Abstract
The mxi-spa locus on the virulence plasmid of Shigella flexneri encodes components of the type III secretion system. mxiE, a gene within this locus, encodes a protein that is homologous to the AraC/XylS family of transcriptional regulators, but currently its role in pathogenesis remains undefined. We characterized the virulence phenotype of a nonpolar mxiE mutant and found that this mutant retained the ability to invade mammalian cells in tissue culture and secrete Ipas (type III effectors required for host cell invasion), although it was less efficient than wild-type Shigella at cell-to-cell spread. Despite its invasive properties in culture, the mxiE mutant was completely avirulent in an animal model. Potential targets for MxiE activation were identified by using promoter-green fluorescent protein fusions, and gene expression was examined under various growth conditions. Six MxiE-regulated genes were discovered: ospB, ospC1, ospE2, ospF, virA, and ipaH9.8. Notably, activation of these genes only occurred within the intracellular environment of the host and not during growth at 37°C in liquid culture. Interestingly, all of the MxiE-regulated proteins previously have been shown to be secreted through the type III secretion system and are putative virulence factors. Our findings suggest that some of these Osp proteins may be involved in postinvasion events related to virulence. Since bacterial pathogens adapt to multiple environments during the course of infecting a host, we propose that Shigella evolved a mechanism to take advantage of a unique intracellular cue, which is mediated through MxiE, to express proteins when the organism reaches the eukaryotic cytosol.
Members of the genus Shigella are gram-negative, facultative intracellular bacteria that are the causative agents of bacillary dysentery in humans. These organisms invade the colonic mucosa, causing severe inflammation, which eventually leads to mucosal destruction and results in blood and mucus in the stools (30). The key aspect of Shigella pathogenesis lies in its ability to invade epithelial cells and spread directly from cell to cell. Epithelial cell invasion occurs by a process similar to phagocytosis and requires major rearrangements of the host cell cytoskeleton (9). Once inside the eukaryotic cell, the bacteria escape from the phagocytic vacuole and are able to multiply in the cytoplasm and spread to adjacent cells via nucleation of host cell actin by the outer membrane protein IcsA (5). The force of the rapid actin polymerization propels the organisms through the cytosol, leading to formation of membrane-bound protrusions, which are then engulfed by neighboring cells. The resulting double membrane-bound vacuole is lysed by bacterial proteins, IpaB and IpaC, allowing the cycle to repeat (51).
The ability of shigellae to invade the host epithelium is dependent upon a 31-kb entry region of the 230-kb virulence plasmid (34). The entry region contains two divergently transcribed loci that encode the genes responsible for invasion of eukaryotic cells, intracellular spread, and production of a type III secretion system. One locus is comprised of the mxi and spa operons, which encodes ca. 20 different proteins that together make up the type III secretion apparatus. Type III, or contact-dependent, secretion systems are present in a wide variety of gram-negative plant and animal pathogens and function to secrete protein effectors out of the bacterium and inject them into the cytosol of eukaryotic host cells (reviewed in reference 27). Effector molecules perform a wide variety of functions, some of which include translocation of other effectors into the host cell and interaction with host cell cytoskeletal components to facilitate entry of the bacteria. Although the effector proteins are unique to each bacterial species, the proteins comprising the structural apparatus are highly conserved even in distantly related organisms.
Some of the events leading to bacterial uptake, such as cytoskeletal rearrangements, are mediated through secreted or translocated effectors. In Shigella, the entry region of the virulence plasmid also contains a cluster of genes encoding the type III secreted effector molecules (the invasion plasmid antigens, IpaA, IpaB, IpaC, and IpaD), which are required for host cell invasion and escape from the phagosome (37). Furthermore, ipgC, which encodes a molecular chaperone that associates with IpaB and IpaC in the bacterial cytoplasm, is found immediately upstream of the ipa genes in this region. While not secreted, IpgC is required for bacterial entry by stabilizing IpaB and IpaC and preventing their premature association (38).
During a natural infection, secretion of presynthesized effectors is triggered by contact with a eukaryotic cell. However, in the laboratory, type III secretion can be artificially induced in the absence of host cell contact by manipulation of the growth medium (e.g., removal of Ca2+ for Yersinia) or by the addition of specific chemicals, such as Congo red for Shigella (3).
Expression of the type III apparatus and its effectors is usually regulated by environmental factors, such as temperature, and often regulated at the level of transcription. In Shigella, the AraC-like protein, VirF, is well established as the primary regulator of virulence gene expression. When the bacteria encounter a 37°C environment, VirF activates transcription of both icsA and the secondary regulatory gene virB, thereby initiating the cascade of virulence gene expression, although exactly how VirF responds to temperature is not yet known (45). This thermal activation is not a result of de novo virF transcription since functional VirF protein is present in the bacterial cytosol even at nonpermissive temperatures but rather is due to changes in the amount of supercoiling at the virB promoter which allows VirF to bind and activate its transcription (55). VirB, in turn, activates transcription of the ipa, mxi, and spa operons, which encode the type III secretion system that is essential for invasion (54).
A common mechanism of gene regulation in bacteria is via regulatory proteins of the AraC/XylS family. To date, this family contains more than 100 members as identified by sequence homology to the AraC protein of Escherichia coli (19). With few exceptions, AraC homologues are transcriptional activators. The defining characteristic of this family of proteins is a C-terminal 99-amino-acid stretch of homology comprising the DNA-binding domain. Importantly, this region contains two helix-turn-helix motifs, at least one of which is responsible for binding to the major groove of DNA. A separate domain (sensor domain) or additional protein acts to bind effector molecules. Members of the AraC/XylS family regulate a broad variety of bacterial functions, which can be divided into three categories: carbon metabolism, stress response, and virulence. The AraC homologues that regulate virulence gene expression have not been shown to bind effectors directly, although they do respond to environmental signals such as temperature, osmolarity, or calcium concentration by an unidentified mechanism (19).
In addition to VirF (mentioned above), the role of several AraC homologues that are involved in bacterial pathogenesis has been the focus of intense research. Many of these proteins function as transcriptional activators of molecules that are secreted through the type III secretion system, their cognate chaperones, and in some cases, type III secretion apparatus structural components as well. The AraC-like InvF protein from Salmonella enterica serotype Typhimurium is required for efficient invasion into cultured epithelial cells and activates the expression of genes whose products are secreted through the type III secretion system (12, 28). InvF is encoded by the first gene of the large inv operon and acts upon the sigDE and sicA-sipBCDA operons (12). SigD and the Sip proteins are secreted through the type III apparatus whereas SigE and SicA function as chaperones for these effectors (13, 26, 29). In contrast to other enteric pathogens, transcription of the type III secretion genes in Salmonella is not regulated by temperature. These organisms respond to other environmental signals, including oxygen concentration, osmolarity, and pH (4).
