Abstract
To identify putative members of a regulon controlled by the H. pylori sensory histidine kinase HP0164 (HK0164), we constructed HK0164 null mutant H. pylori strains and analyzed bacterial gene transcription using DNA arrays. Seven genes were differentially expressed in multiple HK0164 mutant strains compared to their expression in control strains. Strain-dependent variations in differential expression were also detected. These results indicate that the signal transduction circuit utilizing HK0164 controls the transcription of at least seven genes in H. pylori.
An important mechanism by which bacteria detect environmental changes and alter gene expression appropriately is via the use of two-component signal transduction systems (reviewed in references 15 and 18). The sensor histidine kinase of a two-component system monitors environmental changes by means of a domain exposed to the periplasm and, upon detecting an appropriate environmental stimulus, undergoes autophosphorylation of a well-conserved histidine residue in the transmitter domain located on the cytoplasmic side of the membrane. The phosphoryl group is then transferred to a conserved aspartate residue in the receiver domain of the cognate response regulator. Phosphorylation of the response regulator modulates its DNA-binding capability and hence alters expression of specific target genes (18, 19).
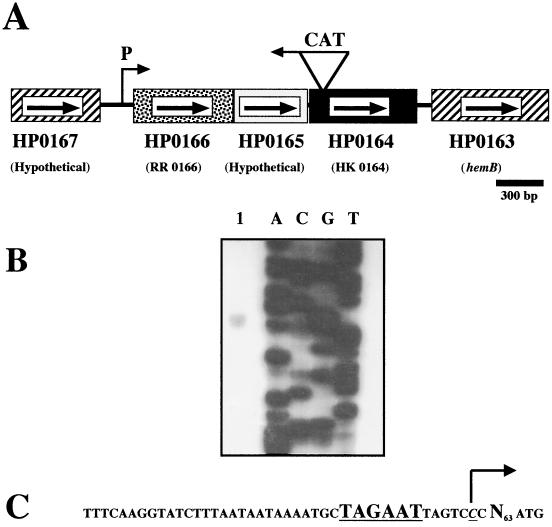
Helicobacter pylori is a gram-negative microaerophilic bacterium that is recognized as a cause of chronic superficial gastritis, peptic ulcer disease, and gastric malignancies in humans (3, 4, 11, 16). The H. pylori genome is predicted to encode four histidine kinases and six response regulator proteins (20). To begin our studies of signal transduction in H. pylori, we selected for analysis the phosphotransfer system encoded by the HP0166, HP0165, and HP0164 genes in the genome of H. pylori strain 26695 (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=ghp) (20). These three genes are found in the same orientation and are predicted to be cotranscribed as an operon (Fig. 1A). Mapping of the transcriptional start point by primer extension analysis allowed the deduction of the HP0166 promoter element in H. pylori strain 26695 (Fig. 1B). The −10 sequence (Fig. 1C) is very consistent with the extended −10 hexamer demonstrated for a subset of H. pylori σ80-dependent promoters (7). As in most other H. pylori promoters (7, 21), there is no consensus between the −35 region of RR0166 and published promoter elements.
FIG. 1.
Organization of the Helicobacter pylori RR0166/HK0164 response regulator/histidine kinase locus and mapping of the RR0166 transcriptional start point. (A) Configuration of the RR0166/HK0164 locus in the genome of H. pylori 26695. The location of the experimentally determined promoter (P) is indicated. The location of the CAT cassette insertion into the histidine kinase gene for HK0164 is also shown. Arrows indicate the direction of transcription of each gene. (B) Primer extension was accomplished with a primer specific for RR0166 (5′ AACTCGCTCAAAAACTCGGC) (lane 1). A sequencing ladder used to determine the location of the transcriptional start point is shown to the right. (C) The sequence of the deduced RR0166 promoter region is shown. The transcriptional start point is underlined and italicized, and the arrow indicates the direction of transcription. The −10 promoter element is underlined, and the number of nucleotides (N) between the transcriptional start point and the translational start codon is indicated.
