical to each other and are collectively referred as ErpA/I/N; likewise, ErpB, ErpJ, and ErpO are identical to each other and are referred to as ErpB/J/O (15, 42, 43, 46). Unless otherwise noted, bacteria were grown at 34°C in Barbour-Stoenner-Kelly (BSK-II) medium supplemented with 6% rabbit serum (Sigma, St. Louis, Mo.).
For temperature shift assays, bacteria were grown at 23°C to mid-logarithmic phase (5 × 107 cells per ml) and were then diluted 1:100 into fresh medium and grown at 34°C (45). For pH effect studies, bacteria were grown to mid-logarithmic phase at a constant 34°C in BSK-II medium buffered with 25 mM HEPES that was adjusted to a pH of either 7.0 or 8.0 (13).
Recombinant proteins.
Recombinant Erp proteins have been previously described (25, 42). Recombinant BppA, BppB, and BppC proteins were constructed using the cre-lox Echo cloning system (Invitrogen, Carlsbad, Calif.). Each recombinant plasmid was sequenced to confirm that the DNA insert was in frame and that no incorrect nucleotide was incorporated during PCR amplification. Polyhistidine-tagged fusion proteins were purified using His-Bind Resin Column Chromatography kits (Novagen, Madison, Wis.). Each protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by staining with Coomassie brilliant blue in order to verify the size and purity of each protein.
Antibodies.
Monoclonal antibodies (MAb) B11 and B5, directed against the B31 ErpA/I/N and OspC proteins, respectively, were produced by hybridomas derived from mice infected with this strain by tick bite (25, 27, 34). Tom Schwan (Rocky Mountain Laboratories, Hamilton, Mont.) provided MAb H9724, specific for the flagellar FlaB subunit of Borrelia spp. (8).
Polyclonal antiserum directed against each Erp protein was previously described (25). Polyclonal antibodies directed against recombinant BppA, BppB, and BppC proteins were each generated by vaccinating rabbits with approximately 30 μg of one purified recombinant protein in complete Freund's adjuvant, followed by booster vaccinations 2 and 4 weeks later with the same dose of protein in incomplete Freund's adjuvant. All rabbits were bled out 2 to 4 weeks after the final boost (Animal Pharm Services, Healdsburg, Calif.).
Some Erp-directed antisera contained antibodies that cross-reacted with other Erp proteins due to their similar amino acid sequences (25). In such an event, cross-reactive antibodies were removed by preadsorption with recombinant Erp proteins and Escherichia coli lysates for 1 h at 37°C (25). Specificities of preadsorbed antisera were then assessed by immunoblot analysis with B. burgdorferi B31 lysates. Each Bpp polyclonal antibody was preadsorbed with an E. coli lysate, and the specificity of each antiserum was assessed by immunoblot with the corresponding recombinant protein.
BALB/c mice were infected by subcutaneous inoculation of 103 culture-grown clonal B. burgdorferi B31 organisms. Three and a half months after infection, sera from 11 mice were collected and used in immunoblot analyses of recombinant BppA, BppB, and BppC proteins.
Indirect immunofluorescence double labeling.
Spirochetes grown at 34°C to mid-logarithmic phase were placed on a 12-mm-diameter circular glass slide, fixed with 100% methanol, and air dried. Slides were washed three times in phosphate-buffered saline (PBS)-0.2% bovine serum albumin plus 0.02% goat serum (Sigma). Each slide was then incubated for 1 h at room temperature with MAb B11, which is specific for ErpA/I/N, immediately followed by incubation for 1 h with a polyclonal antibody specific for ErpB/J/O, ErpG, ErpK, ErpL, ErpM, ErpP, ErpX, or ErpY. After extensive washes, slides were incubated for 1 h at room temperature with a 1:1,000 dilution of Alexa Fluor 488-labeled goat anti-mouse immunglobulin G (IgG), followed by incubation of a 1:1,000 dilution of Alexa Fluor 568-labeled goat anti-rabbit IgG (Molecular Probes, Eugene, Oreg.) (10, 16, 25, 29). Slides were washed three times in PBS and visualized with a Spot Digital Camera and an Axiophote epifluorescence microscope (Zeiss, Hallbergmoos, Germany). Each experiment was repeated three times, and as control experiments, bacteria were treated as described above without the incubation of the secondary antibody or with the secondary antibody in the absence of the primary antibody.
Chloroquine agarose gel electrophoresis and Southern blot analysis.