VirF, an AraC homologue of Yersinia enterocolitica, functions as a transcriptional activator of several plasmid-encoded virulence genes: yopE, yopH, and the lcrGVH-yopBD and yscB-yscM operons (31). These Yops are secreted effectors that facilitate formation of channels in the eukaryotic membrane or are translocated into the eukaryotic cell where they are involved in the modulation of cellular signaling (6, 25, 58). LcrG, LcrV, and LcrH each perform multiple functions ultimately related to the secretion and/or targeting of effector proteins (15, 18) and the yscB-yscM locus encodes type III structural genes (39). VirF itself is under environmental control, and its synthesis is induced when bacteria encounter a 37°C environment (39). Furthermore, recent evidence suggests that temperature regulation of virulence gene expression in Yersinia is modulated through changes in the topology of the virulence plasmid (44).
Clearly, AraC homologues are utilized by several pathogenic bacterial species to regulate expression of type III secretion systems and effectors, which are essential virulence determinants for these organisms. An additional AraC homologue, MxiE, has been described in Shigella, although the genes that it may regulate have not yet been identified (2). The purpose of this study was to determine whether MxiE functions as a transcriptional regulator in Shigella flexneri and to identify targets of its regulation. Here we describe an additional level of virulence gene regulation in Shigella which functions only after host cell invasion and identify six virulence plasmid-encoded genes that are activated by MxiE in the intracellular environment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Bacteria were grown in Luria-Bertani broth at 37°C or tryptic soy broth at 30°C with aeration, unless otherwise stated. Appropriate antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; and chloramphenicol, 10 μg ml−1.
TABLE 1.
Shigella strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| 2457T | Wild-type S. flexneri serotype 2a | 17 |
| BS543 | 2457T/ΔicsA | 48 |
| BS600 | 2457T galU::Tn10 [Δ(recB ptr recC recD)::Placbet exo cat] | 14 |
| BS610 | BS600 mxiE (mxiE::aphA-3) | This work |
| BS611 | 2457T mxiE (mxiE::aphA-3) | This work |
| BS613 | BS611/pRRS13 (PlacmxiE+) | This work |
| SCK9 | 2457T/pRRS13 (PlacmxiE+) | This work |
| SCK45 | mxiE+/pEBD166 | Ampr transformant of 2457T |
| SCK78 | mxiE+/pECK127 (PvirA::gfp) | Ampr transformant of 2457T |
| SCK79 | mxiE::aphA-3/pECK127 (PvirA::gfp) | Ampr transformant of BS611 |
| SCK70 | mxiE+/pECK116 (PipaH9.8::gfp) | Ampr transformant of 2457T |
| SCK71 | mxiE::aphA-3/pECK116 (PipaH9.8::gfp) | Ampr transformant of BS611 |
| SCK39 | mxiE+/pECK94 (PospB::gfp) | Ampr transformant of 2457T |
| SCK42 | mxiE::aphA-3/pECK94 (PospB::gfp) | Ampr transformant of BS611 |
| SCK48 | mxiE+/pECK100 (PospC1::gfp) | Ampr transformant of 2457T |
| SCK52 | mxiE::aphA-3/pECK100 (PospC1::gfp) | Ampr transformant of BS611 |
| SCK49 | mxiE+/pECK102 (PospD2::gfp) | Ampr transformant of 2457T |
| SCK53 | mxiE::aphA-3/pECK102 (PospD2::gfp) | Ampr transformant of BS611 |
| SCK50 | mxiE+/pECK104 (PospE2::gfp) | Ampr transformant of 2457T |
| SCK54 | mxiE::aphA-3/pECK104 (PospE2::gfp) | Ampr transformant of BS611 |
| SCK51 | mxiE+/pECK106 (PospF::gfp) | Ampr transformant of 2457T |
| SCK55 | mxiE::aphA-3/pECK106 (PospF::gfp) | Ampr transformant of BS611 |
| SCK41 | mxiE+/pECK98 (PospG::gfp) | Ampr transformant of 2457T |
| SCK44 | mxiE::aphA-3/pECK98 (PospG::gfp) | Ampr transformant of BS611 |
| SBD150 | mxiE+/pEBD168 (PipgD::gfp) | Ampr transformant of BS543 |
| SBD165 | mxiE::aphA-3/pEBD168 (PipgD::gfp) | Ampr transformant of BS611 |
| Plasmids | ||
| pBluescript SK(+) | Cloning vector, Ampr | Stratagene |
| pUC18K | Vector bearing the aphA-3 cassette, Ampr Kanr | 37 |
| pBAD18 | Arabinose-inducible PBAD expression vector, pBRori, Ampr | 20 |
| pEBD166 | Promoterless gfpmut2 expression vector, colE1ori, Ampr | This work |
| pRRS10 | pBluescript KS(+)::gfpmut2 | This work |
| pRRS11 | 3.6-kb EcoRV fragment of pWR100, containing mxiJ-D, cloned into pBluescript SK(+) | This work |
| pECK4 | 891-bp EcoRI-HindIII (Klenow-treated) fragment of pUC18K, bearing aphA-3, ligated into pRRS11 at ClaI-HindIII (Klenow treated) | This work |
| pECK127 | virA::gfp promoter fusion; 560-bp PCR-generated fragment, extending from 502 bp upstream of the start codon to 58 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK116 | ipaH9.8::gfp promoter fusion; 609-bp PCR-generated fragment, extending from 510 bp upstream of the start codon to 99 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK94 | ospB::gfp promoter fusion; 329-bp PCR-generated fragment, extending from 251 bp upstream of the start codon to 78 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK100 | ospC1::gfp promoter fusion; 610-bp PCR-generated fragment, extending from 510 bp upstream of the start codon to 100 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK102 | ospD2::gfp promoter fusion; 637-bp PCR-generated fragment, extending from 509 bp upstream of the start codon to 128 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK104 | ospE2::gfp promoter fusion; 594-bp PCR-generated fragment, extending from 521 bp upstream of the start codon to 73 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK106 | ospF::gfp promoter fusion; 574-bp PCR-generated fragment, extending from 499 bp upstream of the start codon to 75 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pECK98 | ospG::gfp promoter fusion; 249-bp PCR-generated fragment, extending from 169 bp upstream of the start codon to 80 bp downstream of the start codon, ligated with KpnI-BglII-digested pEBD166 | This work |
| pEBD168 | ipgD::gfp promoter fusion; 936-bp PCR-generated fragment, extending from 556 bp upstream of the ipgD start codon to 380 bp downstream of the start codon, ligated with BglII-digested pEBD166 | This work |
Ampr, ampicillin resistance; Kanr, kanamycin resistance.
The L2 mouse fibroblast cell line was maintained in Dulbecco modified Eagle medium (DMEM; Gibco) containing 10% fetal bovine serum by standard techniques.
Construction of the mxiE mutant strain and promoter-gfp fusions.
Plasmid construction, DNA analysis, and electroporation were performed according to the manufacturer's instructions or previously described standard protocols (46). Oligonucleotide synthesis was performed by using Applied Biosystems automated solid-phase synthesis with standard chemistry at the Uniformed Services University of the Health Sciences (USUHS) Biomedical Instrumentation Center.
