Abstract
The N-terminal repeat (SAC1) of the S-protein of Lactobacillus acidophilus bound efficiently and specifically to cell wall fragments (CWFs) when fused to green fluorescent protein, whereas the C-terminal repeat (SAC2) did not. Treatment of CWFs with hydrofluoric acid, but not phenol, prevented binding. Apparently, SAC1 is necessary and sufficient for cell wall binding. Our data suggest that SAC anchors the S-protein to a cell wall teichoic acid.
S-layers, two-dimensional paracrystalline arrays usually found as the outermost layer of the cell envelope, are composed of a single (glyco)protein. S-layers self-assemble in an entropy-driven process during which multiple, noncovalent interactions between individual S-protein monomers and with the underlying cell surface take place. In some S-proteins, the two types of interaction can be assigned to two separate domains (11, 17).
While relatively little is known about the domains responsible for the interactions between S-protein monomers, a wealth of information about cell wall binding domains (CWBD) has been collected. The surface layer homology (SLH) domain, found in S-proteins of, e.g., Bacillus, Thermophilus, Thermoanaerobacter, and Clostridium species, consists of ∼55 amino acid residues and can be found in 1 to 3 copies in different proteins (3, 9, 12, 15). Originally thought to be a peptidoglycan (PG)-binding domain, the SLH domain is now known to bind PG-associated cell wall polymers. The SLH domain of Bacillus anthracis binds to a pyruvylated PG-associated cell wall polysaccharide through a mechanism that is conserved in related bacteria (13).
In other bacteria, the mechanism by which S-proteins are anchored to the cell wall may be different. The S-protein of Corynebacterium glutamicum, PS2, possesses a C-terminal hydrophobic anchor of approximately 79 amino acid residues that was suggested to interact with a hydrophobic layer composed of mycolic acids present in the cell wall (5). The CWBD of the Lactobacillus acidophilus ATCC 4356 S-layer protein, SAC, consists of a tandemly repeated ∼65-amino-acid sequence with a conserved tyrosine doublet. The two repeats (the N-terminal repeat SAC1 and the C-terminal repeat SAC2) share 26% identical amino acids, most of which are basic and aromatic. SAC shows homology to carbohydrate binding regions of Clostridium difficile toxins and to extracellular glycosyltransferases and cell wall-associated proteinases of lactic acid bacteria, suggesting the involvement of carbohydrates in the cell wall binding mechanism (17).
Here we report on the interaction with cell wall fragments (CWFs) of the individual repeats and on the identity of the receptor for SAC.
Production and purification of HGFP-SAC, HGFP-SAC1, and HGFP-SAC2.
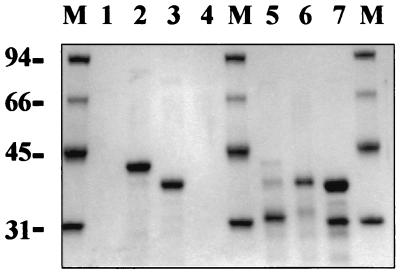
SAC sequences were efficiently produced in Escherichia coli as fusions to the C terminus of His-tagged green fluorescent protein (HGFP). For this purpose, SAC1 and SAC2 sequences were amplified from plasmid pBK1 (1) by PCR using oligonucleotides SP2 (5′ GATCGGATCCCTCGAGATGCACAACGCATACTACTACGA 3′) and SP3 (5′ CCCCAAGCTTTTATTATGCAGCGTTGATGTACTTGTC 3′) for SAC1 and oligonucleotides SP4 (5′ GGGGCTCGAGAACATCGATGGTACTAAGCGTAC 3′) and SP5 (5′ CCCCGGATCCAAGCTTATCGAAGTATCAGAAGATCCTATT 3′)for SAC2. PCR products were cloned in pGEM-T and substituted for SAC in pHGFPSAC (17), yielding pHGFPSAC1 and pHGFPSAC2. Purification of HGFP-SAC1 and HGFP-SAC2 was carried out as described previously for HGFP-SAC (17). The electrophoretic behavior of HGFP-SAC1 and HGFP-SAC2 fully agreed with that of the protein of the predicted size (Fig. 1), suggesting that proteolytic degradation did not occur and that the repeats can fold as separate domains.
FIG. 1.