DNA from B. burgdorferi grown at either 23°C or at 23 shifted to 34°C was purified as previously described (39). Three hundred nanograms of each DNA was heated to 60°C in 1% N-lauroylsarcosine, 10 mM EDTA, 3% Ficoll 400, 0.05 mg of bromphenol blue/ml, and 0.05 mg of xylene cyanol/ml for 2 min; cooled briefly (39); and separated by electrophoresis in a 0.35% agarose gel containing a chloroquine concentration of either 0, 3, 9, or 12 μg/ml (4). Electrophoresis was performed in 0.5× Tris-borate-EDTA buffer (1× Tris-borate-EDTA has 0.089 M Tris [pH 8.0], 0.089 M boric acid, and 0.002 M EDTA) containing the same chloroquine concentration. DNA from each gel was transferred to a Biotrans nylon membrane (ICN, Irvine, Calif.). The experiment was repeated twice, and filter contents were incubated overnight at 55°C with a radiolabeled probe in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0)-0.1% SDS-5 g of nonfat dried milk per liter (26). The membranes were washed once for 20 min and four times for 10 min with 0.2× SSC-0.1% SDS at 55°C. Labeled DNA was visualized using a STORM 860 Phosphor Imager (Molecular Dynamics, Sunnyvale, Calif.)
Probes were produced by PCR using oligonucleotides listed in Table 1, and as template a plasmid containing the appropriate regions of DNA was used. Probe 1 was derived from the ORFC-ORF3 region of cp32-1 and is specific for that plasmid (15). Probe 2 was derived from the promoter region of the erpK gene, a region of DNA that is very well conserved in every erp locus, and thus recognizes all cp32 plasmids (26). Probe 3 was derived from the ospC promoter region encoded on cp26 and is specific for that plasmid (6). PCR conditions for probe production consisted of 25 cycles at 94°C for 1 min, 50°C for 30 s, and 72°C for 1 min. The reaction product was diluted 100-fold in water, subjected to a second round of PCR, and purified in a Centricon-100 (Amicon, Beverly, Mass.). PCR products were examined by agarose gel electrophoresis to ensure that amplification yielded only the appropriate single product (26). Probes were then labeled with [α-32P]dATP (ICN) by random priming (Gibco BRL-Life Technologies, Grand Island, N.Y.).
TABLE 1.
Oligonucleotides used in these studies
| Primer | Sequence (5′ to 3′) | Use for PCR |
|---|---|---|
| ORFD-1 | ACGATAGGGTAATATCAAAAAGG | cp32-1 |
| ORFD-2 | AGTTCATCTAATAAAAATCCCGTG | cp32-1 |
| E-506 | AACTTTTTTTTACATCTTCACCAC | B31 erpK promoter |
| E-513 | CTGTTTGTTAATATGTAATAGCTG | B31 erpK promoter |
| PC-29 | TCTCTAATTCTTCTTGCAATTAGTTG | ospC promoter |
| PC-40 | TCCTGAATTATTACAAGATATAAATA | ospC promoter |
Immunoblot analyses.
B. burgdorferi protein lysates were separated by SDS-PAGE. One gel was stained with Coomassie brilliant blue to confirm loading of equivalent amounts of total protein. A second gel was transferred to a nitrocellulose membrane, followed by overnight blocking with 5% (wt/vol) nonfat dried milk in Tris-buffered saline-Tween. The nitrocellulose membrane was incubated with an appropriate primary antiserum for 1 h at room temperature, followed by 1 h of incubation of conjugated protein A-horseradish peroxidase (Amersham Pharmacia, Piscataway, N.J.). Hybridized bands were visualized using enhanced chemiluminescence (Amersham Pharmacia) and XAR-5 film (Kodak, Rochester, N.Y.).
Effects of coumermycin A1 on protein expression.
Coumermycin A1 (Sigma) was added to cultures of B. burgdorferi grown at 34°C to the following final concentration: 0, 0.001, 0.003, 0.01, 0.03 or 0.1 μg/ml (39). After either 30 min, 4 h, or 4 days of incubation at 34°C, cells were harvested by centrifugation, washed twice in PBS, resuspended in sample buffer, and lysed by boiling for 3 min. Aliquots (5 μg/ml) of each lysate were analyzed by SDS-PAGE and were immunoblotted with MAb B11 (anti-ErpA/I/N), MAb B5 (anti-OspC), and anti-FlaB MAb.
Bacterial fractionation with Triton X-114.
Bacteria were grown to a final concentration of approximately 108 organisms per ml. Bacterial extraction and phase partition were performed using the mild nonionic detergent Triton X-114 (9). Methanol-chloroform precipitation was used to remove detergent contaminants prior to SDS-PAGE (35). The experiment was repeated twice, and control membrane filters were incubated with MAb B11, directed against the outer membrane ErpA/I/N protein, and MAb H9724, directed against the periplasmic flagella (8, 25).