The mxiE-bearing plasmid was constructed by ligating a 3.5-kb EcoRV fragment of pHS4011 (containing mxiD-J from pWR100) into pBluescript SK(+). The nonpolar mxiE deletion and insertion mutant was constructed by removing the 206-bp ClaI-HindIII fragment of mxiE, blunting with Klenow, and ligating an 891-bp EcoRI-HindIII aphA-3 fragment, which also had been blunted with Klenow, into the site to yield pECK4. pECK4 was linearized with NotI and XhoI and used to transform BS600 to form BS610. mxiE::aphA-3 was transduced into 2457T by using P1L4 grown on BS610 to yield BS611.
The promoterless green fluorescent protein (GFP) expression vector, pEBD166, was constructed as follows. The gfpmut2 allele, a mutant of GFP optimized for fluorescence in flow cytometry applications (10), was amplified from pRRS10. The upstream primer was designed to contain restriction sites for KpnI and BglII, followed by a strong ribosome-binding site (AGGAGA), located 7 bp upstream of the GFP start codon. The downstream primer contained a HindIII restriction site. After restriction digestion and agarose gel purification, the GFP-encoding DNA fragment was ligated to KpnI-HindIII-digested pBAD18, forming pEBD166.
Oligonucleotide primer pairs for the promoter sequences of the virA, ipaH9.8, ospB, ospC1, ospD2, ospE2, ospF, ospG, and ipgD genes were designed based on the S. flexneri virulence plasmid sequence and encompassed ca. 500 bp upstream of the start codon of the gene (7). Restriction sites for KpnI and BglII were incorporated into the upstream and downstream primers, respectively, for cloning into pEBD166. Promoter regions were amplified from 2457T at 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min by using 25 cycles. Each PCR product was digested with KpnI and BglII and ligated into pEBD166 that had been similarly digested. Plasmids were electrotransformed into S. flexneri 2457T and BS611.
Virulence assays.
Invasion assays were performed in L2 mouse fibroblasts by using the gentamicin protection modification as previously described (21). Briefly, bacteria at an optical density at 600 nm of 0.72 were washed in DMEM and spun onto subconfluent L2 cell fibroblast monolayers in 35-mm dishes at 6,000 rpm for 10 min. After incubation at 37°C for 30 min, cells were washed with warm phosphate-buffered saline (PBS) three times, and the medium was replaced with warm DMEM containing 50 μg of gentamicin ml−1. After another 30 min incubation at 37°C, the cells were rinsed and treated as described above. Cells were incubated for an additional hour at 37°C and then rinsed three times with warm PBS and treated as described below. To evaluate the formation of protrusions in the eukaryotic cell membrane caused by intracellular movement of the bacteria, infected cells were fixed with a solution of 0.2% glutaraldehyde-2% formaldehyde in PBS for 5 min at 4°C before staining them with Giemsa (47). Plaque assays were performed with confluent L2 fibroblast monolayers as previously described (40). Detection of Ipa synthesis and secretion was as previously described (50). Protein electrophoresis was performed in sodium dodecyl sulfate-10% polyacrylamide gels. Separated proteins were transferred to polyvinylidene difluoride membranes (Schleicher & Schuell, Inc.) and treated with a blocking agent consisting of 5% powdered milk-3% gelatin in Tris-buffered saline. Immunodetection was performed with an anti-IpaB-anti-IpaC-anti-IpaD cocktail and alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G secondary antibody. Blots were visualized by using the chemiluminescent substrate, CDP-Star (Boehringer Mannheim) according to the manufacturer's instructions. Virulence in vivo was assessed with the Serény test as previously described (52).
Flow cytometry.
Flow cytometry experiments were performed on an EPICS XL-MCL flow cytometer (Coulter, Miami, Fla.). Regions were set to analyze 10,000 fluorescent, bacterium-sized particles for each sample. The region comprising the nonfluorescent population was set by using the wild-type strain containing the promoterless gfp plasmid alone (pEBD166) such that this peak is entirely contained within the first decade (mean fluorescence intensity [MFI] = 0.2 to 0.3). Therefore, nonfluorescent populations are represented by MFIs of ≤0.3. All fluorescence intensity values of >1 represent fluorescent bacteria, and populations with MFIs between 0.3 and 1 are weakly fluorescent. Data were analyzed and compiled by using WinList software (Verity Software House, Topsham, Maine). An induction ratio (IR) was calculated by dividing the MFI of the test bacteria (i.e., bacteria grown in liquid culture at 37°C or grown intracellularly at 37°C) by the MFI of bacteria grown in liquid culture at 30°C as described by Lee et al. (32). Growth at 30°C in liquid culture was chosen as the baseline value since virulence genes are known to be repressed at this temperature (35). Based on control experiments, an IR of ≤1.2 reflects a lack of gfp induction under those growth conditions.
Overnight cultures were diluted 1:100 into DMEM containing ampicillin and incubated at either 30 or 37°C with aeration for 2 h. Aliquots were washed with cold PBS and held on ice until analysis. Cells were diluted to allow no more than 1,000 events per s during flow cytometry analysis. To assess promoter activity from bacteria in the intracellular environment, strains were grown to exponential phase in tryptic soy broth containing ampicillin at 37°C and used to infect L2 cells in the gentamicin protection assay as described above with the following modification. After the final incubation, intracellular bacteria were released by the addition of 1 ml of 0.2% Triton X-100 in PBS to each dish and incubation at room temperature for 2 min. Cells were removed by pipetting up and down, and the lysates were held on ice until analysis. Undiluted cell lysates were analyzed by flow cytometry. Samples were run in triplicate, and experiments were repeated at least twice.
RESULTS
Characterization of the virulence phenotype of a mxiE mutant.
We constructed a nonpolar mxiE deletion-insertion mutant such that sections of both the DNA-binding domain and the putative sensor region were removed in order to abolish both activities (Fig. 1A). To this end, the central third of the molecule was excised and replaced with the aphA-3 kanamycin resistance gene cassette (Fig. 1B). The mxiE mutant was able to bind Congo red, an indication that it still has a functional type III secretion system (49). In concordance with this observation, the mxiE mutant was able to synthesize and secrete IpaB, IpaC, and IpaD, as well as the wild-type strain (Table 2 and data not shown). Secretion of IpaB and IpaC is essential both for host cell invasion and for efficient cell-to-cell dissemination (51).
FIG. 1.
(A) Predicted domains of MxiE. The AraC domain (residues 99 to 198) is highlighted in gray, and the helix-turn-helix motifs within this region are indicated by black boxes. The striped boxes denote the putative sensor domain. The region between residues 69 to 138 was deleted in construction of the mxiE mutant. (B) Genetic organization of the ipg-mxi-spa loci adjacent to mxiE. The EcoRV sites denote the fragment that was cloned to form pRRS11. The 206-bp ClaI-HindIII internal fragment of mxiE was replaced with aphA-3 to form strain BS611 as described in Materials and Methods.
