Abstract
Pathogenic Yersinia species employ a type III secretion system (TTSS) to target antihost factors, Yop proteins, into eukaryotic cells. The secretion machinery is constituted of ca. 20 Ysc proteins, nine of which show significant homology to components of the flagellar TTSS. A key event in flagellar assembly is the switch from secreting-assembling hook substrates to filament substrates, a switch regulated by FlhB and FliK. The focus of this study is the FlhB homologue YscU, a bacterial inner membrane protein with a large cytoplasmic C-terminal domain. Our results demonstrate that low levels of YscU were required for functional Yop secretion, whereas higher levels of YscU lowered both Yop secretion and expression. Like FlhB, YscU was cleaved into a 30-kDa N-terminal and a 10-kDa C-terminal part. Expression of the latter in a wild-type strain resulted in elevated Yop secretion. The site of cleavage was at a proline residue, within the strictly conserved amino acid sequence NPTH. A YscU protein with an in-frame deletion of NPTH was cleaved at a different position and was nonfunctional with respect to Yop secretion. Variants of YscU with single substitutions in the conserved NPTH sequence—i.e., N263A, P264A, or T265A—were not cleaved but retained function in Yop secretion. Elevated expression of these YscU variants did, however, result in severe growth inhibition. From this we conclude that YscU cleavage is not a prerequisite for Yop secretion but is rather required to maintain a nontoxic fold.
The pathogenic species of the genus Yersinia (Yersinia pestis, Y. enterocolitica, and Y. pseudotuberculosis) cause infections of highly varied severity in humans. Y. pestis causes plague and is transmitted by flea bites or infectious aerosols, whereas Y. enterocolitica and Y. pseudotuberculosis are enteric pathogens that cause gastroenteritis after the ingestion of contaminated food or water (for reviews, see references 9 and 35). Still, the virulence mechanisms of the different species show a lot of similarities. One such similarity is the ability to inhibit phagocytosis, which enables the pathogens to replicate in lymphoid tissues. This is conferred by an ca. 70-kb plasmid that is required for virulence in all three species. The plasmid encodes a type III secretion system (TTSS) that delivers antihost proteins or virulence effectors called Yops (Yersinia outer proteins) into the cytosol of eukaryotic cells (12, 13). Yop secretion is normally triggered by eukaryotic cell contact (36, 37), but it can also be induced in vitro by growing the bacteria in calcium-depleted medium at 37°C (13).
TTSSs are found in several gram-negative animal and plant pathogens (20, 39). The overall mechanism of secretion appears to be conserved in the different systems. Typically, 20 to 25 proteins are required to assemble a functional secretion system. Nine of these proteins are conserved not only in the TTSSs of different pathogens but also in the bacterial flagellar export apparatus (for reviews, see references 1, 20, and 25). For several animal pathogens, the type III secretion organelle, also referred to as the secreton, has been isolated and analyzed (5, 6, 21, 22, 40, 44). The basal body of this structure possesses two sets of rings resembling the flagellar basal body. The components common to the virulence associated and the flagellar TTSS are believed either to associate with the cytoplasmic face of the basal body-like structure or to form a pore in the inner membrane ring (15, 42). A common feature of secretons isolated so far is a needle-like structure that protrudes from the ring structure located in the outer membrane. This needle is required for secretion, suggesting that the combination of the basal portion and needle extension (needle complex) constitutes an intact secretion organelle (5, 6, 21, 22, 40, 44). In Yersinia spp. the needle-like structure is comprised of the YscF protein and localizes to the bacterial cell surface prior to eukaryotic cell contact (18; P. Edqvist, J. Olsson, M. Lavander, L. Sundberg, Å. Forsberg, H. Wolf-Watz, and S. Lloyd, unpublished data).
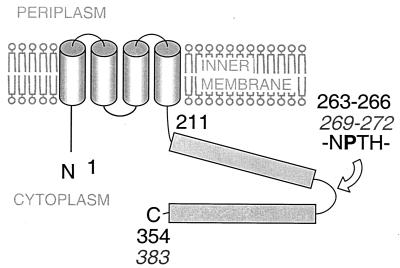
The proteins forming the actual secretion apparatus are believed to somehow identify the type III secretion substrates to enable their secretion through the basal body-like structure. One key protein in the export of flagellar components is FlhB, a membrane protein with a large cytoplasmic C-terminal domain (29, 30). YscU, the corresponding protein of the TTSS of Yersinia spp. has also been shown to localize to the cytoplasmic membrane (3). The flagellum is a tripartite organelle consisting of the membrane spanning basal body, the propulsive flagellar filament, and the hook which joins the two former (for reviews, see references 2 and 26). During flagellar assembly, one critical event is when the hook is completed and the secretion system switches from secreting hook components to filament components. FliK, a protein secreted by the flagellar secretion system during hook assembly, somehow senses when the hook is completed and communicates with the secretion system to switch the export specificity to flagellin (23, 28, 33, 45). As a consequence, fliK mutants have two distinct mutant phenotypes: they are disabled (i) in the terminating hook assembly and (ii) in the initiating filament assembly, leading to the formation of so-called “poly-hook” structures (17, 34, 43). Interestingly, extragenic suppressor mutants that allow secretion and assembly of filament-type substrates have been isolated and mapped to flhB, which indicates that FliK and FlhB interact to control the order in which flagellar components are secreted (23, 45). It has been suggested that FliK, upon the completion of hook assembly, switches the substrate specificity of the flagellar export apparatus by altering the conformation of FlhB in order to promote the export of the filament component, flagellin. Minamino and Macnab also demonstrated that FlhB is proteolytically cleaved at position proline-270 within a region of four amino acids, i.e., NPTH (Fig. 1), which is conserved in all FlhB/YscU homologues (31).
FIG. 1.
Schematic drawing of the structure of the homologous proteins YscU and FlhB adapted from the studies by Allaoui et al. (3) and Minamino and Macnab (31). Black numbers indicate amino acid positions of YscU, and numbers in italics (gray) represent those of FlhB. The sequence NPTH (amino acids 263 to 266 and amino acids 269 to 272) is conserved through the YscU/FlhB family and contains the proline-270 where FlhB has been shown to be cleaved (31).
A phenomenon corresponding to the assembly of poly-hooks in the flagellar TTSS has been reported by Galan and coworkers in the type III secretion system encoded by Salmonella pathogenicity island 1. Specifically, an invJ mutant assembles a type III secreton with abnormally long needles (22) and is unable to secrete effector proteins (10), which suggests that it is defective in substrate specificity switching. In addition, InvJ and corresponding proteins of other secretons show low levels of homology to FliK, arguing for a similar switching mechanism between different secretion substrates also in other TTSSs.