In situ protease analysis.
Protease sensitivity assays were performed as previously described (10, 25). Briefly, B. burgdorferi was pelleted by centrifugation, followed by resuspension in PBS. Bacteria were then incubated at room temperature in PBS containing either 40 μg of proteinase K (Sigma)/ml or 0.05 μg of pronase (Boehringer Mannheim, Indianapolis, Ind.)/ml for 30 min or 1 or 2 h. The activity of either protease was inhibited after each time point by addition of 10 μl of inhibitor cocktail, which consisted of 0.8 mM PefaBloc SC (Boehringer Mannheim), 10 mM phenylmethylsulfonyl fluoride (Sigma), and 0.5 mM EDTA. Cells were harvested by centrifugation, washed twice in PBS, resuspended in sample buffer, and lysed by boiling for 3 min. Aliquots (15 μg/ml) of each lysate were analyzed by SDS-PAGE and immunoblotted with polyclonal antiserum directed against BppC. The experiment was repeated twice, and control aliquots of bacteria were incubated in buffer for 2 h at room temperatures, without added protease, followed by the addition of inhibitors, sample buffer, and boiling as for protease-treated bacteria.
Induction of cp32-encoded bacteriophage.
This procedure was performed as previously described (24). Briefly, bacteria were grown to mid-logarithmic phase, pelleted, and then resuspended to the original culture volume in fresh medium. The culture was divided into two equal aliquots. To one aliquot, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) was added to a concentration of 10 μg/ml. Both the treated and untreated cultures were incubated at 34°C for 2 h. The cultures were centrifuged and resuspended in an equal volume of fresh BSK-II medium, followed by incubation for 60 h at 34°C. After the cells had recovered, bacteria were harvested by centrifugation, washed twice in PBS, and lysed by boiling. Lysates of both untreated and treated cells were separated by SDS-PAGE. One gel was stained with Coomassie brilliant blue to ensure equal loading of total proteins. The other gel was immunoblotted with either MAb B11 or a polyclonal antiserum directed against either BppA or BppC.
RESULTS
Coexpression of Erp proteins.
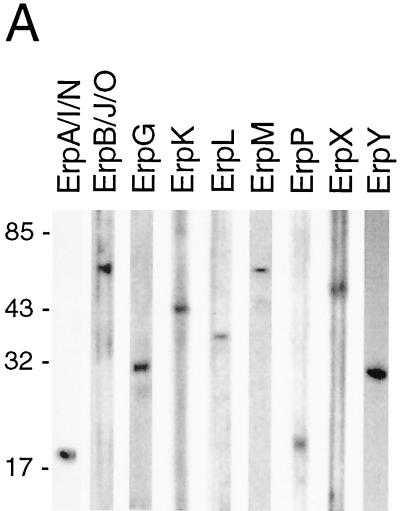
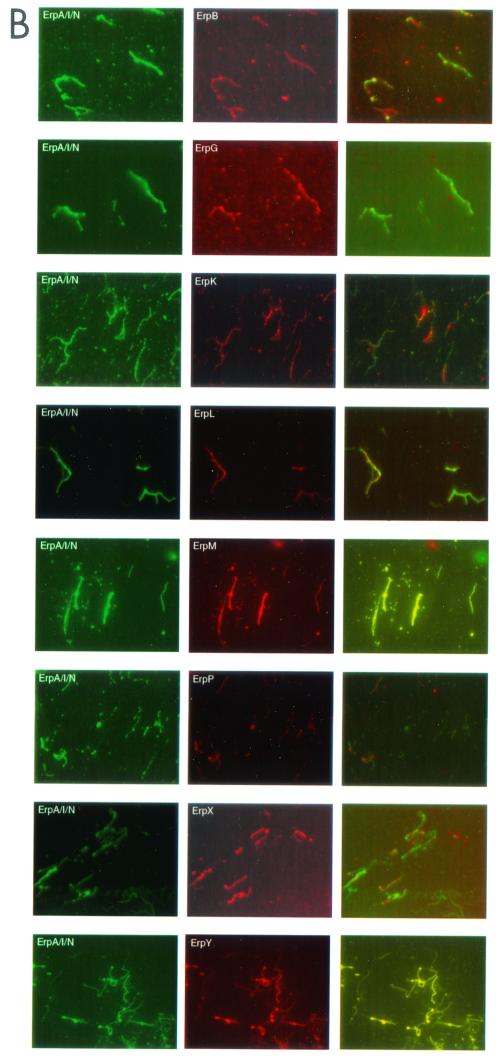
It has been suggested that B. burgdorferi cannot simultaneously coexpress its entire repertoire of Erp proteins (1, 3, 18, 29). We examined this possibility by double-label immunofluorescence assay (IFA), in which bacteria were simultaneously incubated with a murine MAb specific for ErpA/I/N and a rabbit polyclonal antiserum specific for one different Erp protein. Prior to use, the specificity of each rabbit antiserum was first confirmed by immunoblot analysis (Fig. 1A). Following incubation with these).