TABLE 2.
Virulence properties of a mxiE mutant strain
| Strain | Description | % Invasiona | % Plaquing efficiencyb | Plaque diam (mm)c | Serény test resultd | Ipa synthesis and secretione |
|---|---|---|---|---|---|---|
| 2457T | Wild type | 0.39 | 0.47 | 1.33 | + | + |
| BS611 | mxiE::aphA-3 | 0.41 | 0.45 | 0.87 | − | + |
| BS613 | mxiE/PlacmxiE+ | 0.59 | 0.68 | 0.84 | + | ND |
| SCK9 | wt/PlacmxiE+ | 0.50 | 0.71 | 1.29 | NDf | ND |
Calculated by dividing the number of bacteria recovered from gentamicin-treated cells, assayed in triplicate, by the number of bacteria present in the inoculum and then multiplying the result by 100.
Calculated by dividing the number of plaques formed on confluent L2 monolayers at 72 h, assayed in triplicate, by the number of bacteria present in the inoculum and then multiplying the result by 100.
Calculated by measuring the diameter of the zone cleared on L2 monolayers. Each value represents the average of 10 plaques measured from each dish, assayed in triplicate in three separate experiments.
Keratoconjunctivitis in the guinea pig eye (52).
Detection of IpaB, IpaC, and IpaD in culture supernatants or cell-associated pellets by immunoblotting after induction of secretion using Congo red as described in Materials and Methods.
ND, not determined.
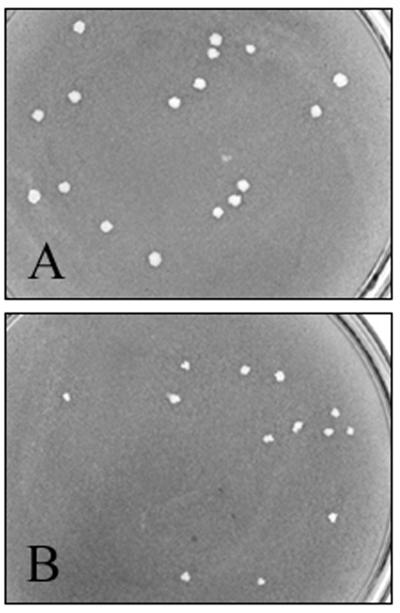
Upon initial characterization of the mxiE mutant, we noticed that this strain produced smaller plaques than the wild-type strain (Table 2). Moreover, the plaque morphology of the mutant strain was irregular compared to the wild-type plaques (Fig. 2). Although the small plaque size indicated that the mxiE mutant was somewhat compromised in its ability to spread from cell to cell, it was able to invade cultured cells and form protrusions just as efficiently as wild-type Shigella (Table 2). The defect in the mxiE mutant that leads to the reduced plaque size phenotype was also evident in the Serény test, which measures a strain's ability to invade eukaryotic cells and spread from cell to cell within an animal host (52). The mxiE mutant was completely avirulent in the Serény test (Table 2). Pathology as severe as that seen with wild-type Shigella could be restored by complementation of the mutant with full-length mxiE driven from a constitutive promoter. Clearly, in assays that measure postinvasion events, the mxiE mutant strain was partially (plaque formation) or fully (Serény test) attenuated. These results led us to examine the influence of MxiE on genes that may be needed postinvasion for a productive disseminated infection in the host.
FIG. 2.
Effect of the mxiE mutation on plaque formation in L2 monolayers. Plaques were visualized by staining with neutral red 72 h postinfection. (A) 2457T, wild-type parent; (B) BS611 (mxiE::aphA-3). Magnification, ×2.3.
Analysis of virA and ipaH9.8 expression.
Given that MxiE shows homology with members of the AraC/XylS family of transcriptional regulators and that mxiE is found within the region encoding the type III secretion system, we strongly suspected that MxiE would play a role in the regulatory cascade of virulence factors. Moreover, MxiE is quite homologous to InvF of Salmonella, which is required for expression of genes that are secreted through the type III secretion machinery (12). Since the results above demonstrated that MxiE is essential for Shigella virulence and appears to be involved in postinvasion events, we examined the influence of MxiE on the expression of genes that may be involved in postinvasion processes.
VirA and IpaH9.8 are two of at least 15 proteins that are secreted through the Shigella type III secretion system (16, 42, 57). Demers et al. suggested that transcription of both virA and ipaH9.8 is regulated by the type III secretion machinery, although the transcriptional regulator was not identified (16). To test whether MxiE is the type III system component responsible for virA activation, a transcriptional fusion of the virA promoter to gfp, the gene encoding GFP, was constructed, and expression of this fusion was examined in both the mxiE mutant and wild-type strains. The GFP reporter system is at least as sensitive as the β-galactosidase system (33) and has the advantage of allowing us to quantify the amount of GFP expression from individual bacteria by using a flow cytometer. GFP expression from bacteria grown in liquid culture (extracellular) at 30 or 37°C was compared to that of bacteria that were allowed to invade cultured cells and replicate intracellularly. Gene expression was examined after bacterial invasion of eukaryotic cells since transcription of virA and ipaH9.8 was reported to be transiently activated after entry into host cells (16). An IR was calculated by dividing the MFI of bacteria grown at 37°C, either in liquid culture or intracellularly, by the MFI of bacteria grown at 30°C in liquid culture as described in Materials and Methods.