We examined here the function of the YscU protein in the Yersinia type III secretion system. We demonstrate that YscU is cleaved at the same position, proline-264 (Fig. 1), as was previously shown for FlhB, which results in a C-terminal fragment of ca. 10 kDa. The different domains of YscU were found to influence Yop expression and secretion differently. High levels of full-length YscU resulted in lowered Yop expression, whereas expression of the C-terminal 10-kDa domain in a wild-type bacterial background instead resulted in increased Yop secretion. Deletion of the conserved four amino acids, NPTH (amino acids 263 to 266), rendered YscU nonfunctional in Yop secretion, and in this case the proteolysis occurred at a different position. Single substitutions of the conserved residues prevented cleavage of YscU, but the protein was still functional in secretion, which suggests that proteolytic cleavage of YscU is not essential for its function in the TTSS. Instead, it seems that the cleavage of YscU is essential for the survival of the bacteria, since the uncleaved form of YscU was highly toxic.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmid used in this study are listed in Table 1. Yersinia strains were grown in either Luria broth (LB) or brain heart infusion (BHI; Oxoid). BHI was supplemented with 2.5 mM CaCl2 (BHI plus Ca2+) or 5 mM EGTA, 20 mM MgCl2, and 0.1% Triton X-100 (BHI minus Ca2+). For solid media Yersinia selective agar base (YSAB; Difco) or blood agar base (BAB; Merck) was used. To test the growth phenotype (MOX-test), BAB plates were supplemented with 20 mM Na oxalate, 20 mM MgCl2, and 0.2% glucose (MOX plates, Ca2+ free) or with 2.5 mM Ca2+. Escherichia coli strains were grown in LB or on BAB plates.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Y. pseudotuberculosis | ||
| YPIII(pIB102) | Wild-type, parental strain; Kmr | 8 |
| YPIII(pIB75) | pIB102 yscU in-frame deletion; Kmr | This study |
| YPIII | Virulence plasmid-cured strain | 7 |
| E. coli | ||
| S17-1λpir | RP4-2 Tc::Mu-Km::Tn7 (λpir) | 14 |
| TOP10 | Commercial One Shot competent cells | Invitrogen |
| BL21(DE3) | IPTG-inducible T7 RNA polymerase | 41 |
| Plasmids | ||
| pCR2.1 | Commercial cloning vector; Ampr | Invitrogen |
| pMMB66EH | Ptac expression vector; Ampr | 16 |
| pET-22b | T7 overexpression vector; Ampr | Novagen |
| pKK223-3 | Ptac expression vector; Ampr | Amersham Pharmacia Biotech |
| pDM4 | Suicide vector carrying sacB; Cmr | 27 |
| pLS13 | 669-bp PCR fragment of ΔyscU25-329 on pDM4 | This study |
| pLS21 | yscU cloned under tac promoter of pMMB66EH | This study |
| pPE33 | yscU cloned under tac promoter of pKK223-3 | Edqvist et al., unpublished |
| pPE40 | yscU(Δ263-266) cloned under tac promoter of pKK223-3 | This study |
| pPE41 | yscU(N263A) cloned under tac promoter of pKK223-3 | This study |
| pPE42 | yscU(P254A) cloned under tac promoter of pKK223-3 | This study |
| pPE43 | yscU(T265A) cloned under tac promoter of pKK223-3 | This study |
| pPE5 | His6yscU(202-354) under T7 promoter of pET-22b | This study |
| pML11 | yscU(211-354) cloned under tac promoter of pMMB66EH | This study |
| pML12 | yscU(211-354)-Flag cloned under tac promoter of pMMB66EH | This study |
| pML13 | yscU-Flag fusion cloned under tac promoter of pMMB66EH | This study |
| pML14 | yscU(264-354) cloned under tac promoter of pMMB66EH | This study |
| pML16 | yscU(Δ263-266) Flag cloned under tac promoter of pMMB66EH | This study |
| pML17 | yscU(N263A) Flag cloned under tac promoter of pMMB66EH | This study |
| pML18 | yscU(P254A) Flag cloned under tac promoter of pMMB66EH | This study |
| pML19 | yscU(T265A) Flag cloned under tac promoter of pMMB66EH | This study |
Kmr, Kanamycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
When appropriate, antibiotics were used at the following final concentrations: kanamycin, 50 μg/ml; ampicillin-carbenicillin, 100 μg/ml; and chloramphenicol, 20 μg/ml.
DNA methods.
Preparation of plasmid DNA, restriction enzyme digests, separation by gel electrophoresis, ligation, and transformation of E. coli strains were performed according to standard methods (38). Certain DNA fragments were purified from agarose gels by using GeneClean Kit (Bio 101) or GeneElute Minus EtBr Spin Columns (Sigma). DNA sequencing reactions were performed by using the ThermoSequenase dye terminator cycle sequencing kit and analyzed with a SEQ4×4 sequencer (Amersham Pharmacia Biotech).
Sequence analysis.
A similarity search of protein sequence databases was conducted by using the basic local alignment search tool (BLAST) server program with default parameters (4). The MegAlign program from DNASTAR with default parameters was used to perform protein alignments.
Construction of the yscU in-frame deletion strain.
The yscU in-frame deletion strain, YPIII(pIB75) was constructed as follows. A double PCR was performed with the primer pairs YscU3/YscU4 and YscU5/YscU6 (Table 2), respectively, by using the virulence plasmid pIB102 as a template, rendering one fragment from the upstream and one from the downstream region of yscU. These fragments were ligated by PCR with the primer pair YscU3/YscU6. The resulting PCR product was digested with XbaI and SacI and cloned into the same restriction sites of pDM4, yielding plasmid pLS13. This plasmid was transformed into E. coli S17-1λpir, from which it was introduced by conjugation into the recipient Yersinia strain YPIII(pIB102). Clones with pLS13 integrated into pIB102 by a single recombinant event were selected on YSAB plates containing kanamycin and chloramphenicol. To select for bacteria having lost the suicide vector sacB-dependent sucrose sensitivity (27) was employed. Sucrose-resistant colonies were PCR amplified to confirm the presence of the deletion. The resulting strain, YPIII(pIB75), carries a yscU in-frame deletion of amino acids 25 to 329.
TABLE 2.
Primer sequencesa
| Primer | Sequence |
|---|---|
| YscU3 | 5′-GCT GAT CTA GAT TAA TCG CCG CTG TAT TGG C-3′ |
| YscU4 | 5′-CTC TAT TTG TTT CGC TAC CTG TCC CTT TTT-3′ |
| YscU5 | 5′-GTA GCG AAA CAA ATA GAG GCC ACA GCT GAA-3′ |
| YscU6 | 5′-GCT CAC GAG CTC GCA CAG GAG AAA TAC AAT TAC C-3′ |
| YscU7 | 5′-GCT CAG AAT TCT GAT CCC TGT TTT GGA GAA GT-3′ |
| YscU8 | 5′-GC TCA CTG CAG ACC ATA TTC CTA GTT ACA TTG C-3′ |
| YscU19 | 5′-GCT CAC GAA TTC ATG AGC GGA GAA AAG ACA GA-3′ |
| YscU21 | 5′-GCT CAC GAA TTC ATT AAG GAA CTT AAA ATG AGC A-3′ |
| YscU23 | 5′-CGA CAG CTG CAG TTA TTT ATC GTC ATC ATC TTT ATA ATC TAA CAT TTC GGA ATG TTG TTT CT-3′ |
| YscU37 | 5′-CGC TCA TCA GTG GTG GAA TTC ATG CCG ACC-3′ |
| PE9 | 5′-GCC GCC CAT ATG AGC AAG GAT GAG ATC AAA-3′ |
| PE10 | 5′-GGC CTC GAG TAA CAT TTC GGA ATG TTG TTT-3′ |
| PE34 | 5′-CCA ATA GCA ATA GCT ACC ACC AC-3′ |
| SL99 | 5′-GTG GTG GTA GCT ATT GCT ATT GG-3′ |
| PE73 | 5′-GTG GTG GTA GCT GCT CCG ACC CAT ATT GC-3′ |
| PE74 | 5′-GTG GTA GCT AAT GCG ACC CAT ATT GC-3′ |
| PE75 | 5′-GTA GCT AAT CCG GCC CAT ATT GCT ATT G-3′ |
| PE102 | 5′-GCA ATA TGG GTC GGA GCA GCT ACC ACC AC-3′ |
| PE103 | 5′-GCA ATA TGG GTC GCA TTA GCT ACC AC-3′ |
| PE104 | 5′-CAA TAG CAA TAT GGG CCG GAT TAG CTA-3′ |
The primer sequences are based on Y. pseudotuberculosis yscU GenBank accession no. L25667, nucleotide positions 4452 to 5516.