FIG. 1.
Double-labeling IFAs of B. burgdorferi. (A) Specificity of MAb B11 for ErpA/I/N and of each polyclonal antibody raised against ErpB/J/O, ErpG, ErpK, ErpL, ErpM, ErpP, ErpX, and ErpY. Bacteria were cultured at 34°C, and lysates were analyzed by immunoblot with each antibody preparation. Number of kilodaltons is given on left. (B) IFA of fixed B. burgdorferi labeled with MAb B11 (left column) and labeled with a polyclonal antibody against a specific Erp protein (middle column). Both IFAs were analyzed (right column).
There were occasions when double labeling was difficult to detect in a small percentage of a bacterial population. These results are similar to those reported previously by other researchers and have been cited as evidence that B. burgdorferi does not coexpress Erp proteins (29). However, we performed an additional experiment where we serially diluted MAb B11 in double-labeling IFA. Two significant observations were made during these experiments. Firstly, the more diluted that the MAb became, the harder it was to detect green-labeled bacteria. For example only 40% of the bacteria in a population were labeled upon incubation with a 107-fold dilution of MAb B11, while all were labeled with the undiluted MAb (data not shown). Secondly, incubation with the diluted MAb yielded stronger IFA signals from the polyclonal antibodies. Thus, the titer of one antibody can affect not only its own IFA signal strength but may also cause quenching of other IFA signals. Double-labeling IFA with fourfold- diluted anti-ErpA/I/N MAb resulted in labeling of all bacteria.
Supercoiling state of DNA from cultures grown at 23 and 34°C.
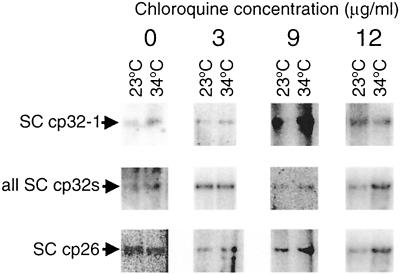
It has been suggested that the differences in erp expression levels at different temperatures are due to plasmid supercoiling (39). For that purpose, DNA from bacteria grown at 23°C and from those grown at 34°C was purified and analyzed by electrophoresis in chloroquine-agarose gels followed by Southern blotting. Different concentrations of chloroquine, a DNA intercalator, were added to each gel, as this unwinds the double helix and slows down the migration of negatively supercoiled circular DNA (4, 20, 31, 37, 39). Neither the cp32-1-specific probe 1 nor the generic cp32-labeling probe 2 detected any differences in the migration pattern of supercoiled plasmid DNA extracted from cultures grown at 23°C or from cultures shifted from 23 to 34°C (Fig. 2).
FIG. 2.
Effect of culture temperature on supercoiled circular DNA. Plasmid DNAs from bacteria grown at 23 or 34°C were separated on agarose gels containing either 0, 3, 9, or 12 μg of chloroquine/ml, followed by Southern blot analysis. Each experiment was repeated twice, and arrows indicate supercoiled (SC) DNA. In some panels, linearized plasmid is also visible. Top row, membranes probed with probe derived from ORF3-ORFC region encoded on cp32-1 (15), which is specific for that plasmid. Middle row, membranes probed with the generic cp32 probe 2, which recognizes every erp promoter region (26). Lowest row, membranes probed with probe 3, derived from the ospC promoter region of cp26 (6).
In addition to the cp32s, we also analyzed DNA supercoiling of an unrelated circular plasmid, cp26, which encodes the 24-kDa outer surface lipoprotein OspC (33, 38). Similar to the case for Erp proteins, OspC synthesis is increased when B. burgdorferi is cultured at 34°C rather than 23°C (41). Southern blot analysis with a cp26-specific probe indicated that supercoiling of this plasmid was not detectably affected by temperature (Fig. 2).
Effects of coumermycin A1 on protein expression.