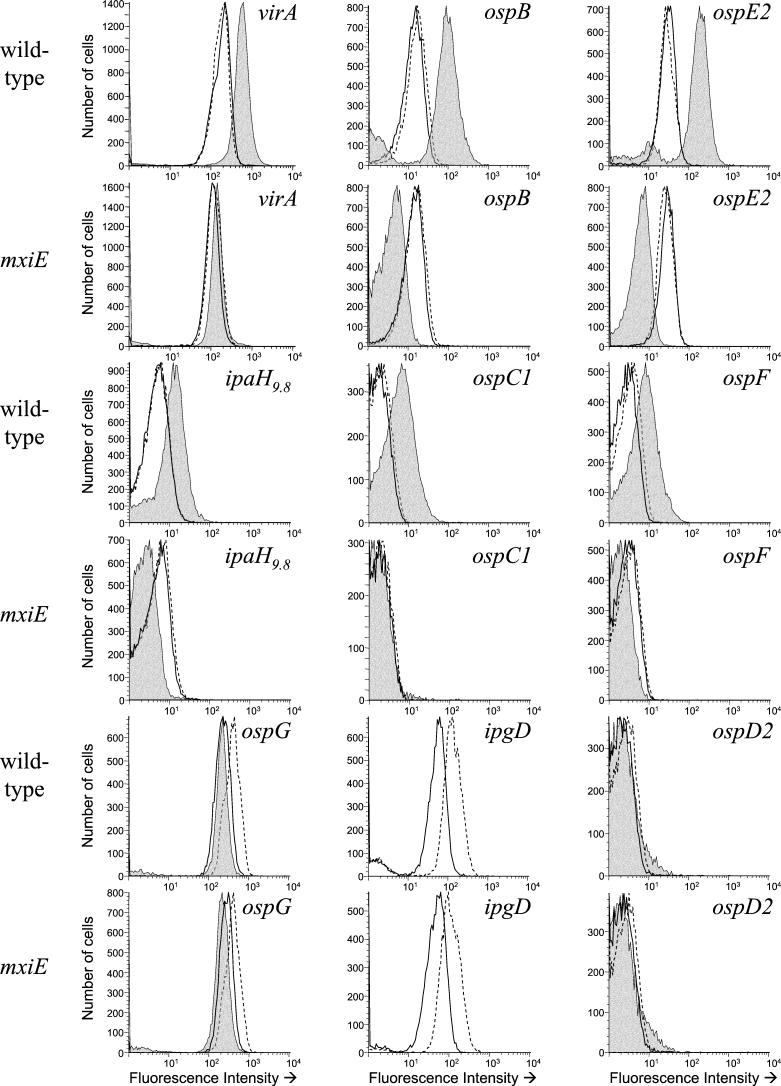
The level of GFP expression from the virA promoter in both the wild-type strain and the mxiE mutant was the same whether the organisms were grown at 30 or 37°C in liquid culture (Table 3 and Fig. 3). These results indicate that virA expression is not temperature dependent since no induction was seen at 37°C in liquid culture compared to 30°C, where virulence genes are repressed (35). Moreover, gene expression was not strictly secretion dependent because activation of type III secretion using Congo red, a dye known to induce type III secretion in the absence of host cell contact (3), also did not influence expression of virA (data not shown). In contrast, when bacteria were recovered from the intracellular environment and analyzed, GFP expression from the virA promoter was found to be induced in the wild-type strain but remained near baseline levels in the mxiE mutant (IRs of 2.9 and 1.3, respectively [Table 3]). Therefore, virA was only induced in the intracellular environment and required functional MxiE for full induction.
TABLE 3.
MFIs of promoter-GFP fusion constructs under differing growth conditionsa
| Promoter | Presence of MxiE | Avg MFI ± SDb at:
|
Intracellular IRc | Fold inductiond | ||
|---|---|---|---|---|---|---|
| 30°C (liquid) | 37°C (liquid) | 37°C (intracellular) | ||||
| virA | + | 20.00 ± 0.17 | 18.83 ± 0.06 | 57.77 ± 3.79 | 2.9 | |
| − | 11.93 ± 0.15 | 13.33 ± 0.06 | 15.40 ± 0.27 | 1.3 | 2.2 | |
| ospB | + | 1.53 ± 0.02 | 1.82 ± 0.006 | 10.70 ± 0.46 | 7.0 | |
| − | 1.52 ± 0.02 | 1.66 ± 0.006 | 0.52 ± 0.01 | 0.3 | 23.3 | |
| ospE2 | + | 3.27 ± 0.03 | 3.09 ± 0.02 | 18.73 ± 0.23 | 5.7 | |
| − | 3.06 ± 0.12 | 2.83 ± 0.006 | 0.73 ± 0.001 | 0.2 | 28.5 | |
| ipaH9.8 | + | 0.60 ± 0.004 | 0.61 ± 0.02 | 1.61 ± 0.03 | 2.7 | |
| − | 0.61 ± 0.006 | 0.64 ± 0.002 | 0.35 ± 0.004 | 0.6 | 4.5 | |
| ospC1 | + | 0.25 ± 0.003 | 0.25 ± 0.001 | 0.96 ± 0.084 | 3.8 | |
| − | 0.23 ± 0.001 | 0.24 ± 0.002 | 0.23 ± 0.001 | 1.0 | 3.8 | |
| ospF | + | 0.34 ± 0.00 | 0.40 ± 0.003 | 1.04 ± 0.014 | 3.1 | |
| − | 0.35 ± 0.003 | 0.38 ± 0.004 | 0.26 ± 0.001 | 0.7 | 4.4 | |
| ospG | + | 24.87 ± 0.06 | 42.27 ± 0.25 | 22.37 ± 0.15 | 0.9 | |
| − | 28.93 ± 0.15 | 41.27 ± 0.35 | 22.67 ± 0.50 | 0.8 | 1.1 | |
| ipgD | + | 5.61 ± 0.09 | 13.63 ± 0.06 | 20.73 ± 7.28 | 3.7 | |
| − | 5.14 ± 0.03 | 12.10 ± 0.10 | 24.43 ± 4.86 | 4.8 | 0.8 | |
| ospD2 | + | 0.28 ± 0.001 | 0.33 ± 0.005 | 0.26 ± 0.003 | 0.9 | |
| − | 0.30 ± 0.007 | 0.34 ± 0.003 | 0.26 ± 0.005 | 0.9 | 1.0 | |
| Vector alonee | + | 0.24 ± 0.002 | 0.25 ± 0.001 | 0.23 ± 0.001 | 1.0 | |
| − | 0.24 ± 0.01 | 0.23 ± 0.006 | 0.22 ± 0.005 | 0.9 | 1.1 | |
A graphical depiction of a representative of each sample is shown in Fig. 3.
The MFI was measured by using a flow cytometer and is based on a logarithmic scale. Values represent the means ± the standard deviations of triplicate determinations from one experiment. Experiments were repeated at least twice for each promoter, and trends were the same in each experiment.
The IR was calculated by dividing the MFI of bacteria grown intracellularly by the MFI of bacteria grown in liquid at 30°C, as described in Materials and Methods. An IR between 0.8 and 1.2 indicates no difference in gfp induction compared to background (30°C [liquid]).
Fold induction indicates the level of gfp induced in the wild type compared to that in the mxiE mutant. The fold induction was calculated by dividing the IR of the wild-type strain by the IR of the mxiE mutant strain.
Nonfluorescent bacteria are represented by an MFI of ≤0.3.
FIG. 3.
Flow cytometry analysis of S. flexneri 2457T (wild-type) and the mxiE mutant (mxiE::aphA-3) containing promoter-gfp fusions. Each promoter examined is indicated above the histogram. The histograms denote GFP expression from extracellular bacteria grown in liquid culture at 30°C (solid black lines), in liquid culture at 37°C (dotted lines), and intracellular bacteria (gray shaded). Histograms depict a representative of each sample. The average MFIs for each sample set are given in Table 3.