Construction of yscU derivatives.
Derivatives of the yscU gene were cloned under the tac promoter of pMMB66EH, generating plasmids as follows: pLS21 (yscU; amino acids 1 to 354), pML13 (yscUFlag; amino acids 1 to 354 plus a C-terminal Flag epitope; DYKDDDDK [19]), pML11 (yscU211-354; amino acids 211 to 354), pML12 (yscU211-354-Flag; amino acids 211 to 354 plus a C-terminal Flag epitope), and pML14 (yscU264-354; amino acids 264 to 354) were engineered by using the primer pairs YscU7/YscU8, YscU19/YscU23, YscU21/YscU8, YscU21/YscU23, and YscU37/YscU8 (Table 2), respectively, in a PCR with YPIII(pIB102) as a template.
Amplified DNA fragments were ligated with the linearized pCR2.1-TOPO cloning vector and transformed into E. coli TOP10 One Shot chemically competent cells, all according to the manufacturer's instructions (Invitrogen). Plasmids were purified from TOP10 cells and restricted with EcoRI and PstI. The excised fragments were purified from a 1% SeaKem GTG agarose gel by using GeneClean Kit (Bio 101). Ligation with pMMB66EH, restricted with EcoRI and PstI, was followed by electroporation into E. coli S17-1λpir. A positive clone was used for introduction of the constructs into the recipient Yersinia strains by conjugation. Transformants were plated onto YSAB plates selecting for both the virulence plasmid and pMMB66EH. Positive clones were identified by PCR, plasmid preparation, and restriction of these plasmids with PstI and EcoRI.
Construction of mutations in the cytoplasmic domain of YscU.
The template used for deletion of the four amino acid conserved site, NPTH (amino acids 263 to 266), was pLS21 (Table 1). To introduce substitutions at residues N263, P264, and T265 the previously described plasmid pPE33 (Table 1; Edqvist et al.) was used as a template. The site-directed mutagenesis was carried out according to the procedures in the QuickChange Mutagenesis Kit (Stratagene). The mutagenic oligonucleotides used (Table 2) were as follows: NPTH deletion PE34/SL99, N263A mutation PE73/PE102, P264A mutation PE74/PE103, and T265A mutation PE75/PE104, resulting in the construction of plasmids pPE40, pPE41, pPE42, and pPE43, respectively. The mutant versions of yscU were Flag tagged and introduced into pMMB66EH by using the pPE40-43 plasmids as templates in PCR with primers YscU19 and YscU23, essentially as described above when Flag tagging the wild-type yscU. The resulting plasmids were denoted pML16, pML17, pML18, and pML19, respectively (Table 1).
Analysis of LCR phenotypes: MOX test.
The strains tested were grown over night and then diluted in 0.9% NaCl. The serial dilutions were spread on Ca2+ containing or MOX plates. The plates were incubated at 26 or 37°C overnight. The number of colonies was counted for determination of growth phenotypes.
Definition of growth phenotypes.
Strains unable to grow at 37°C without the addition of Ca2+ are defined as calcium dependent, which is the wild-type growth phenotype for Y. pseudotuberculosis. Calcium-independent mutants grow at 37°C irrespective of the Ca2+ concentration, a phenotype usually coupled to impaired secretion. A strain unable to grow at 37°C is defined as temperature sensitive and usually secretes Yops irrespective of calcium.
Viable count.
Strains expressing YscU variants that were not proteolytically cleaved, i.e., N263A, P264A, or T265A, showed a decreased growth rate in liquid culture. To further investigate the growth deficiency viable count in the absence and presence of IPTG (isopropyl-β-d-thiogalactopyranoside) was performed. The strains containing the different YscU variants were cultured overnight and then diluted in 0.9% NaCl. The serial dilutions were spread on BAB plates with added antibiotics, as well as plates with both antibiotics and 0.4 mM IPTG. Colony formation was monitored for 3 days, and the colonies were counted for calculation of the viability after 3 days of incubation.
Yop secretion assay.
Overnight cultures of Yersinia strains grown at 26°C were diluted (1:10) in fresh media (BHI plus Ca2+or BHI minus Ca2+) and grown for 1 h at 26°C prior to a temperature shift to 37°C and growth for an additional 3 h. For induction of the tac promoter, IPTG was added to a final concentration of 0.4 mM. After measuring the optical density at 600 nm (OD600), the bacteria were harvested by centrifugation and dissolved in 400 μl of 2× sodium dodecyl sulfate (SDS) sample buffer. The culture supernatants were filtrated through a 0.45-μm (pore-size) filter to rid them of any remaining bacteria. Then, 400 μl of the supernatants was removed and diluted in 400 μl of 2× SDS sample buffer, to be separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by enhanced chemiluminescence (ECL) Western blot as described below. The rest of the supernatant proteins were precipitated with 10% trichloroacetic acid and collected by centrifugation. The proteins were dissolved in 500 μl of 2% SDS at 37°C, precipitated with acetone, and pelleted again by centrifugation. The secreted proteins were finally dissolved in 400 μl of 2× SDS sample buffer to be separated by SDS-PAGE and visualized by Coomassie brilliant blue staining.
Immunoblotting.
Samples were routinely boiled for 5 min prior to separation by denaturating SDS-PAGE (12 to 15% [wt/vol] acrylamide). Electroblotting of proteins to Immobilon-P Transfer Membranes (Millipore) was performed by using Trans-Blot Semi-Dry transfer cell (Bio-Rad) and buffers as described by Laemmli (24). Membranes were blocked overnight in Tris-buffered saline (TBS) with 5% nonfat dry milk.
Yop proteins were detected with a polyclonal rabbit antiserum raised against the secreted Yops. The Flag-tagged derivatives of YscU were detected with anti-Flag M2 monoclonal mouse antibodies (Sigma).
After three 10-min washes in TBS with 0.1% Tween (TBS-T), the membranes were probed with a secondary antibody conjugated to horseradish peroxidase and directed against mouse or rabbit (Amersham Pharmacia Biotech). Visualization of the proteins was performed with ECL detection (Amersham Pharmacia Biotech).