The HP0166 gene, referred to herein as RR0166, is predicted to encode a response regulatory protein. Previous studies have classified the H. pylori HP0166 gene product as a member of the OmpR family of response regulators (2, 15, 20). Two previous reports concluded that the RR0166 gene is essential in H. pylori (2, 12). In H. pylori strain 26695, open reading frames (ORFs) HP0164 and HP0165 are predicted to encode two separate portions of a histidine kinase (20), whereas in a second sequenced strain of H. pylori, J99, the corresponding histidine kinase sequences are fused into a single ORF (designated JHP0151) (http://scriabin.astrazeneca-boston.com/hpylori/) (1). The possible significance of such strain-specific variations in histidine kinase structure has not yet been determined. In vitro phosphorylation studies utilizing a recombinant histidine kinase, HP0164/HP0165, derived from a third strain (H. pylori G27) have demonstrated that HP0164/HP0165 and HP0166 represent a cognate histidine kinase and response regulator (2).
As a first step in identifying genes regulated by the HK0164/RR0166 phosphorelay system in H. pylori, we constructed an H. pylori mutant strain in which HK0164 was disrupted by insertion of an antibiotic cassette. To do this, we amplified a 2,117-bp segment of the H. pylori strain 26695 chromosome (containing the RR0166 and HP0165 ORFs and the majority of the HK0164 ORF) using primers VU-0167-F (5′ CTTGCAATACCAATTGAGCAC) and VU-0164-R (5′ CAAAACCATGGGCTTGGCTG) and cloned this DNA into pGEM-T to yield a plasmid designated pHK0164. The cloned HK0164 gene then was disrupted by inserting the chloramphenicol acetyltransferase (CAT) gene of Campylobacter coli into a BclI site within the HK0164 ORF. The resulting plasmid (pHK0164::CAT), which is unable to replicate in H. pylori, was introduced by natural transformation (17) into H. pylori strain 26695, and chloramphenicol-resistant colonies were selected. Proper placement of CAT within the chromosomal copy of the HK0164 gene in H. pylori via a double-crossover event was confirmed by PCR (7, 8). The resulting H. pylori HK0164::CAT strain was designated H. pylori 26695/HK0164−. As a control for the presence of the CAT gene, H. pylori strain 26695 was transformed with pCTB2CAT (6, 7), which allowed the placement of the CAT gene in the intergenic region between cysS and vacA. The resultant strain was designated H. pylori 26695 CAT Control.
We then compared gene transcription in H. pylori 26695 CAT Control with transcription in the isogenic HK0164 ORF mutant strain (H. pylori 26695/HK0164−). The use of this control strain eliminated the possibility that any differential transcriptional events observed were due to potential trans-acting properties of the CAT protein. H. pylori strains were grown for 24 h at 37°C in 5% CO2 on blood agar plates and then harvested for RNA isolation. Total RNA (30 μg) from each strain was used to generate 33P-labeled cDNA using 5 μg of random hexamers (Gibco/BRL) in the presence of an RNase inhibitor (Promega); 10 mM dithiothreitol; 0.5 mM (each) dGTP, dCTP, and dTTP; 5 μM dATP; 50 μCi of [33P]dATP; and 0.25 U of reverse transcriptase (Superscript II; Gibco/BRL). cDNA synthesis was accomplished over 2 h at 42°C. Radiolabeled cDNA from each strain then was hybridized with identical H. pylori DNA arrays (http://www.eurogentec.be; Eurogentec, Seraing, Belgium). These DNA arrays on nylon membranes contain features representing >95% of the predicted ORFs in the sequenced H. pylori strain 26695, and nearly all features correspond to entire ORFs. Hybridized arrays were analyzed with a phosphorimager (Fuji Inc.), and quantification of hybridization signals was accomplished with ImageQuant software (Fuji Inc.). Signals from duplicate spots on each array were combined to yield an average signal intensity for each array feature. All 33P cDNA quantifications were normalized by determining percentages of the total amount of 33P bound to the array. In this way, we were able to control for the minor differences between arrays due to variations in the specific activities of individual cDNA labeling reactions and variations in hybridization or washing conditions.