Bacteria containing the ipaH9.8 promoter-gfp fusion were very weakly fluorescent when grown extracellularly at either 30 or 37°C (Table 3). There was no difference in reporter expression between the wild-type and mxiE mutant strains when they were grown in liquid culture at either temperature. When the bacteria were allowed to invade eukaryotic cells, gfp was induced from the ipaH9.8 promoter in the wild-type strain (IR of 2.7) (Fig. 3 and Table 3). In contrast, GFP levels were reduced in the mxiE mutant under these conditions (IR of 0.6) and, in fact, the MFI of the mxiE mutant bacteria recovered from the cytoplasmic environment was even lower than that of the bacteria grown in liquid culture. These results demonstrate that induction of gfp from the ipaH9.8 promoter was dependent upon the presence of MxiE; the fluorescence intensity decreased nearly fivefold in its absence. Furthermore, as seen with virA, no induction from the ipaH9.8 promoter could be detected after growth in the presence of Congo red (data not shown).
Analysis of osp expression.
In addition to virA and ipaH9.8, we chose to examine expression of a recently described set of virulence plasmid-encoded proteins of unknown function, termed Osps (outer Shigella proteins). The Osps were identified through N-terminal sequencing of proteins collected from the culture supernatant of a type III secretion constitutive mutant (7). Six unrelated Osp groups were identified (B to G), some of which have multiple homologous members possibly due to gene duplication. We tested the ability of MxiE to induce expression from the promoters of the osp genes under differing growth conditions by using promoter-gfp fusions as described above.
Four distinct gfp expression profiles could be identified from the flow cytometry results by using osp promoter fusions. In the first expression group (group 1), which includes ospB and ospE2, the organisms displayed a moderate, but equivalent, level of fluorescence when grown in liquid culture (extracellularly) at either 30 or 37°C (Table 3 and Fig. 3). In contrast, the MFI increased by a factor of 6 to 7 when the bacteria were recovered from the intracellular environment compared to growth in liquid culture. Full, intracellular induction of gfp required MxiE. In the absence of MxiE, the MFI of bacteria grown inside eukaryotic cells was reduced compared to growth in liquid culture. In contrast, GFP expression from bacteria grown in liquid culture was not affected by the presence or absence of MxiE. The expression of genes in group 1 was not temperature dependent since no induction was seen at 37°C in liquid culture compared to 30°C, nor was gene expression dependent upon type III secretion because Congo red also did not influence their expression (data not shown). Thus, expression from the group 1 promoters ospB and ospE2 was only upregulated in the intracellular environment and this was dependent on the presence of MxiE (ca. 25-fold induction in the presence of MxiE compared to without MxiE).
The expression profiles of the second group of promoters, which includes ospC1 and ospF, were very similar to that of group 1 (Fig. 3). The main difference between group 2 promoters and those described above is that these promoters were nonfluorescent or only very weakly fluorescent when grown extracellularly but displayed an increase in fluorescence intensity when the bacteria were allowed to invade eukaryotic cells (Table 3). As seen with group 1 promoters, intracellular induction of gfp was dependent on the presence of MxiE. Neither growth at 37°C in liquid culture nor the addition of Congo red to the medium was able to induce expression of the group 2 promoters (Table 3 and data not shown). The expression profile of ipaH9.8 is consistent with those in group 2.
The third promoter group was temperature regulated, i.e., upregulated at 37°C. Both ospG and ipgD fall into this group. The amount of GFP detected from these strains nearly doubled when the organisms were grown at 37°C compared to growth at 30°C (Table 3). This induction was the same whether or not the bacteria contained functional MxiE. Furthermore, in the ospG reporter strain this induction was confined to growth in liquid culture. GFP levels were similar to baseline levels when bacteria were recovered from the intracellular environment (IRs of 0.9 and 0.8 for the wild type and mxiE mutant, respectively). The ipgD promoter was used as an internal control of the flow cytometry system. Expression of IpgD, a type III protein, is well characterized and known to be significantly upregulated at 37°C (1). As expected, gfp expression from the ipgD promoter was temperature dependent, with an IR of 2.4 at 37°C in liquid culture. Moreover, ipgD also was induced in the intracellular environment (a 37°C environment), although the presence of MxiE did not affect this induction (Table 3). Similar to the other promoter groups, the addition of Congo red to the culture medium had no effect on GFP expression in these strains (data not shown).
Finally, group four promoters were unaffected by any of the growth conditions tested. This group includes ospD1 and ospD2. The ospD2 promoter fusion was completely inactive in this system (Fig. 3). Strains containing the ospD2-gfp promoter fusion were practically nonfluorescent under all conditions tested, and the MFIs were nearly identical whether tested in the presence or absence of MxiE (Table 3). Similar results were obtained by using an ospD1-gfp promoter fusion (data not shown).
DISCUSSION
In this study we analyzed the role of MxiE, a member of the AraC/XylS family of transcriptional regulators and part of the Mxi-Spa type III secretion system. A deletion-insertion mutant of mxiE rendered the bacteria avirulent in an animal model (the Serény test). The wild-type phenotype could be restored by complementation with mxiE driven from a constitutive promoter, demonstrating that mxiE is an essential virulence gene for Shigella. The mxiE mutant appeared not to be compromised in invasion events in tissue culture and was still able to spread from cell to cell, albeit less efficiently than did the wild-type bacteria. Ipa proteins, important for entry into host cells and translocation of effectors, were still synthesized and secreted as efficiently in a mxiE mutant as the wild-type strain. Cell-to-cell spread was not affected at least shortly after invasion because the mxiE mutant strain was able to form protrusions. These protrusions presumably would be functional in invading neighboring cells since this mutant is also able to form plaques.
The virulence defect of the mutant and the homology to AraC suggested that the role of MxiE might be to regulate expression of factors involved in postinvasion steps of infection. We initially examined the effect of MxiE on the expression of virA and ipaH9.8. Based on the work of Demers et al., both of these genes were shown to require a member of the type III secretion machinery for expression and were expressed transiently inside eukaryotic cells (16). Furthermore, VirA is not required for invasion, and a virA mutant displays a virulence phenotype similar to that seen with the mxiE mutant, namely, the virA mutant is as invasive as the wild-type strain and the production and secretion of Ipa proteins is not affected (16). In addition, the virA mutant forms small plaques and is attenuated in the Serény test. Using a promoterless gfp reporter system to analyze potential MxiE-regulated genes by flow cytometry, we demonstrated that MxiE is required to activate transcription from both the virA and the ipaH9.8 promoters. Notably, these genes were only activated in the intracellular environment and required MxiE for activation. Furthermore, the addition of Congo red to the medium had no effect on expression of these genes. These results are in contrast to a study by Demers et al. (16), who demonstrated the transcription of a virA-lacZ fusion in response to Congo red in the growth medium. This discrepancy may be explained by the amount of Congo red used. In our assays, Congo red was added at a concentration of 35 μg/ml, which is already 5 to 10 times higher than the amount needed to induce type III secretion in culture (3). Demers et al. used a 100-μg/ml concentration of Congo red; this concentration is far greater than is necessary to stimulate secretion. The biological effect of such high concentrations of Congo red on secretion needs to be examined more closely.