Purification of Flag-tagged constructs for N-terminal sequencing.
To establish the locus for cleavage of YscU, Flag-tagged constructs were used. Overnight cultures of the Y. pseudotuberculosis strains YPIII(pIB75+pML13) and YPIII(pIB102+pML12) were diluted 1:20 in 50 ml of fresh medium and cultured at 26°C to an OD600 of 0.5. Expression of the YscU derivatives was induced by addition of 0.4 mM IPTG, and the bacteria were grown for an additional 3 h at 37°C. Cells were harvested by centrifugation, and the pellet was dissolved in 5 ml of TBS (pH 7.4) with 0.1 mM phenylmethylsulfonyl fluoride. Bacteria were lysed by sonication and, after pelleting the debris by centrifugation, the supernatant was incubated overnight with anti-Flag M2 affinity gel beads (Sigma) at 4°C. Upon removal of the supernatant, the gel beads were washed repeatedly with TBS (pH 7.4). The Flag fusion proteins were eluted by competitive elution with 150-μl volume aliquots of a solution containing 100 μg of Flag peptide/ml in TBS (pH 7.4). The proteins in the resulting eluates were separated by SDS-PAGE and blotted to polyvinylidene difluoride Immobilon-P transfer membranes (Millipore). Upon Coomassie brilliant blue staining, the YscU fragments were excised and N-terminally sequenced by Per-Ingvar Ohlsson of the Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, Sweden.
Preparation of YscU antiserum.
YPIII(pIB102) genomic DNA was PCR amplified with the primer pair PE9/PE10. The resulting PCR product, which consists of codons 202 to 354 (encoding the cytoplasmic domain of YscU), was digested with NdeI and XhoI and cloned into the same sites of the pET-22B vector. This places a six-histidine tag on the C terminus of YscU. This construct, pPE5, was transformed into E. coli BL21(DE3). Cells were grown overnight at 37°C in LB containing 100 μg of carbenicillin ml−1. The cultures were diluted 100-fold into 500 ml of fresh medium and were grown to an OD600 of 0.5. Expression of the YscU cytoplasmic domain was induced by the addition of 0.4 mM IPTG, and the cells were grown for an additional 3 h at 37°C. Cells were then centrifuged and resuspended in 10 ml of lysis buffer (50 mM Tris, pH 8.0; 25 mM NaCl; 0.1 mM EDTA). Lysozyme was added to a final concentration of 0.1 mg ml−1, and the cells were left on ice for 30 min. Cells were then lysed by sonication and were centrifuged for 20 min at 20,800 × g to pellet unlysed cells and insoluble material. The insoluble pellet, containing the cytoplasmic domain of YscU, was washed with 1% Triton X-100-1 mM EDTA and then resuspended in lysis buffer containing 8 M urea. The resuspension was centrifuged as described above to remove insoluble material, and the soluble supernatant was passed over a nickel-nitrilotriacetic acid column (Qiagen). The column was washed with 10 volumes of lysis buffer containing 8 M urea, and the cytoplasmic domain of YscU was eluted stepwise with lysis buffer-urea containing increasing concentrations of imidazole. Purified YscU was diluted to 4 M urea and antibodies against purified YscU were generated in rabbits by AgriSera AB, Vännäs, Sweden.
RESULTS
YscU is essential for Yop secretion.
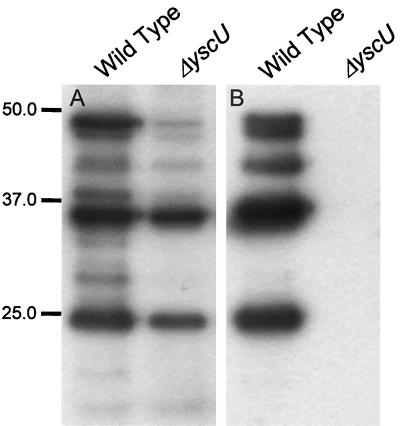
In order to facilitate studies of the role of YscU in Yop secretion, a yscU mutant strain, denoted YPIII(pIB75), was constructed in Y. pseudotuberculosis. The yscU mutant did not secrete any Yops during inducing conditions in vitro, i.e., growth in Ca2+-depleted medium at 37°C (Fig. 2B). Like other previously described type III secretion mutants, the yscU mutant strain also showed decreased Yop expression compared to the wild-type strain YPIII(pIB102) (Fig. 2A). In agreement with the secretion-negative phenotype, the yscU strain showed a calcium-independent growth phenotype (data not shown); i.e., in contrast to the wild-type strain, the yscU mutant did not require calcium for growth at 37°C. The phenotype of the yscU strain of Y. pseudotuberculosis was essentially as previously described for a yscU mutant of Y. enterocolitica by Allaoui et al. (3).
FIG. 2.
Yop secretion is blocked and Yop expression diminished in the yscU-null mutant. Yop expression and secretion was analyzed in the yscU-null mutant YPIII(pIB75) and the wild-type strain YPIII(pIB102) by ECL Western blot, with polyclonal antiserum raised against the secreted Yops whole-cell fractions (A) and supernatant fractions (B), respectively. The bacteria were cultured in BHI medium depleted for calcium at 37°C. For details, see Materials and Methods. Molecular mass markers are shown to the left in kilodaltons.
High levels of YscU downregulate Yop expression and secretion.
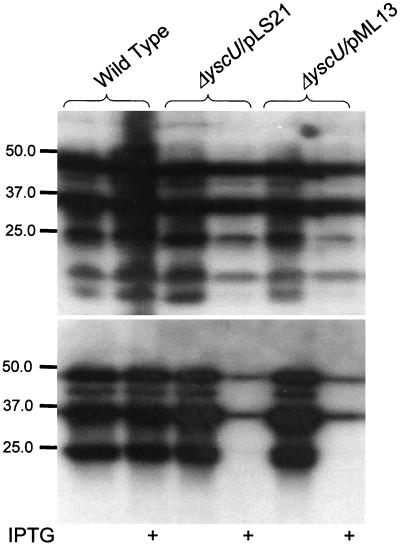
To verify that the defect in secretion in the yscU mutant strain YPIII(pIB75) was due to lack of a functional YscU protein, the yscU mutant was complemented with a plasmid, pLS21, expressing a wild-type copy of yscU under the control of the tac promoter. Secretion, as well as expression, levels of Yops were restored by the low levels of YscU expressed from the leaky tac promoter in the absence of the inducer IPTG (Fig. 3). Induction of YscU expression by the addition of IPTG resulted in lowered Yop secretion and Yop expression (Fig. 3). YscU is a membrane protein (3) and therefore it was possible that overexpression of YscU affected the growth rate and thereby indirectly affected Yop expression and secretion. However, no significant effect of different YscU expression levels on growth rate could be observed (data not shown). Thus, only low levels of YscU are required for functional type III secretion, whereas higher levels of YscU have a negative effect on the expression and secretion of Yop proteins.
FIG. 3.