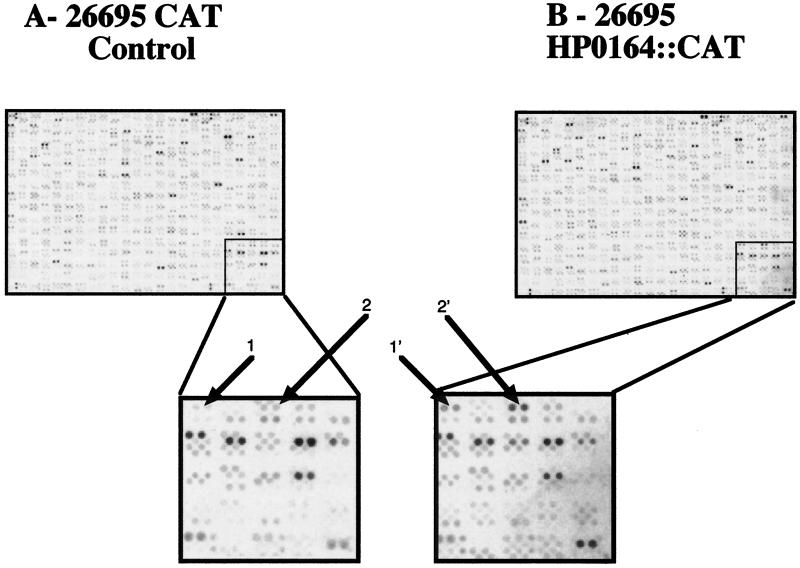
The DNA array analysis was performed three different times, each time with different DNA arrays (Eurogentec) and cDNA preparations from independent cultures of both H. pylori 26695 CAT Control and 26695/HK0164−. A representative array is shown in Fig. 2. Quantification of bound 33P allowed the identification of array features that hybridized differentially between control 26695 cDNA and 26695/HK0164− cDNA. Previous work using vacA::xylE operon fusions has demonstrated that placement of the CAT cassette in the cysS-vacA intergenic region has the effect of moderately decreasing vacA transcription (6). The decreased vacA transcription resulting from placement of the CAT gene in the cysS-vacA intergenic region was indeed detected in the present array analysis (the normalized signal intensity decreased 2.16-fold [±0.6-fold] in H. pylori 26695 CAT Control compared to that in 26695/HK0164−). This finding supports the validity of the DNA array analysis. An independent DNA array analysis using a wild-type H. pylori 26695 strain (without the CAT gene present in the cysS-vacA intergenic region) indicated that there was no significant difference in vacA transcription between the wild-type strain and the HK0164− strain (data not shown).
FIG. 2.
Whole-genome transcription profiles of Helicobacter pylori 26695 CAT Control and an isogenic HK0164 null mutant. Identical DNA arrays were probed with 33P-labeled cDNA from H. pylori 26695 CAT Control (A) and H. pylori 26695/HK0164− (B). Enlargements of the indicated regions of the arrays illustrate the differential levels of transcription of two of the genes identified as members of the HK0164/RR0166 regulon. 1 and 1′, HP0681; 2 and 2′, HP0682. These genes were differentially transcribed in these two H. pylori strains in three independent DNA array experiments. The results shown in this figure are typical of three separate experiments.
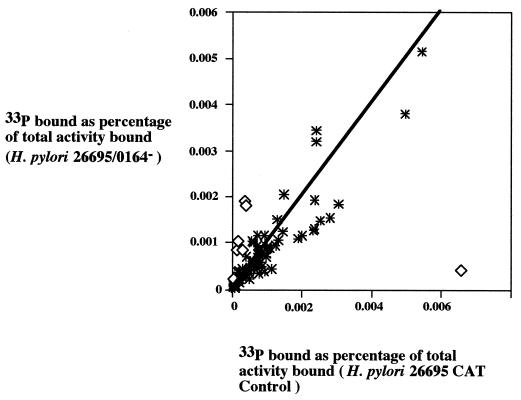
We next analyzed a panel of 81 arbitrarily selected putative housekeeping genes. The mean ratio of normalized signal intensities for array features corresponding to these housekeeping genes was 0.923 ± 0.507 (in H. pylori 26695/HK0164− compared to that in H. pylori 26695 CAT Control). We then endeavored to identify potentially differentially transcribed genes. Differences in levels of gene expression between the H. pylori 26695/HK0164− strain and the H. pylori 26695 CAT Control strain were expressed as induction ratios; i.e., levels in the HK0164− mutant strain were derepressed (positive numbers) or repressed (negative numbers). Genes with mean induction ratios of >2 or <−2 (corresponding to more than 2 standard deviations outside the range for housekeeping genes) were defined as potentially differentially transcribed genes. Based on these criteria, we identified at least six genes whose expression levels were consistently derepressed in H. pylori 26695/HK0164− relative to levels of expression in the control strain and one gene that was consistently repressed (Table 1 and Fig. 3).