The lack of MxiE activity during growth in liquid culture is not due to a lack of transcription of mxi-spa. mxiE is found within a large operon that encodes structural components of the type III secretion system (Fig. 1B). Since gene products found both up- and downstream of mxiE are required for host cell invasion and the mxiE mutant is still able to invade host cells as well as the wild-type strain, this demonstrates that the mxi-spa operon is transcribed in the mutant strain. Furthermore, transcription mapping experiments identified mxiE as part of a larger mxi transcript (K. A. Lampel, unpublished data). Finally, although there is an intergenic space of 130 bp between mxiE and the upstream gene, mxiM, no promoter activity could be demonstrated in this region (data not shown). These results imply that transcription of mxiE is under the same control as the rest of the mxi operon and therefore presumably regulated by VirB. Since mxi and spa genes are transcribed in response to growth in a 37°C environment, there must be either some sort of posttranscriptional or posttranslational regulation of MxiE activity until it is needed or else there is a cofactor for MxiE that is available only when the bacteria reach the intracellular environment.
In addition to virA and ipaH9.8, we examined the effect of MxiE on the expression of several Osps. These proteins were identified in the culture supernatant of a constitutive secretion mutant (7). The original grouping of the osp gene families was based solely on amino acid homologies, and the different families are not related to each other. Based on results presented here, the osp family groupings may need to be reconsidered. One representative of each of the six osp gene families was tested and four genes (ospB, ospC1, ospE2, and ospF) were found to be activated by MxiE. As we saw with virA and ipaH9.8, all of the MxiE-regulated osp genes were activated only within the intracellular environment and not in liquid culture. Adding Congo red to the liquid medium also had no effect. Two of the osp genes, ospB and ospE2, were very highly activated by MxiE in the intracellular environment, demonstrating an approximately 25-fold increase in fluorescence compared to the mxiE mutant. gfp induction in these strains was greater than that seen for the remaining MxiE-regulated targets, ospC1, ospF, virA, and ipaH9.8, which displayed a two- to fivefold increase in fluorescence compared to the mxiE mutant.
The two other osp genes tested, ospD2 and ospG, were not regulated by MxiE. The ospG reporter was activated by growth at 37°C in liquid culture but not by the intracellular environment. The ospD2 reporter was not activated under any conditions tested. The fact that no ospD2 expression was observed in this study is not surprising. The three members of the OspD family show various degrees of homology to ShET-2, a Shigella enterotoxin. Enterotoxins induce fluid secretion while the organism is in the intestinal lumen and would not be expected to be needed by the pathogen once inside the intracellular environment. Perhaps other environmental cues (e.g., iron starvation) are required to initiate transcription of the ospD genes.
The remarkable finding of the present study is the absolute dependence upon a cytosolic cue for the expression of MxiE-regulated genes. Expression of bacterial virulence factors is tightly regulated during infection of a host to ensure that these proteins are only expressed when and where they are needed. Expression of MxiE-activated genes during initial entry steps may hinder productive infections, whereas expression within the proper environment (inside the host cell) may be essential for later steps of the infection. With this in mind, the MxiE-regulated putative effectors would most likely target eukaryotic proteins or processes. The role of the MxiE-regulated Osps in postinvasion events related to virulence is currently under investigation.
Recently, Toyotome et al. demonstrated that secretion of IpaH9.8 occurred later than that of IpaB, IpaC, and IpaD (56). IpaB, IpaC, and IpaD could be detected 30 min after Congo red induction of the type III secretion system, but IpaH9.8 was not detected until 2 h postinduction. IpaH9.8 is also delayed in its secretion into the cytoplasm but, once secreted, IpaH9.8 is targeted to the host cell nucleus (56). IpaH is homologous to YopM of Yersinia pestis, which is trafficked to the host cell nucleus via a vesicular pathway, although its activity in the nucleus is not yet known (53). We identified ipaH9.8 as a MxiE-regulated gene. The fact that it is delayed in secretion through the type III apparatus is consistent with our hypothesis that MxiE regulates expression of effectors involved in postinvasion events. Moreover, since all of the MxiE-activated genes identified in this study previously have been shown to be secreted by the type III secretion system, MxiE may function as a central regulator of proteins that are secreted through the type III system after invasion of the host cell.
It is intriguing that of the six MxiE-regulated genes reported here, only virA was identified in previous screens for virulence factors. This may be due to the methods used to detect virulence genes. Since MxiE regulation only occurs within the intracellular environment and the mxiE mutant retains the ability to invade and form plaques, a screen for genes involved in later steps of infection may require use of an animal model. Induction of genes whose expression is unique to the intracellular environment has been demonstrated in both Salmonella and Shigella (8, 23, 43). Headley and Payne detected six proteins that were induced in S. flexneri when these organisms were recovered from the cytoplasm of cultured cells, although these six proteins were not identified (23). Furthermore, different protein profiles were observed when the organisms were labeled in tissue culture cells compared to liquid medium. Consequently, it is likely that no single method may be sufficient to detect all of the genes that are induced in the intracellular environment. However, by using a variety of approaches, the full complement of Shigella genes that are activated intracellularly should be able to be elucidated. The results presented here are an initial step toward that goal.
While this study was under review, Mavris et al. published a study demonstrating that MxiE, in conjunction with IpgC, activates transcription of both virA and ipaH9.8 (36). These results agree with our findings, but we have, in addition, identified several other MxiE-regulated targets, namely, ospB, ospC1, ospE2, and ospF. Moreover, we have characterized the virulence properties of a mxiE mutant and shown that mxiE is an essential virulence gene in vivo. Importantly, the work presented here extends the observations of Mavris et al. to demonstrate that MxiE activation of its targets occurs only in the intracellular environment. Several possibilities might explain the fact that we did not see MxiE-dependent activation of target promoters in vitro as described by Mavris et al. These researchers demonstrated MxiE-regulated gene activation in the presence of very high concentrations of Congo red, in a constitutive secretion mutant of Shigella, or by using mxiE on a multicopy plasmid expressed in E. coli. Our system employed a wild-type Shigella strain in a tissue culture model, which more closely mimics the normal environment encountered by the bacteria during infection. Nevertheless, based on work presented here and that of Mavris et al., it seems likely that the eukaryotic cytoplasm provides a constitutive secretion environment that is the intracellular cue required for MxiE activity in vivo.
The fact that MxiE regulates postinvasion events and that MxiE-regulated targets are, for the most part, previously uncharacterized, is intriguing. Other enteric pathogens, such as Salmonella and Yersinia, possess two or more distinct type III secretion systems. These systems are differentially regulated and the effectors are involved at different steps of infection. The SPI1-encoded type III secretion system of Salmonella is required for bacterial entry into host cells, whereas the SPI2-encoded type III secretion system is involved in intracellular survival and replication (11, 24). In Yersinia, the virulence plasmid-encoded type III secretion system (Ysc) is required early in infection for the organism to resist macrophage killing, whereas a newly described type III secretion system (Ysa) found on the Yersinia chromosome is believed to play a role late in infection (22). Shigella has only one known type III secretion system. The role of this system in invasion is well characterized, although its effectors also are required for some postinvasion steps of infection (41, 51). Perhaps shigellae have evolved unique mechanisms of virulence gene regulation to adapt its single type III secretion system to function in both invasion and postinvasion events, whereas other enteric pathogens evolved separate secretion systems to perform these functions. One of these unique regulation mechanisms is through the intracellular induction of proteins secreted through the type III system in response to MxiE activation.