Transcomplementation of the yscU null mutant strain. ECL Western blot, with polyclonal antiserum raised against the secreted Yops was used to study complementation of the yscU-null mutant YPIII(pIB75) with pLS21 or pML13 in trans expressing wild-type or a Flag-tagged variant of YscU, respectively, whole-cell fractions (A) and supernatant fractions (B), respectively. The bacteria were cultured in BHI lacking calcium at 37°C, with or without 0.4 mM IPTG, as implied in the figure. Molecular mass markers are shown to the left in kilodaltons.
The C-terminal cytoplasmic domain of YscU promotes elevated secretion.
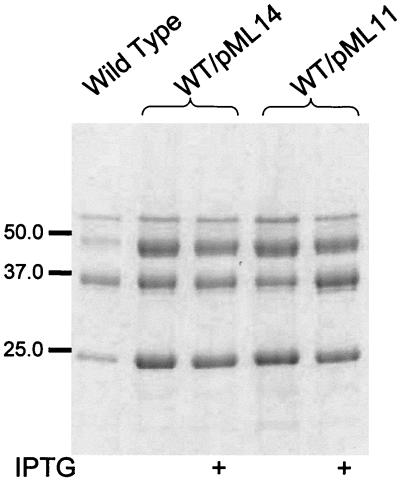
A study by Allaoui et al. (3) showed that the N-terminal part of YscU contains four membrane-spanning domains, whereas the C-terminal part of the protein resides in the cytoplasm. We reasoned that the mechanism by which YscU downregulated Yop expression and secretion could be via the C-terminal cytoplasmic domain. Therefore, the C-terminal cytoplasmic part of YscU (amino acids 211 to 354) was cloned under control of the tac promoter of pMMB66EH. The resulting plasmid, denoted pML11, was introduced into the wild-type strain YPIII(pIB102). Interestingly, overexpression of the cytoplasmic part of YscU resulted in increased secretion of Yops under inducing conditions, i.e., growth in calcium-depleted media at 37°C (Fig. 4). However, the level of Yop expression did not differ from the wild-type level (data not shown). Overexpression of the cytoplasmic domain in the wild-type strain, YPIII(pIB102), under conditions that do not promote Yop secretion, i.e., room temperature or growth at 37°C in the presence of calcium, had no effect on secretion (data not shown). Therefore, our results indicated that YscU expression levels somehow influence Yop secretion and/or expression. The C-terminal domain, YscU (amino acids 211 to 354), has a positive effect on Yop secretion, whereas high levels of full-length YscU result in lowered secretion and expression of Yops.
FIG. 4.
The cytoplasmic domain of YscU stimulates Yop secretion. Secretion of Yops by strain YPIII(pIB102) with pML14 or pML11 expressing the C-terminal cytoplasmic domain of YscU, amino acids 264 to 354 or amino acids 211 to 354, respectively. Separation of proteins in the supernatant fractions was performed by SDS-PAGE, followed by Coomassie brilliant blue staining. The bacteria were cultured in BHI lacking calcium at 37°C, with or without 0.4 mM IPTG, as indicated in the figure. Molecular mass markers are shown to the left in kilodaltons.
YscU is cleaved at position proline-264 in the C-terminal domain.
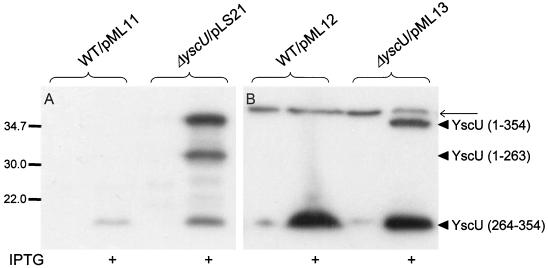
A recent study of FlhB, the YscU homologue of the flagellar assembly-secretion system, has revealed that FlhB expressed in E. coli is proteolytically cleaved in the C-terminal cytoplasmic part at proline-270 (31). Since the sequence around the cleavage site, NPTH (Fig. 1), is conserved in all of the FlhB homologues of TTSSs, we sought to determine whether YscU, like FlhB, is also cleaved in its C terminus. The antiserum raised against the cytoplasmic domain of YscU recognized protein bands of three different sizes (40, 30, and 10 kDa) in strains where YscU expression was induced by IPTG (Fig. 5A). This indicated that also YscU, similar to FlhB, was proteolytically cleaved. Unfortunately, the quality of the antiserum was too poor to detect YscU or any variants of the protein when expressed at low levels (Fig. 5A). To facilitate the monitoring of C-terminal fragments of YscU, we tailed the full-length YscU protein with a C-terminal Flag epitope (19). The Flag-tagged derivative was cloned into pMMB66EH. The resulting plasmid was denoted pML13 and was verified to complement the yscU mutant; i.e., full complementation was achieved without IPTG induction, whereas overexpression after addition of IPTG resulted in the downregulation of Yop expression and secretion (Fig. 3). Strains expressing the Flag-tagged YscU were then analyzed by Western blot with the anti-Flag monoclonal antibody. In the absence of IPTG, a 10-kDa fragment was the only protein recognized by the monoclonal antibodies. When IPTG was added, a 40-kDa protein corresponding to full-length YscU also reacted with the antibodies (Fig. 5B). This demonstrated that YscU is cleaved within the cytoplasmic domain to yield a 10-kDa C-terminal fragment and a 30-kDa N-terminal fragment. To identify the site for proteolytic cleavage, the C-terminal part of the Flag-tagged YscU was purified with anti-Flag antibodies coupled to agarose beads. Amino acid sequencing of the purified protein revealed that cleavage occurred at proline-264. Alignment of YscU and FlhB showed that this position corresponds to position Pro-270 of FlhB (31) (Fig. 1). This indicates that the two proteins are cleaved at identical sites. The cleavage of YscU appeared to be essentially complete at low YscU expression levels, which also appear to be optimal for efficient Yop secretion. However, when YscU was overexpressed, proteolytic cleavage was incomplete (Fig. 5). Interestingly, in this case Yop expression and secretion are lowered. This indicates that the full-length form of YscU negatively affects expression, whereas the small 10-kDa C-terminal fragment of YscU upregulates secretion. To test this hypothesis, we utilized plasmid pML12, where the cytoplasmic domain of YscU (amino acids 211 to 354) was Flag-tagged at its C terminus. Similar to the nontagged construct, expression of the flag-tagged cytoplasmic domain of YscU in the wild-type strain YPIII(pIB102) resulted in elevated secretion of Yops (data not shown). This part of YscU was found to be cleaved at the same position as full-length YscU, i.e., at Pro-264 (verified by N-terminal sequencing; Fig. 5), indicating that the 10-kDa cytoplasmic cleavage product of YscU was responsible for the increased Yop secretion. When the cytoplasmic domain of YscU, corresponding to the fragment generated by cleavage (amino acids 264 to 354), was expressed from plasmid pML14, in strain YPIII(pIB102) (Fig. 4), this also turned out to be the case, i.e., elevated secretion was seen under conditions known to induce Yop secretion in vitro. This suggested that cleavage of YscU at position proline-264 is important for function and possibly also influences Yop secretion levels.
FIG. 5.