TABLE 1.
Gene products regulated via the H. pylori histidine kinase HK0164
| Gene product designation (26695)a | Function | Induction ratiob |
|---|---|---|
| HP0681c | Hypothetical, H. pylori specific | 4.96 ± 2.6 |
| HP0682c | Hypothetical, H. pylori specific | 4.08 ± 1.53 |
| HP0725d | hopP, outer membrane protein, contingency protein | 3.34 ± 1.78 |
| HP1288e | Hypothetical, H. pylori specific | 3.88 ± 2.89 |
| HP1289e | Hypothetical, H. pylori specific | 4.11 ± 1.6 |
| HP1432 | Histidine- and glutamine-rich protein | −15.64 ± 12.31 |
| HP1399 | rocF, arginase | 2.45 ± 0.86 |
Gene product designations are based on the H. pylori 26695 genome nomenclature of Tomb et al. (20).
33P signals from individual array features were calculated as percentages of the total amount of 33P bound to the array (normalized signals). Induction values are ratios of normalized signals in the arrays probed with H. pylori 26695/HK0164− cDNA to normalized signals in arrays probed with control cDNA. Results are means ± standard deviations based on results of three independent microarray experiments.
HP0681 and HP0682 are predicted to be members of the same operon and are members of the paralogous gene families 11 and 12, respectively (20). The only other members of these paralogous families are HP1289 and HP1288.
HP0725 is a contingency gene predicted to be out of frame based on CT dinucleotide repeat deletions near the 5′ end of the gene (20).
HP1288 and HP1289 are predicted to be members of the same operon and are members of the paralogous gene families 11 and 12, respectively (20). The only other members of these paralogous families are HP0682 and HP0681.
FIG. 3.
Correlation between message abundance in H. pylori 26695 CAT Control and the isogenic HK0164 null mutant. Identical arrays were probed with 33P-labeled cDNA from H. pylori 26695 CAT Control and an isogenic HK0164 mutant strain, as shown in Fig. 2. The scatter plot depicts normalized 33P signals for 88 H. pylori genes. Signals obtained with cDNA from the H. pylori CAT Control strain are shown on the horizontal axis (as a percentage of total 33P bound to the array), and signals with cDNA from H. pylori 26695/HK0164− are shown on the vertical axis. The diagonal line represents the ratio of the two signals as 1. Asterisks depict signals for 81 arbitrarily selected housekeeping genes that were transcribed at similar levels in H. pylori 26695 CAT Control and H. pylori 26695/HK0164−. Open diamonds depict signals for genes identified as differentially transcribed in the two strains. The results shown here are representative of results of three independent experiments.