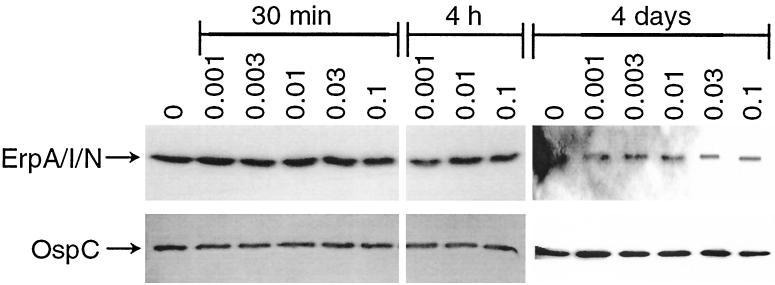
Previous studies have shown that the antibiotic coumermycin A1 induces relaxation of DNA supercoiling in B. burgdorferi via inhibition of DNA gyrase (21, 39). With this in mind, we further investigated the effect of this drug on erp expression. However, we observed constant expression levels of ErpA/I/N, regardless of the concentration of the antibiotic used (Fig. 3). In addition, bacteria did not exhibit alteration in expression of the circular plasmid-encoded OspC protein either.
FIG. 3.
Effects of coumermycin A1 on protein expression. B. burgdorferi was incubated with 0.001, 0.003, 0.01, 0.03, and 0.1 μg of coumermycin A1/ml for 30 min, 4 h, and 4 days, followed by Western blot analysis. Top row, membranes incubated with MAb B11 directed against ErpA/I/N. Lower row, membranes incubated with MAb B5 directed against OspC.
Independence of Erp expression from that of other cp32-encoded proteins.
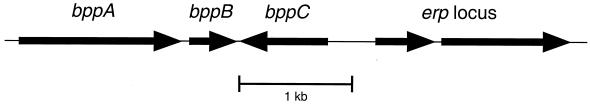
If B. burgdorferi regulates synthesis of Erp proteins by a plasmid-wide mechanism such as alteration of supercoiling, then we expect other cp32-encoded proteins to be regulated similarly. For that purpose we characterized the expression profiles of three proteins encoded by genes located immediately 5′ of all erp loci (Fig. 4). We named the open reading frames bppA, bppB, and bppC (Borrelia plasmid protein, previously referred to as ORF8/7, ORF10 and ORF6, respectively) (46). These are also members of the paralog families PF35, PF36, and PF37, respectively (14). Each gene and protein were given an allelic designation that corresponds with the cp32 plasmid on which the gene is located (e.g., bppA1, bppB1, and bppC1 are located on cp32-1). We determined that BppA encodes a 51-kDa protein, BppB encodes a 17-kDa protein, and BppC encodes a 32-kDa protein. Increasing the significance of characterizing these three genes and their protein products is the fact that the start codon of each bppC gene is located 430 bp from the start codon of its adjacent erp locus, in the opposite orientation (46).
FIG. 4.
Orientation of the erp, bppA, bppB, and bppC loci on B. burgdorferi cp32 (drawn to scale). erp loci may be either mono- or bicistronic. The start codon of bppC is located approximately 430 bp from the start codon of its adjacent erp locus. bppA encodes a 51-kDa protein, bppB encodes a 17-kDa protein, and bppC encodes a 32-kDa protein.
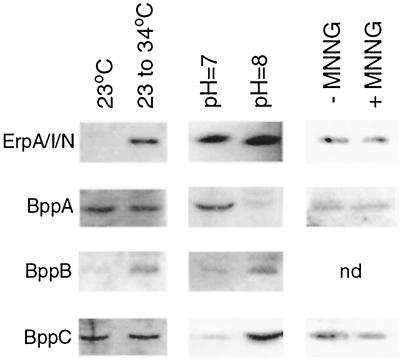
First, we examined the effects of culture temperature on BppA, BppB, and BppC synthesis and compared those results with those of the Erp proteins. As previously reported, synthesis of ErpA/I/N increased with a shift in temperature (Fig. 5). However BppA and BppC levels were not detectably affected by culture temperature, while that of BppB increased slightly at the higher temperature.
FIG. 5.
Western blot analysis of ErpA/I/N, BppA, BppB, and BppC expression by B. burgdorferi. Left column, bacteria cultured at 23°C or shifted from 23 to 35°C. Middle column, bacteria cultured at pH 7.0 or 8.0. Right column, bacteria treated with MNNG (+) or left untreated (−). nd, not done.
Expression of some proteins of B. burgdorferi is also affected by culture pH (13). With this in mind, we studied the expression of BppA, BppB, and BppC by bacteria grown at different pHs. As previously reported (6, 13) ErpA/I/N protein synthesis was not affected by pH (Fig. 5). However, expression levels of BppA, BppB, and BppC were influenced by pH. BppA was produced at increased levels by bacteria grown at pH 7.0, compared to production by those grown at pH 8.0. Synthesis of both BppB and BppC showed opposite patterns, being expressed at higher levels in bacteria cultured at pH 8.0 than in those cultured at pH 7.0. Results from these studies strongly suggest that expression of Erp, BppA, BppB, and BppC proteins is also regulated through separate mechanisms, rather than a single, plasmid-wide mechanism such as supercoiling.