The cytoplasmic domain of YscU is proteolytically cleaved at proline-264. The cleavage pattern of full-length YscU was analyzed in the yscU-null mutant strain YPIII(pIB75) with pLS21 or pML13 expressing a wild-type or a Flag-tagged form of YscU, respectively. For analysis of the cleavage of the cytoplasmic domain (amino acids 211 to 354) of the protein, the wild-type strain YPIII(pIB102) with pML11 or pML12 was utilized, expressing a nontagged and a Flag-tagged variant of this domain of YscU, respectively. Proteins were visualized by using ECL Western blot to whole-cell fractions with antiserum raised against the cytoplasmic domain of YscU (amino acids 202 to 354) (A) or monoclonal antibodies directed against the Flag epitope (B). The different forms of YscU indicated in the figure are as follows: the full-length protein (amino acids 1 to 354), the N-terminal cleavage product (amino acids 1 to 263), and the C-terminal cleavage product (amino acids 264 to 354). The bacteria were cultured in BHI depleted for calcium at 37°C, with or without 0.4 mM IPTG, as indicated in the figure. The thinner arrow points to a nonspecific protein band recognized by the anti Flag-antibodies, which also appears in negative controls, i.e., in Yersinia strains that do not carry Flag-tagged YscU constructs. Molecular mass markers are shown to the left in kilodaltons.
The proteolytic cleavage of YscU is not required for function.
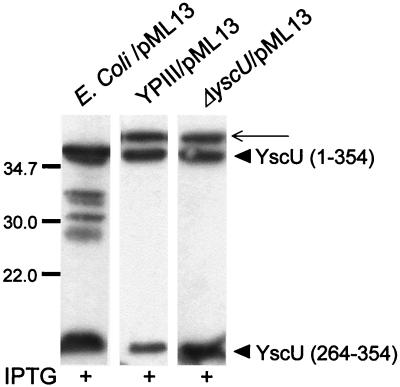
We then investigated the kinetics of YscU cleavage in different genetic backgrounds and growth conditions. The different strains with Flag-tagged yscU in trans under the control of the tac promoter were grown at room temperature or at 37°C in media containing or depleted of calcium. YscU cleavage occurred irrespective of growth conditions, indicating that the cleavage itself is not a signal for TTSS assembly and secretion. Furthermore, YscU was also proteolytically cleaved in various bacterial backgrounds. The fact that YscU was cleaved also in the virulence plasmid-cured Y. pseudotuberculosis strain YPIII (TTSS-negative), as well as in E. coli K-12, indicates that the protease responsible for cleavage is not specific for the TTSS (Fig. 6).
FIG. 6.
The proteolytic cleavage of YscU is independent of a functional TTSS. ECL Western blot with monoclonal antibodies directed against the Flag epitope was employed to visualize the cleavage of YscU in whole-cell fractions in different bacteria: E. coli K-12 strain S17-1λpir, the plasmid-cured strain YPIII, and the yscU-null mutant YPIII(pIB75), all containing pML13 expressing full-length YscU tailed with a Flag tag. The different forms of YscU indicated in the figure are the full-length protein YscU (amino acids 1 to 354) and the C-terminal cleavage product YscU (amino acids 264 to 354). The thinner arrow points to a protein band recognized by the Flag antibodies that appears also in negative controls, i.e., Yersinia strains where no Flag-tagged construct is present. The Yersinia strains were cultured at 37°C in calcium-depleted medium, and the E. coli strain was grown in LB at 37°C; all cultures were supplemented with 0.4 mM IPTG. Molecular mass markers are shown to the left in kilodaltons.
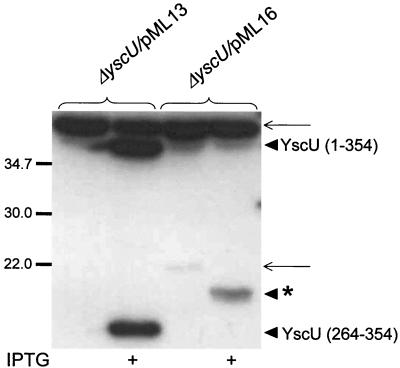
To determine whether proteolytic cleavage of YscU is critical for function, we utilized plasmid pML16 expressing a mutated form of YscU with an in-frame deletion of the conserved sequence NPTH (amino acids 263 to 266). These residues are highly conserved in all YscU and FlhB homologues and overlap the proteolytic cleavage site. Flag-tagged YscU with the ΔNPTH deletion and wild-type YscU were expressed in trans in the yscU mutant strain. Bacteria were grown at 37°C in medium lacking calcium, which normally induces Yop secretion. Western blots of cell pellet fractions revealed that the ΔNPTH mutant protein was also cleaved, but in this case the C-terminal fragment was larger than that observed for wild-type YscU (Fig. 7). This suggests that the cytoplasmic domains of the ΔNPTH mutant have an altered conformation. Significantly, no Yop secretion was seen in the ΔNPTH mutant (Fig. 8C), indicating that the conformation and correct cleavage of the cytoplasmic domains are critical for the function of YscU in type III secretion by Yersinia.
FIG. 7.
The YscU ΔNPTH mutant shows a different cleavage pattern compared to the wild-type protein. The C-terminally Flag-tagged proteins YscU or YscU ΔNPTH were expressed in the yscU-null mutant strain YPIII(pIB75) from plasmids pML13 and pML16, respectively. The cleaved forms of YscU were visualized by ECL Western blot with monoclonal antibodies directed against the Flag epitope. The different forms of YscU indicated in the figure are as follows: the full-length protein YscU (amino acids 1 to 354), the C-terminal cleavage product, YscU (amino acids 264 to 354), and the ca 15-kDa C-terminal fragment of the ΔNPTH mutant protein which is indicated with an asterisk. The thinner arrows point to two nonspecific bands that also appear in negative controls with Yersinia strains that do not carry any Flag-tagged YscU constructs. The bacteria were cultured in BHI depleted of calcium at 37°C, with or without 0.4 mM IPTG, as indicated in the figure. Molecular mass markers are shown to the left in kilodaltons.
FIG. 8.
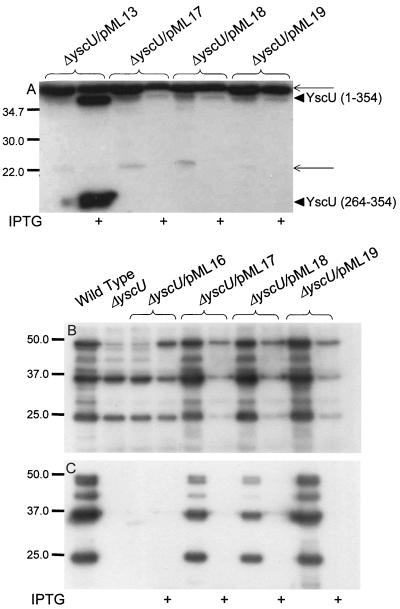
YscU cleavage is not required for functional Yop secretion. Four forms of YscU with different mutations (ΔNPTH, N263A, P264A, and T265A) were expressed from plasmids pML16, pML17, pML18, or pML19, respectively, in the yscU-null mutant strain YPIII(pIB75) and compared to the wild-type strain YPIII(pIB102). (A) ECL Western blot with monoclonal antibodies directed against the Flag epitope to visualize the Flag epitope containing fragments of YscU. The different forms of YscU indicated are, the full-length protein YscU (amino acids 1 to 354), the C-terminal cleavage product, YscU (amino acids 264 to 354). The thinner arrows point to two nonspecific bands which also appear in negative controls with Yersinia strains that do not carry any Flag-tagged constructs. ECL Western blot with polyclonal antiserum raised against the secreted Yops to analyze Yop expression and secretion was performed on whole-cell fractions (B) and supernatant fractions (C), respectively. The bacteria were cultured in BHI lacking calcium at 37°C, with or without 0.4 mM IPTG, as indicated in the figure. Molecular mass markers are shown to the left in kilodaltons.