To test the validity of these initial results, we hybridized independent preparations of cDNA from these same two H. pylori strains with H. pylori DNA arrays from a different manufacturer (Sigma-Genosys). In addition, to verify that the differential levels of transcription described above resulted specifically from the HK0164 mutation rather than from an unrecognized mutation elsewhere in the genome, we constructed an independent HK0164 mutant derivative of H. pylori strain 26695 and compared the level of gene transcription in this strain with that in wild-type H. pylori strain 26695 (rather than the 26695 CAT Control strain), using arrays from Sigma-Genosys. The Panorama H. pylori Gene Arrays (Sigma-Genosys) contain 1,681 PCR-amplified ORFs, representing all putative H. pylori strain 26695 protein-coding genes (1,590 genes) and 91 protein-coding genes of H. pylori strain J99 that are unique to this strain (http://www.sigma-genosys.com/epp_microbia_hpyl_generalinfo.asp). In contrast to features on the DNA arrays from Eurogentec, most of the features on arrays from Sigma-Eurogentec were designed to represent a region of the ORF that has as little homology to other ORFs as possible. The results of these array hybridizations were quantified using Array Vision software (Imaging Research, St. Catharines, Ontario, Canada). These experiments confirmed that seven genes identified in the initial analysis (Table 1) were differentially transcribed in 26695/HK0164− mutant strains and control strains. When gene transcription in H. pylori 26695 CAT Control was compared with transcription in the initial 26695/HK0164− mutant, the mean induction ratio of the six depressed genes was significantly higher than the mean induction ratio for all genes on the array (P = 0.014, two-tailed Student's t test with unequal levels of variance) Similarly, when gene transcription in wild-type H. pylori 26695 was compared with transcription in the second 26695/HK0164− mutant strain, again the mean induction ratio of the six derepressed genes was significantly higher than the mean induction ratio for all genes on the array (P = 0.028).
To examine possible strain-specific differences in levels of transcriptional regulation, we also constructed an independent HK0164 mutant derivative of a different H. pylori strain, B128. The genome-wide transcriptional profiles of this mutant strain and the corresponding wild-type control then were analyzed by genome-wide transcriptional profiling, using arrays from Eurogentec. These experiments confirmed that most of the genes derepressed in the original H. pylori 26695/HK0164− mutant (Table 1) were derepressed in the new H. pylori B128/HK0164− mutant strain. In addition, the levels of expression of a pair of paralogous genes (HP1408 and HP0427) and multiple downstream paralogous genes were noted to be strongly derepressed in the H. pylori B128/HK0164− mutant strain relative to levels of expression in the B128 wild-type strain. The levels of transcription of these putative operons in the 26695/HK0164− mutant were not different from those in the 26695 control strain (data not shown). Further work will be required to understand the apparent strain-specific differences in the transcriptional regulation at these loci.
With the sole exception of HP1399, all of the identified derepressed genes are H. pylori specific (i.e., absent from the genomes of other bacteria analyzed thus far) and do not have known functions. HP1399 (rocF) encodes arginase, an enzyme in the H. pylori urea cycle that hydrolyzes l-arginine to l-ornithine and urea (13, 14). H. pylori arginase activity also inhibits production of nitric oxide by macrophages, which may enhance the capacity of the bacteria to evade phagocytic killing (9). HP0725 is predicted to encode an outer membrane protein. However, based on genome sequence data, HP0725 is predicted to be unexpressed in H. pylori strain 26695 due to deletions of CT dinucleotide repeats near the extreme 5′ end of the gene and the associated premature termination of the ORF (20). Expression of this gene is likely regulated by a slip-strand mispairing mechanism. Other genes identified as derepressed in the HK0164 mutant strain of H. pylori are HP0682 and HP0681. These genes are predicted to constitute a bicistronic operon. Interestingly, these genes are members of two different paralogous gene families (20) that each contain only two genes (HP1288 and HP1289), and these paralogs, HP1288 and HP1289, are similarly derepressed in the H. pylori 26695 HK0164 null mutant. In contrast to the six genes discussed above that are derepressed in the HK0164 mutant strain, the HP1432 gene (encoding a histidine- and glutamine-rich protein) is transcribed in significantly higher quantities in the H. pylori 26695 CAT Control strain than in the HK0164 null mutant strain. High-level transcription of this gene apparently requires the action of a phosphorylated response regulator, RR0166.