Virulence gene regulation in Shigella is a complex process involving multiple gene products and a variety of environmental signals. Temperature regulation of virulence factors is well characterized in Shigella. Here we have identified an essential virulence gene involved in a novel, additional level of regulation of type III secreted proteins in the intracellular environment. Elucidation of the molecular mechanism of regulation by MxiE should yield interesting insights into the evolution of Shigella as a facultative intracellular pathogen.
Acknowledgments
We thank Ed Oaks for monoclonal IpaB, IpaC, and IpaD antibodies. We also thank Michael N. Flora and especially Karen M. Wolcott of the USUHS Biomedical Instrumentation Center for excellent technical assistance with oligonucleotide synthesis and flow cytometry, respectively.
This work was supported by grant AI24656 from the National Institute of Allergy and Infectious Diseases. C.D.K. is supported by a National Research Service Award from the National Eye Institute (EY07002).
The opinions or assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
REFERENCES
- 1.Allaoui, A., R. Menard, P. J. Sansonetti, and C. Parsot. 1993. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61:1707-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol. Microbiol. 7:59-68. [DOI] [PubMed] [Google Scholar]
- 3.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 5.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bliska, J. B., K. Guan, J. E. Dixon, and S. Falkow. 1991. A mechanism of bacterial pathogenesis: tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. USA 88:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 8.Burns-Keliher, L. L., A. Portteus, and R. Curtiss III. 1997. Specific detection of Salmonella typhimurium proteins synthesized intracellularly. J. Bacteriol. 179:3604-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerc, P., and P. J. Sansonetti. 1987. Entry of Shigella flexneri into HeLa cells: evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect. Immun. 55:2681-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, K. H., L. S. Robinson, and V. L. Miller. 2001. SigE is a chaperone for the Salmonella enterica serovar Typhimurium invasion protein SigD. J. Bacteriol. 183:1452-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day, W. A., Jr., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBord, K. L., V. T. Lee, and O. Schneewind. 2001. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 183:4588-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demers, B., P. J. Sansonetti, and C. Parsot. 1998. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 17:2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 19.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale, T. L., and S. B. Formal. 1981. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 23.Headley, V. L., and S. M. Payne. 1990. Differential protein expression by Shigella flexneri in intracellular and extracellular environments. Proc. Natl. Acad. Sci. USA 87:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 25.Holmstrom, A., J. Olsson, P. Cherepanov, E. Maier, R. Nordfelth, J. Pettersson, R. Benz, H. Wolf-Watz, and A. Forsberg. 2001. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol. Microbiol. 39:620-632. [DOI] [PubMed] [Google Scholar]
- 26.Hong, K. H., and V. L. Miller. 1998. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J. Bacteriol. 180:1793-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaniga, K., J. C. Bossio, and J. E. Galán. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 29.Kaniga, K., S. C. Tucker, D. Trollinger, and J. E. Galán. 1995. Homologues of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaBrec, E. H., H. Schneider, T. J. Magnani, and S. B. Formal. 1964. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J. Bacteriol. 88:1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator VirF and temperature in the expression of the pYV plasmid genes of Yerinia enterocolitica. Mol. Microbiol. 6:379-388. [PubMed] [Google Scholar]
- 32.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lissemore, J. L., J. T. Jankowski, C. B. Thomas, D. P. Mascotti, and P. L. deHaseth. 2000. Green fluorescent protein as a quantitative reporter of relative promoter activity in Escherichia coli. BioTechniques 28:82-89. [DOI] [PubMed] [Google Scholar]
- 34.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavris, M., A.-L. Page, R. Tournebize, B. Demers, P. J. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 37.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menard, R., P. J. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-525. [DOI] [PubMed] [Google Scholar]
- 39.Michiels, T., J. C. Vanooteghem, C. Lambert de Rouvroit, B. China, M. P. Sory, A. Gustin, and G. R. Cornelis. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J. Bacteriol. 173:4994-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page, A.-L., H. Ohayon, P. J. Sansonetti, and C. Parsot. 1999. The secreted IpaB and IpaC invasions and their cytoplasmic chaperone IpgC are required for intercellular dissemination of Shigella flexneri. Cell. Microbiol. 1:183-193. [DOI] [PubMed] [Google Scholar]
- 42.Parsot, C., R. Menard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 43.Pfeifer, C. G., S. L. Marcus, O. Steele-Mortimer, L. A. Knodler, and B. B. Finlay. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect. Immun. 67:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde, J. R., X. S. Luan, H. Rohde, J. M. Fox, and S. A. Minnich. 1999. The Yersinia enterocolitica pVY virulence plasmid contains multiple intrinsic DNA bends which melt at 37°C. J. Bacteriol. 181:4198-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai, T., C. Sasakawa, S. Makino, and M. Yoshikawa. 1986. DNA sequence and product analysis of the virF locus responsible for Congo red binding and cell invasion in Shigella flexneri 2a. Infect. Immun. 54:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sandlin, R. C., M. B. Goldberg, and A. T. Maurelli. 1996. Effect of O side-chain length and composition on the virulence of Shigella flexneri 2a. Mol. Microbiol. 22:63-73. [DOI] [PubMed] [Google Scholar]
- 48.Sandlin, R. C., and A. T. Maurelli. 1999. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect. Immun. 67:350-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuch, R., and A. T. Maurelli. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect. Immun. 65:3686-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuch, R., and A. T. Maurelli. 2001. Spa33, a cell surface-associated subunit of the Mxi-Spa type III secretory pathway of Shigella flexneri, regulates Ipa protein traffic. Infect. Immun. 69:2180-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway in intercellular dissemination of Shigella flexneri. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 52.Serény, B. 1957. Experimental keratoconjunctivitis shigellosa. Acta Microbiol. Acad. Sci. Hung. 4:367-376. [PubMed] [Google Scholar]
- 53.Skrzypek, E., C. Cowan, and S. C. Straley. 1998. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol. Microbiol. 30:1051-1065. [DOI] [PubMed] [Google Scholar]
- 54.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 55.Tobe, T., M. Yoshikawa, and C. Sasakawa. 1995. Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J. Bacteriol. 177:1094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 34:32071-32079. [DOI] [PubMed] [Google Scholar]
- 57.Uchiya, K.-I., T. Tobe, K. Komatsu, T. Suzuki, M. Watarai, I. Fukuda, M. Yoshikawa, and C. Sasakawa. 1995. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol. Microbiol. 17:241-250. [DOI] [PubMed] [Google Scholar]
- 58.Von Pawel-Rammingen, U., M. V. Telepnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]