Prior to these studies, nothing was known about these three proteins. Our interest in learning more about them increased greatly as a consequence of the differences in expression regulation detected between them and the Erp proteins.
Recent studies have shown that the cp32 plasmids may be packaged into bacteriophage-like particles (23, 24). These plasmids also encode proteins that exhibit holin-like functions (17). In addition, BLAST-P analysis suggested that a domain of BppC is homologous to that of bacteriophage integrases (data not shown). With this mind, we added the bacteriophage-inducing agent MNNG (23, 24) to growing spirochetes. However, immunoblot analyses did not reveal any detectable differences in Erp, BppA, or BppC protein expression levels as results of this treatment (Fig. 5), suggesting that neither protein is involved with production of bacteriophage particles.
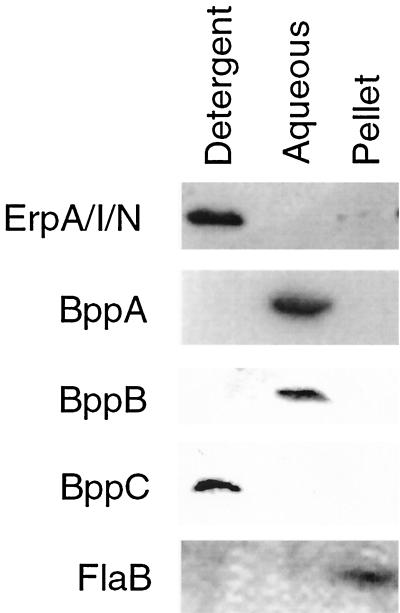
We further examined the BppA, BppB, and BppC proteins by determining their cellular localization within B. burgdorferi. After Triton X-114 extraction, BppA and BppB were found in the aqueous phase (Fig. 6), suggesting that they are periplasmic proteins. BppC was found in the Triton X-114 detergent phase, suggesting that it is located in a bacterial membrane. Control experiments with MAbs directed against FlaB and ErpA/I/N indicated that these proteins are located with the insoluble pellet protoplasmic cylinder and detergent (membrane) fraction, respectively, as expected.
FIG. 6.
Triton X-114 extraction of B. burgdorferi cells. Fractions were separated into three different phases. The detergent phase contains the outer membrane proteins. The aqueous phase contains periplasmic proteins. The pellet contains the protoplasmic cylinder (cytoplasm, the inner membrane, and the anchored periplasmic flagella).
The results obtained from Triton X-114 extraction prompted us to examine whether BppC was embedded in the inner or outer membrane. We therefore examined susceptibility of BppC to proteolysis in situ. Cultured bacteria were incubated with protease, with the idea that surface-exposed proteins of intact bacteria will be degraded, whereas those below the surface will remain untouched. Since a number of borrelial outer surface proteins are resistant to some proteolytic enzymes (10, 12, 22, 25, 50), two different enzymes were used in these studies. The results of these experiments showed that neither protease degraded BppC (data not shown). Control experiments indicated that bacterial outer membranes were not damaged, since no proteolysis of the periplasmic flagellum was observed. Furthermore, enzymes were functionally active, as detected by proteolysis of ErpA/I/N (data not shown). These results indicate that BppC does not appear to be surface exposed in B. burgdorferi or is in some manner protected from proteolysis.
As a final step in characterizing BppA, BppB, and BppC, we examined whether they are produced by B. burgdorferi during mammalian infection. Sera collected from mice infected with B. burgdorferi contained antibodies directed against all three proteins: 54% recognized BppA recombinant protein, 20% recognized BppB, and 64% recognized BppC (data not shown). Similarly, all Erp proteins are also produced during mammalian infection (47).
DISCUSSION
We hypothesized that all Erp proteins are coexpressed and that circumstances resulting in transcription of one erp locus will have similar effects on other erp's. Our hypothesis is based on the following observations: firstly, every erp locus contains a highly conserved promoter region (47). Secondly, they are all similarly regulated by the same environmental conditions in vitro (47). Finally, DNA-binding proteins specifically interact with all tested erp promoter regions (6). The results from the IFA double-labeling studies presented herein further support this hypothesis, as all Erp proteins were observed to be simultaneously expressed by individual bacteria.