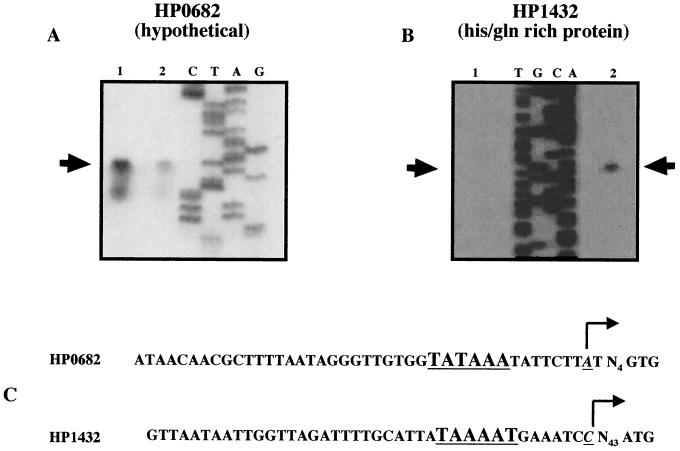
As an independent confirmation of the results detected by genome-wide transcriptional analyses, we performed quantitative primer extension analysis for two of the genes determined to be regulated by the histidine kinase HK0164. We have previously shown that quantitative primer extension analysis of H. pylori gene transcription correlates well with the results of complementary transcriptional reporter assays (6). Total cellular RNA was extracted and primer extension analyses were performed as previously described (6) using 32P-labeled oligonucleotides specific for target genes. We selected HP0682, for which the level of transcriptional activity in the absence of a functional histidine kinase was consistently increased, as well as HP1432, for which the level of transcriptional activity in the absence of functional histidine kinase was consistently repressed in the 26695 genetic background. In agreement with the DNA array analyses, quantitative primer extension analysis indicated that HP0682 signal intensity is greater in H. pylori 26695/HK0164− than in the H. pylori 26695 CAT Control strain (Fig. 4A). The primer used in this experiment, WM0682-X (5′ CCACTACACAACTAAATGTTC), was designed such that its 3′ end lay within an HP0682-specific region of the gene and thereby prevented mispriming from the highly similar HP1288. Consistent with the data from DNA array analysis, primer extension analysis provided evidence that the transcription of HP0682 is indeed regulated by the HK0164 signal transduction system. At present, it is not known whether HP1288 is similarly regulated or whether the DNA array results reflect cross-hybridization of differentially transcribed HP0682 cDNA to HP1288 features on the array. Primer extension analysis, using WM1432-X (5′ CCGTAGTAATGGTGGTGGTGCG) as the primer, also confirmed the repression of HP1432 transcription in the HK0164 mutant strain (Fig. 4B). Thus, primer extension analyses confirmed the pleiotropic nature of the regulation mediated by the HK0164/RR0166 signal transduction pathway, negatively regulating one set of genes while positively regulating expression of at least one gene. Examination of the promoter sequences for HP0682 and HP1432, deduced from the transcriptional start points (Fig. 4C), reveals that the −10 hexamers are well conserved and are very typical of −10 hexamers seen in other H. pylori σ80 promoters (7). Examination of adjacent sequences in the regions of these promoter elements failed to reveal any strikingly conserved sequences that could be suggested as cis-acting sequences responsible for the binding of the RR0166. Potentially, this may be due to the presence of RR0166 binding sites of different affinities and, hence, different sequences associated with each of these genes.
FIG. 4.
(A and B) Quantitative primer extension analysis of two genes identified as putative members of the H. pylori HK0164/RR0166 regulon. Transcriptional start points, indicated by arrows, were mapped for genes HP0682 (A) and HP1432 (B). Primers specific for each gene (5′ CCACTACACAACTAAATGTTC and 5′CCGTAGTAATGGTGGTGGTGCG, respectively) were annealed to identical quantities of RNA from H. pylori 26695/HK0164− (lanes 1) and H. pylori 26695 CAT Control (lanes 2). Sequencing ladders used to determine the exact transcriptional start point are shown as well. Each primer extension was repeated with independently derived RNA, and similar results were obtained. The differences in observed primer extension signal intensities were consistent with results obtained by analysis of genome-wide transcriptional profiles. (C) The deduced promoter sequences for HP0682 and HP1432 are shown. The transcriptional start points are underlined and italicized, and the bent arrow indicates the direction of transcription. The −10 promoter elements are underlined, and the number of nucleotides (N) between the transcriptional start point and the translational start codons are indicated.