Interaction of HGFP-SAC (lanes 2 and 5), HGFP-SAC1 (lanes 3 and 6), and HGFP-SAC2 (lanes 4 and 7) with purified native L. acidophilus CWFs. Lane 1, native CWFs only; lanes 2 to 4, CWF-bound material; lanes 5 to 7, remaining nonbound material. Lanes M contain Mr reference proteins. To minimize nonspecific binding of E. coli proteins to purified CWFs, all binding experiments were performed in 50 mM Tris-HCl (pH 7.5)-150 mM NaCl.
Interaction of HGFP-SAC, HGFP-SAC1, and HGFP-SAC2 with native L. acidophilus CWFs.
To determine the interaction of the SAC repeats with the cell wall, native CWFs were prepared by sonification of L. acidophilus cells, collected by centrifugation (20 min, 20,000 × g, 4°C), resuspended in 1% sodium dodecyl sulfate, and incubated at 100°C for 30 min. This process was repeated three times, followed by incubation with 25 μg of RNase A/ml and 25 μg of DNase I/ml for 30 min at 37°C. Finally, CWFs were washed five times with deionized water and lyophilized. Figure 1 shows that HGFP-SAC and HGFP-SAC1 bound quite well to L. acidophilus CWFs whereas HGFP-SAC2 did not, implying that SAC1 is necessary and sufficient for binding in vitro. An explanation for the absence of SAC2 binding is that it is not properly folded. SAC2 contains 12 lysine and 5 glycine residues, compared to 7 and 3 for SAC1, and the absence of a central proline residue in SAC2 results in a more hydrophilic and possibly less rigid structure for SAC2. Structural rigidity may be needed for proper receptor binding, and its absence suggests that SAC2 could have a different role in cell wall binding, e.g., strengthening of electrostatic interactions with the cell wall.
At NaCl concentrations greater than 150 mM, binding of HGFP-SAC1 was severely reduced, while NaCl concentrations greater than 250 mM were needed to reduce the binding of HGFP-SAC (data not shown). These results indicate that binding of SAC with native CWFs through electrostatic interactions is stronger than that of SAC1, supporting a cooperative role for SAC2. None of the peptides bound to purified Lactobacillus casei CWFs (data not shown), as was previously observed for HGFP-SAC (17).
Binding of HGFP-SAC and HGFP-SAC1 to HF- and phenol-extracted L. acidophilus CWFs.
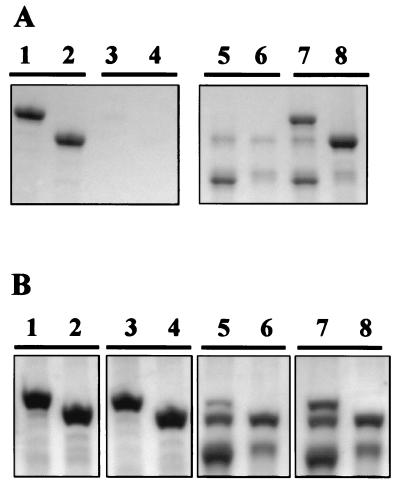
To investigate the cell wall receptor for SAC, chemical extraction experiments of L. acidophilus CWFs were performed. L. acidophilus CWFs were subjected to hydrofluoric acid (HF) extraction (40% [wt/vol] for 96 h at 4°C), which dissociates PG and PG-associated components (8). HF treatment of CWFs, which resulted in a loss in mass of ∼16%, completely interfered with the binding of HGFP-SAC and HGFP-SAC1 (Fig. 2A), indicating that a PG-associated polymer, and not PG itself, is responsible for binding of SAC and SAC1.
FIG. 2.
Interaction of HGFP-SAC (lanes 1, 3, 5, and 7) and HGFP-SAC1 (lanes 2, 4, 6, and 8) with native HF-extracted CWFs (A) and phenol-extracted CWFs (B) from L. acidophilus. Lanes 1 to 4, CWF-bound material; lanes 5 to 8, nonbound material.
Almost no HF-extracted material could be recovered as ethanol-precipitable material, indicating that the PG-associated polymer was degraded. This suggests that acid-labile compounds such as lipoteichoic acid (LTA) and teichoic acid (TA), rather than polysaccharide and teichuronic acid, which are all removed from PG by HF treatment, are involved in the binding process, although an acid-labile modification of PG cannot be excluded (7, 16). Phenol treatment (80%, 65°C, 1 h) of CWFs, which extracts LTA but not TA from the cell wall of gram-positive bacteria (10), did not affect binding of HGFP-SAC and HGFP-SAC1 (Fig. 2B). This result suggests that PG-associated TA rather than LTA is the cell wall receptor for SAC.