However, there were occasions when it appeared that a small percentage of the bacteria in a population was not expressing both tested Erp proteins. Similar results were reported previously (29), leading to a suggestion that synthesis of some Erp proteins may be regulated by additional factors. However, our studies indicate that inability to detect an Erp protein by IFA can be a result of differences in concentrations of the antisera used. MAb B11, directed against ErpA/I/N, appeared to contain a higher titer of antibodies than did the polyclonal antisera. As would be expected, serial dilution of MAb B11 significantly weakened its signal to the point at which an IFA signal was undetectable. Furthermore, as the signal from MAb B11 weakened, the signal from the polyclonal antiserum increased. Use of diluted MAb B11 resulted in double-labeling IFA of all tested bacteria. These results indicate that (i) dilute antibody preparations exhibit weak IFA signals and that (ii) antibody preparations with a high titer can mask IFA signals from antisera with a weak titer. We conclude that incomplete double labeling was due to antibody titers, not to the inability of the bacteria to simultaneously coexpress all Erp proteins.
It has also been suggested that the temperature-mediated effects on circular plasmid-encoded genes are due to changes in DNA supercoiling (39). DNA supercoiling is used as a sensor for environmental changes for many pathogenic and nonpathogenic organisms (4, 20, 28, 31, 37). Raising the culture's temperature can induce a reduction in DNA twists, while changing to a lower temperature may increase the number of DNA twists (28, 37). However, our studies detected no differences in supercoiled DNA migration pattern from cultures grown at 23 or 34°C. The similarities in supercoiling were perhaps to be expected, since bacteria were cultured for 2 or 3 days following temperature shift, ample time for the bacteria to adjust any immediate supercoiling changes (41, 45). In addition, if the temperature-related differences in erp gene regulation are caused by changes in supercoiling, inhibition of DNA gyrase activity with coumermycin A1 should alter the expression levels of Erp proteins and of other circular plasmid-encoded proteins. Yet our studies demonstrated that inhibiting this enzyme did not cause any detectable differences in protein expression levels. Thus, we conclude that the effect of temperature on Erp expression is not significantly due to changes in DNA supercoiling.
While size of the plasmids, chloroquine concentrations used, or other factors may have influenced the results described above, we expected that other cp32-encoded proteins would be regulated similarly if expression of such genes is regulated through DNA supercoiling. For this reason, we examined the expression patterns of three proteins encoded by genes adjacent to the erp loci, BppA, BppB, and BppC. Expression of these proteins appears to be regulated by mechanisms independent of those controlling Erp synthesis, since the expression levels of all four protein types were influenced differently by environmental factors.
The unique expression pattern of BppA, BppB, and BppC led us to characterize these proteins further. Immunoblot analyses of Triton X-114 fractions indicated that both BppA and BppB are soluble proteins. BppC was found in the bacterial outer membrane, while the protease sensitivity assay suggested that BppC is not surface exposed. Previous studies (13) observed that the pH of the culture medium caused differential expression of many membrane proteins. One unidentified protein with an apparent molecular mass of 34 kDa showed increased expression in alkaline conditions (13). Based on the size, predicted pI, and expression profile, this protein could be the 32-kDa membrane protein BppC. Additionally, the BppA, BppB, and BppC proteins are all produced by B. burgdorferi during mammalian infection, as sera from infected mice contained antibodies directed against each protein. Continued characterization of these proteins will shed additional light on the function of these proteins and their roles in B. burgdorferi biology.
Several mechanisms appear to be involved in the regulation of Erp protein expression by B. burgdorferi, including responses to temperature and undetermined soluble chemicals in the environment (1, 6, 45). Results of the present studies expanded on these earlier conclusions. All Erp proteins were demonstrated to be simultaneously expressed by individual bacteria. Culture temperature change did not detectably change the topology of the plasmid DNA, and inhibition of DNA gyrase did not influence Erp expression. Additionally, we found that mechanisms altering expression of the Erp proteins were locus specific and did not influence proteins encoded by nearby loci. These data strengthen the hypothesis that all erp genes are similarly regulated and produced by B. burgdorferi at the same point(s) in the spirochete's vertebrate-tick infectious cycle. As most, if not all, Erp proteins bind host complement regulatory factor H (30, 44), such coexpression is to be expected for the survival of these bacteria in nature. Further studies of the Erp and other cp32 phage-encoded proteins will continue to increase understanding of the regulatory mechanisms controlling protein synthesis and function in B. burgdorferi.
Acknowledgments
This study was funded by National Institutes of Health grant RO1-AI44254 to B. Stevenson.