Recently, Dietz et al. reported the use of a magnetocapture assay to identify several promoter sequences interacting with the RR0166 response regulator (5). RR0166 was reported to act as a transcriptional activator for two paralogous H. pylori operons (HP1408 to HP1412 and HP0427 to HP0423), as a well as for a family of paralogous genes exemplified by HP0119. In addition, it was reported that HP0166 negatively autoregulated its own expression. Interestingly, in our present studies of H. pylori 26695, we were unable to detect regulation of any of these genes by the RR0166 two-component system and the 20-bp region reported to serve as a binding site for RR0166 is not apparent near either of the promoter regions experimentally analyzed in the present study. However, we did detect very clear derepression of the HP1408 and HP0427 operons when analyzing the H. pylori B128/HK0164− mutant. These findings suggest that there are differences in the transcriptional regulatory systems of different H. pylori strains.
A recent comparative genomic approach to the study of sensory histidine kinases in prokaryotes revealed that the vast majority of bacterial and archaeal histidine kinases can be classified into one of five families based on the phylogeny and secondary structures of the proteins (10). Interestingly, the histidine kinase HK0164 of H. pylori could not be classified by this scheme. In fact, among the histidine kinases of the pathogenic bacteria sequenced to date, H. pylori and the closely related species Campylobacter jejuni had the greatest numbers of nontypeable histidine kinases: two and five, respectively (10). This may suggest a unique aspect of the biology of two-component signal transduction pathways in these pathogens.
Acknowledgments
This work was supported by NIH grant DK53623 and the Medical Research Service of the Department of Veterans Affairs (T.L.C.) and by a grant from The Jeffress Memorial Trust (J-602) (M.H.F.). J.D.H. was supported by a Pfizer undergraduate research fellowship.
We thank Antoaneta Necheva and James Graham for valuable assistance with DNA array methodology and analysis.
REFERENCES
- 1.Alm, R. A., L. L. Ling, D. T. Moir, et al. 1999. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 4.Cover, T. L., and M. J. Blaser. 1996. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv. Intern. Med. 41:85-117. [PubMed] [Google Scholar]
- 5.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsyth, M. H., J. C. Atherton, M. J. Blaser, and T. L. Cover. 1998. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect. Immun. 66:3088-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsyth, M. H., and T. L. Cover. 1999. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J. Bacteriol. 181:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsyth, M. H., and T. L. Cover. 2000. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect. Immun. 68:3193-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98:13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, D. J., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147:1197-1212. [DOI] [PubMed] [Google Scholar]
- 11.Labigne, A., and H. deReuse. 1996. Determinants of Helicobacter pylori pathogenicity. Infect. Agents Dis. 5:191-202. [PubMed] [Google Scholar]
- 12.McDaniel, T. K., K. C. Dewalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15-23. [DOI] [PubMed] [Google Scholar]
- 13.McGee, D. J., F. J. Radcliff, G. L. Mendz, R. L. Ferrero, and H. L. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendz, G. L., and S. L. Hazell. 1996. The urea cycle of Helicobacter pylori. Microbiology 142:2959-2967. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno, T. 1998. His-Asp phosphotransfer signal transduction. J. Biochem. 123:555-563. [DOI] [PubMed] [Google Scholar]
- 16.Mobley, H. L. 1996. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am. J. Med. 100:25-115. [DOI] [PubMed] [Google Scholar]
- 17.Nedenskov-Sorensen, P., G. Bukholm, and K. Bovre. Natural competence for genetic transformation in Campylobacter pylori. J. Infect. Dis. 161:365-366. [DOI] [PubMed]
- 18.Stock, A. L., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 19.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction systems. ASM Press, Washington, D.C.
- 20.Tomb, J. F., O. White, A. R. Kerlavage, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 21.Vanet, A., L. Marsan, A. Labigne, and M. F. Sagot. 2000. Inferring regulatory elements from a whole genome. An analysis of Helicobacter pylori σ80 family of promoter signals. J. Mol. Biol. 297:335-353. [DOI] [PubMed] [Google Scholar]