We also constructed three YscU variants with point mutations in the conserved amino acids overlapping the cleavage site of YscU, N263A, P264A, and T265A. All of these mutant forms of YscU were tailed with C-terminal Flag epitopes to monitor cleavage. The mutated versions of YscU complemented the yscU-null mutant with respect to Yop expression and secretion (Fig. 8B and C). The T265A mutant complemented Yop secretion essentially to wild-type levels. Induction of higher expression levels of the three point mutant variants of YscU resulted in a block in Yop expression and secretion (Fig. 8B and C). Surprisingly, none of these forms of YscU were proteolytically cleaved (Fig. 8A), indicating that these three residues are critical for cleavage to occur. Importantly, from this we can conclude that cleavage per se is not essential for the function of YscU in Yop secretion. However, the single site mutations of YscU had severe effects on the growth of Y. pseudotuberculosis especially after expression was induced by IPTG. Monitoring growth in liquid culture revealed that growth ceased within two divisions after addition of IPTG and after prolonged incubation the OD of these cultures decreased. The effect on growth occurred irrespective of the growth temperature and in both Y. pseudotuberculosis strains and E. coli K-12 (data not shown). Plating overnight cultures containing clones with the YscU single-site mutations yielded a >1,000-fold reduction in colony formation on plates containing IPTG compared to plates lacking IPTG. No significant effect on growth was seen for strain YPIII(pIB75) expressing wild-type YscU or the ΔNPTH mutant protein in trans, whereas already low expression levels of the point mutated YscU variants affected growth rate. Colonies of strain YPIII(pIB75) expressing the single-site-mutated proteins were not visible until after 2 days of growth compared to 1 day for the strain expressing wild-type YscU. This shows that the expression of these variants of YscU, which retained function in Yop secretion but could not be cleaved, was highly toxic. Therefore, one important role of cleavage of YscU is to prevent the toxic affect of the full-length YscU protein on the bacterial cell.
DISCUSSION
Structurally, the type III secreton resembles the basal body of the flagellar type III export apparatus and the two systems have also been suggested to share a secretion mechanism involving channeling of virulence effectors and filament proteins, respectively, through the membrane-spanning structures. The common secretion system is postulated to consist of nine proteins, which are conserved among all TTSS sequenced so far including that of the flagella. These proteins are believed to localize in the cytoplasm or at the inner ring structure of the secretion organelles and suggested to function as a secretion pore. The membrane protein FlhB is one of the key factors required for secretion of flagellar components (29). FlhB acts in concert with the soluble protein FliK to control the substrate switch from hook to filament proteins (23, 28, 33, 45).
In this work we have studied the role of the FlhB homologue YscU of Y. pseudotuberculosis in type III secretion of Yop effector proteins. We confirm that YscU, as previously shown in Y. enterocolitica (3), is essential for Yop secretion in Y. pseudotuberculosis. A yscU-null mutant was unable to secrete Yops during inducing conditions in vitro, and the mutant also showed decreased Yop expression compared to the wild-type strain. This is a common phenomenon in type III secretion mutants and is believed to be the result of a feedback inhibition mechanism (11).
Only low levels of YscU were required to fully restore Yop expression and secretion when the yscU mutant was transcomplemented, whereas higher expression levels of YscU resulted in lowered Yop expression as well as secretion. This initial observation suggested that YscU, like FlhB, can influence protein secretion levels. Previous work by Allaoui et al. (3) had revealed that a large C-terminal domain of YscU localizes to the cytoplasm, and hence we hypothesized that it could be involved in controlling Yop secretion. In line with this, overexpression of the C-terminal cytoplasmic domain of YscU (amino acids 211 to 354) in the wild-type strain resulted in elevated Yop secretion. Importantly, Yop expression levels were not affected, indicating that the C-terminal cytoplasmic part of YscU preferentially acts at the level of secretion.
The YscU/FlhB protein family is one of the most highly conserved of the TTSSs, and the homologies of the cytoplasmic domains are particularly high. Aligning the proteins of the YscU/FlhB family reveals the presence of a four-amino-acid sequence in the middle of the C-terminal cytoplasmic domain, NPTH, which is conserved among the FlhB/YscU homologues. Interestingly, it was recently shown that FlhB was unstable when expressed in E. coli and that proteolytic cleavage occurred at position Pro-270 within the conserved NPTH sequence, yielding a 10-kDa C-terminal fragment (31). Using a C-terminally Flag-tagged variant of YscU, we could establish that YscU was also cleaved at the corresponding site, Pro-264, during conditions promoting Yop secretion. Cleavage at the same position also occurred when the Flag-tagged C-terminal cytoplasmic domain YscU (amino acids 211 to 354) was expressed in the wild-type strain. This result, together with our observation that overexpression of the whole C-terminal cytoplasmic portion of YscU leads to increased secretion, suggested that it might be the C-terminal cleavage product YscU (amino acids 264 to 354) that promotes Yop secretion. By expressing this variant of YscU (amino acids 264 to 354) in trans in the wild-type strain we could show that the 10-kDa C-terminal fragment resulting from the proteolytic cleavage is biologically active and promotes elevated Yop secretion.
These data strongly argued that the observed cleavage at Pro-264 was important for YscU function in type III secretion. However, YscU cleavage is not mediated by other components directly involved in TTSS, since efficient cleavage also occurred in a Y. pseudotuberculosis strain lacking the entire TTSS, as well as in E. coli K-12 lacking a functional TTSS. This indicated that the cleavage per se is not a key regulatory event in induction and assembly of TTSS.
To further investigate the significance of YscU proteolysis in relation to Yop secretion, we constructed a variant of YscU where the conserved residues NPTH (amino acids 263 to 266) were deleted. Deletion of these four amino acids resulted in the loss of YscU function in Yop secretion, and YscU was now cleaved at a different position, yielding an approximately 15-kDa C-terminal fragment. Minamino and Macnab (31) recently showed that the 10-kDa cytoplasmic domain of FlhB resulting from the proteolytic cleavage at Pro-270 interacted with the other cytoplasmic domain, corresponding to YscU amino acids 211 to 263 (Fig. 1). Since proteolysis of the ΔNPTH mutant appears to result in disruption of the domain between amino acids 211 and 263, it would abolish any potential interaction between the two cytoplasmic domains of YscU. This could then explain why the ΔNPTH mutant is nonfunctional in Yop secretion. Furthermore, the different cleavage pattern observed for the ΔNPTH mutant indicates that deletion of the conserved four-amino-acid motif affects the conformation of the cytoplasmic domain of YscU, which in turn also could render the protein nonfunctional.