Chemical composition of native and HF-extracted L. acidophilus CWFs and HF-extracted material.
Determination of the phosphorus content of native CWFs showed that they contained 1.12 μmol of phosphorus per mg (dry weight), while in HF-extracted CWFs, only 0.01 μmol/mg was detected (6). This result is in agreement with the conclusion that teichoic acids (TA and/or LTA) present in native CWFs are removed by HF treatment. The phosphorus was recovered in the nonprecipitable fraction of the HF extract, confirming that the extracted polymer was degraded. Only one-third of the phosphorus could be extracted from CWFs with hot phenol (0.71 versus 1.12 μmol/mg), in agreement with the conclusion that phenol extracts LTA but not TA (10). Since phenol treatment had no effect on binding, we suggest that TA is the receptor for SAC.
Repeats in other CWBD.
Although the SAC repeats and SLH have similar sizes, they do not show amino acid sequence similarity. In addition, secondary-structure determinations predict a beta-stranded structure for SAC1 and SAC2 but a helix-loop-helix structure for SLH. Interestingly, SLH domain-containing proteins showed strongly diminished cell wall binding properties and increased proteolytic sensitivity when 1 or 2 copies of the SLH domain were C-terminally deleted, suggesting that a structure comprising three SLH repeats represents the functional domain (2, 14). Multiple repeats are also required for binding of the streptococcal proteins PspA and LytA, while for the lactococcal autolysin, AcmA, one repeat suffices (4, 18). Our studies show that SAC2 can be deleted without compromising cell wall binding capacity or proteolytic resistance. This indicates that SAC1 is both a structural and a functional unit. SAC2 might provide strength to the cell wall interaction of SAC without possessing a direct binding capacity itself.
REFERENCES
- 1.Boot, H. J., C. P. A. M. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brechtel, E., and H. Bahl. 1999. In Thermoanaerobacterium thermosulfurigenes EM1 S-layer homology domains do not attach to peptidoglycan. J. Bacteriol. 181:5017-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brechtel, E., M. Matuschek, A. Hellberg, E. M. Egelseer, R. Schmid, and H. Bahl. 1999. Cell wall of Thermoanaerobacterium thermosulfurigenes EM1: isolation of its components and attachment of the xylanase XynA. Arch. Microbiol. 171:159-165. [DOI] [PubMed] [Google Scholar]
- 4.Buist, G. 1997. Ph.D. thesis. Groningen University, Groningen, The Netherlands.
- 5.Chami, M., N. Bayan, J. L. Peyret, T. Gulikkrzywicki, G. Leblon, and E. Shechter. 1997. The S-layer protein of Corynebacterium glutamicum is anchored to the cell wall by its C-terminal hydrophobic domain. Mol. Microbiol. 23:483-492. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P. S., T. Y. Toribara, and H. Warner. 1956. Microdetermination of phosphorus. Anal. Chem. 28:1756-1758. [Google Scholar]
- 7.Egelseer, E. M., K. Leitner, M. Jarosch, C. Hotzy, S. Zayni, U. B. Sleytr, and M. Sára. 1998. The S-layer proteins of two Bacillus stearothermophilus wild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical chemical composition. J. Bacteriol. 180:1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekwunife, F. S., J. Singh, J. K. G. Taylor, and R. J. Doyle. 1991. Isolation and purification of a cell wall polysaccharide of Bacillus anthracis (Sterne). FEMS Microbiol. Lett. 82:257-262. [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, W., H. U. Koch, and R. Haas. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523-530. [DOI] [PubMed] [Google Scholar]
- 11.Jarosch, M., E. M. Egelseer, D. Mattanovich, U. B. Sleytr, and M. Sára. 2000. S-layer gene sbsC of Bacillus stearothermophilus ATCC 12980: molecular characterization and heterologous expression in Escherichia coli. Microbiology 146:273-281. [DOI] [PubMed] [Google Scholar]
- 12.Matuschek, M., K. Sahm, A. Zibat, and H. Bahl. 1996. Characterization of genes from Thermoanaerobacterium thermosulfurigenes EM1 that encode two glycosyl hydrolases with conserved S-layer-like domains. Mol. Gen. Genet. 252:493-496. [DOI] [PubMed] [Google Scholar]
- 13.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are noncovalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 15.Mesnage, S., E. Tosicouture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 16.Ries, W., C. Hotzy, I. Schocher, U. B. Sleytr, and M. Sára. 1997. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J. Bacteriol. 179:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterization of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 18.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]