We thank Don Cohen for technical assistance with epifluorescence microscopy; Karl Drlica, Waimun Huang, and Scott Samuels for constructive comments concerning DNA supercoiling techniques; Tom Schwan for providing MAb H9724; and Kelly Babb, Melissa Hines, Natalie Mickelsen, Jennifer Miller, and Julie Stewart for technical assistance, helpful comments on experimental procedures, and comments on the manuscript.
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Alice, A. F., and C. Sanchez-Rivas. 1997. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr. Microbiol. 35:309-315. [DOI] [PubMed] [Google Scholar]
- 5.Anguita, J., S. Samanta, B. Revilla, K. Suk, S. Das, S. W. Barthold, and E. Fikrig. 2000. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect. Immun. 68:1222-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babb, K., N. El-Hage, J. C. Miller, J. A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G., W. Burgdorfer, S. F. Hayes, O. Peter, and A. Aeschlimann. 1983. Isolation of a cultivable spirochete from Ixodes ricinus ticks of Switzerland. Curr. Microbiol. 8:123-126. [Google Scholar]
- 8.Barbour, A. G., S. F. Hayes, R. A. Heiland, M. E. Schrumpf, and S. L. Tessier. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt, M. E., B. S. Riley, J. D. Radolf, and M. V. Norgard. 1990. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect. Immun. 58:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunikis, J., and A. G. Barbour. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect. Immun. 67:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 12.Carroll, J. A., N. El-Hage, J. C. Miller, K. Babb, and B. Stevenson. 2001. The Borrelia burgdorferi RevA antigen is a surface-exposed outer membrane protein whose expression is regulated in response to environmental temperature and pH. Infect. Immun. 69:5286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 15.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 93:7973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damman, C. J., C. H. Eggers, D. S. Samuels, and D. B. Oliver. 2000. Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J. Bacteriol. 182:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Silva, A. M., and E. Fikrig. 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am. J. Trop. Med. Hyg. 53:397-404. [DOI] [PubMed] [Google Scholar]
- 20.Drlica, K. 1992. Control of bacterial supercoiling. Mol. Microbiol. 6:425-433. [DOI] [PubMed] [Google Scholar]
- 21.Drlica, K., and M. Snyder. 1978. Superhelical Escherichia coli DNA: relaxation by coumermycin. J. Mol. Biol. 120:145-154. [DOI] [PubMed] [Google Scholar]
- 22.Dunn, J. J., B. N. Lade, and A. G. Barbour. 1990. Outer surface protein A (OspA) from the Lyme disease spirochete, Borrelia burgdorferi: high level expression and purification of a soluble recombinant form of OspA. Protein Expr. Purif. 1:159-168. [DOI] [PubMed] [Google Scholar]
- 23.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggers, C. H., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 26.El-Hage, N., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmore, R. D., Jr., and M. L. Mbow. 1999. Conformational nature of the Borrelia burgdorferi B31 outer surface protein C protective epitope. Infect. Immun. 67:5463-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein, E., and K. Drlica. 1984. Regulation of bacterial DNA: supercoiling plasmid linking numbers vary with growth temperature. Proc. Natl. Acad. Sci. USA 81:4046-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 31.Krispin, O., and R. Allmansberger. 1995. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 134:129-135. [DOI] [PubMed] [Google Scholar]
- 32.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mbow, M. L., R. D. Gilmore, Jr., and R. G. Titus. 1999. An OspC-specific monoclonal antibody passively protects mice from tick-transmitted infection by Borrelia burgdorferi B31. Infect. Immun. 67:5470-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nally, J. E., J. F. Timoney, and B. Stevenson. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruss, G. J., and K. Drlica. 1989. DNA supercoiling and prokaryotic transcription. Cell 56:521-523. [DOI] [PubMed] [Google Scholar]
- 37.Rohde, J. R., J. M. Fox, and S. A. Minnich. 1994. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol. Microbiol. 12:187-199. [DOI] [PubMed] [Google Scholar]
- 38.Sadziene, A., B. Wilske, M. S. Ferdows, and A. G. Barbour. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuels, D. S., and C. F. Garon. 1993. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob. Agents Chemother. 37:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson, B., W. R. Zückert, and D. R. Akins. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2:411-422. [PubMed] [Google Scholar]
- 48.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J. Y., and M. Syvanan. 1992. DNA twist as a transcriptional sensor for environmental changes. Mol. Microbiol. 6:1861-1866. [DOI] [PubMed] [Google Scholar]
- 50.Zückert, W. R., T. A. Kerentseva, C. L. Lawson, and A. G. Barbour. 2001. Structure conservation of neurotropism-associated VspA within the variable Borrelia Vsp-OspC lipoprotein family. J. Biol. Chem. 276:457-463. [DOI] [PubMed] [Google Scholar]