Interestingly, substitutions of individual amino acids of the NPTH motif of YscU, i.e., N263A, P264A, and T265A, resulted in YscU derivatives that were not cleaved, demonstrating that these three residues are critical for cleavage to occur. However, when expressed at low levels, these YscU variants complemented the yscU-null mutant strain with respect to type III secretion. Thus, specific cleavage at the conserved amino acid residue was not, as initially anticipated, required for YscU function in Yop secretion. Instead, induction of higher levels of transcription of these uncleaved but functional variants of YscU was toxic not only to Y. pseudotuberculosis but also to E. coli K-12. Thus, for functional type III secretion the conformation of the cytoplasmic domain of YscU appears to be critical, whereas the specific cleavage at the conserved proline-264 is required for survival of the bacterium since the uncleaved functional form of YscU is highly toxic to the bacterium.
Cleavage of YscU and FlhB occurs also in E. coli (31), and we show here that cleavage occurs in a strain lacking TTSS. This indicates that the protease promoting cleavage could be common to most gram-negative bacteria. A conserved protease might be essential to these bacteria, and the toxic effect seen during high expression levels of the YscU variants (N263A, P264A, and T265A) could be the result of out-titration of the protease; therefore, the toxic effect would be indirect and not directly dependent on the expression of the noncleavable variants of YscU. However, cleavage was almost complete also when the wild-type form of YscU was expressed at high levels and, in addition, the toxic effect on the bacterial cell was also seen at low expression levels of the different YscU variants. Therefore, we favor the idea that the toxic effect is directly mediated by the YscU variants that were not cleaved and was the result of a different conformation of these proteins.
During the sequential assembly of the bacterial flagellum a key event is the switch from export of hook-rod substrates to filamental substrates. In this event, FlhB and FliK play a central role. fliK mutants display a poly-hook phenotype, i.e., they are disabled in the step where export of hook-rod proteins normally is exchanged for export of filament protein (17, 34, 43). The poly-hook phenotype of a fliK mutant can be suppressed by second-site mutations in the cytoplasmic domain of FlhB (23, 45). This clearly suggests a mechanism where FlhB and FliK, directly or indirectly, interact to somehow control secretion of the different substrates.
Recent work (Edqvist et al.) has demonstrated that this role in the regulation of the type III system is shared by the FlhB homologue in Y. pseudotuberculosis TTSS, YscU, and the protein corresponding to FliK, YscP. A yscP mutant exported higher levels of the needle component YscF to the bacterial surface and was impaired in Yop secretion, thus indicating that this mutant is deficient in the switch from assembly of the substrates that constitute the type III export machinery to secretion of Yop proteins. In analogy to the flagellar system, second-site mutations in the C-terminal cytoplasmic domain of YscU suppress the phenotype of the yscP mutant (Edqvist et al.).
Interestingly, YscP shows low levels of homology to InvJ of Salmonella enterica serovar Typhimurium, which has been found by Galan and coworkers to control the length of the needle part of the type III secreton in Salmonella (22). This suggests that different pathogens share a common mechanism of TTSS whereby a critical step involves a switching mechanism from secreting needle components to effector proteins. The cytoplasmic part of FlhB resulting from specific cleavage has been shown to interact with secretion substrates including rod-hook, filament proteins, as well as FliK (31, 32). Therefore, it is conceivable that YscU also interacts with the type III secretion substrates YscF, Yops, and YscP. As suggested for FlhB, it is likely that initially the affinity of YscU is highest for the rod-hook analogue YscF but that upon needle completion conformational changes in YscU are induced by protein-protein interactions, possibly via YscP, to increase the affinity of YscU for Yop substrates to allow switching to effector protein secretion.
Acknowledgments
This work was supported by grants from the Swedish Research Council and the Foundation for Strategic Research.
REFERENCES
- 1.Aizawa, S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S. I. 1996. Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19:1-5. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui, A., S. Woestyn, C. Sluiters, and G. R. Cornelis. 1994. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J. Bacteriol. 176:4534-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Blocker, A., P. Gounon, E. Larquet, K. Niebuhr, V. Cabiaux, C. Parsot, and P. Sansonetti. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 7.Bolin, I., L. Norlander, and H. Wolf-Watz. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolin, I., and H. Wolf-Watz. 1984. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 43:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G., J. C. Vanootegem, and C. Sluiters. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans-acting pYV and chromosomal genes. Microb. Pathog. 2:367-379. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., and H. Wolf-Watz. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 15.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 16.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Hirano, T., S. Yamaguchi, K. Oosawa, and S. Aizawa. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoiczyk, E., and G. Blobel. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. USA 98:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopp, T. P., K. S. Prickett, V. L. Price, R. T. Libby, C. J. March, D. P. Cerretti, D. L. Urdal, and P. J. Conlon. 1988. A short polypeptide marker sequence useful for recombinant protein identification and purification. Biotechnology 6:1204-1210. [Google Scholar]
- 20.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 22.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsukake, K., T. Minamino, and T. Yokoseki. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176:7625-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. A. 1997. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 5:148-156. [DOI] [PubMed] [Google Scholar]
- 26.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 27.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamino, T., B. Gonzalez-Pedrajo, K. Yamaguchi, S. I. Aizawa, and R. M. Macnab. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295-304. [DOI] [PubMed] [Google Scholar]
- 29.Minamino, T., T. Iino, and K. Kutuskake. 1994. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176:7630-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minamino, T., and R. M. MacNab. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 33.Muramoto, K., S. Makishima, S. I. Aizawa, and R. M. Macnab. 1998. Effect of cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J. Mol. Biol. 277:871-882. [DOI] [PubMed] [Google Scholar]
- 34.Patterson-Delafield, J., R. J. Martinez, B. A. Stocker, and S. Yamaguchi. 1973. A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype-superhooks. Arch. Mikrobiol. 90:107-120. [DOI] [PubMed] [Google Scholar]
- 35.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf-Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231-1233. [DOI] [PubMed] [Google Scholar]
- 37.Rosqvist, R., K. E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schesser, K., M. S. Francis, A. Forsberg, and H. Wolf-Watz. 2000. Type III secretion systems in animal- and plant-interacting bacteria, p. 239-263. In P. Cossart, P. Boquet, S. Normark, and P. Rappuoli (ed.), Cellular microbiology. American Society for Microbiology Press, Washington, D.C.
- 40.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studier, F. W., and B. A. Moffat. 1986. Use of bacteriophage T-7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, H., K. Yonekura, K. Murata, T. Hirai, K. Oosawa, and K. Namba. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J. Struct. Biol. 124:104-114. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, T., and T. Iino. 1981. Role of the flaR gene in flagellar hook formation in Salmonella spp. J. Bacteriol. 148:973-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, A. W., S. Yamaguchi, F. Togashi, S. I. Aizawa, I. Kawagishi, and R. M. Macnab. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]