Abstract
The way in which the genes involved in cysteine biosynthesis are regulated is poorly characterized in Bacillus subtilis. We showed that CysL (formerly YwfK), a LysR-type transcriptional regulator, activates the transcription of the cysJI operon, which encodes sulfite reductase. We demonstrated that a cysL mutant and a cysJI mutant have similar phenotypes. Both are unable to grow using sulfate or sulfite as the sulfur source. The level of expression of the cysJI operon is higher in the presence of sulfate, sulfite, or thiosulfate than in the presence of cysteine. Conversely, the transcription of the cysH and cysK genes is not regulated by these sulfur sources. In the presence of thiosulfate, the expression of the cysJI operon was reduced 11-fold, whereas the expression of the cysH and cysK genes was increased, in a cysL mutant. A cis-acting DNA sequence located upstream of the transcriptional start site of the cysJI operon (positions −76 to −70) was shown to be necessary for sulfur source- and CysL-dependent regulation. CysL also negatively regulates its own transcription, a common characteristic of the LysR-type regulators. Gel mobility shift assays and DNase I footprint experiments showed that the CysL protein specifically binds to cysJ and cysL promoter regions. This is the first report of a regulator of some of the genes involved in cysteine biosynthesis in B. subtilis.
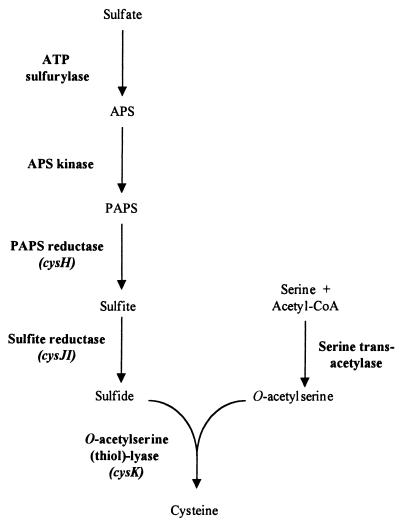
All living organisms require sulfur for the synthesis of proteins and essential cofactors. Sulfur can be assimilated either from inorganic sources, such as sulfate and thiosulfate, or from organic sources, such as sulfate esters, sulfamates, and sulfonates. In Escherichia coli, sulfate is transported into the cell via an ATP-binding cassette-type sulfate-thiosulfate transport system (5, 11). Sulfate is subsequently reduced to sulfide by a series of enzymatic steps involving ATP sulfurylase, adenosine 5′-phosphosulfate kinase, 3′-phosphoadenosine 5′-phosphosulfate (PAPS) sulfotransferase, and sulfite reductase (Fig. 1). An O-acetyl-l-serine thiol-lyase condenses sulfide and O-acetylserine to form cysteine. In E. coli and Salmonella enterica serovar Typhimurium, at least 22 genes are required for the transport and reduction of sulfate and for its incorporation into cysteine. Most of these genes are coordinately regulated in the cysteine regulon (11). The high-level expression of these genes requires CysB, a LysR-type transcriptional activator, the inducer N-acetylserine, and sulfur-limiting conditions (11, 26). The CysB protein binds as a tetramer just upstream of the −35 promoter region of the positively regulated cys genes. The interaction of CysB with the inducer causes the activator to undergo a conformational change, allowing it to interact with the activation sites of the cysJ, cysK, and cysP promoters (7, 23). l-Cysteine, sulfide, and thiosulfate downregulate l-cysteine biosynthesis (11, 12).
FIG. 1.
Biosynthesis of l-cysteine in bacteria. The different enzymes involved in sulfate assimilation and cysteine biosynthesis are indicated. The B. subtilis genes used in this study are in parentheses. APS, adenosine 5′-phosphosulfate; CoA, coenzyme A.
In Bacillus subtilis, the assimilation of sulfate and the biosynthesis of cysteine may occur via similar pathways (Fig. 1). Indeed, the enzymes and the genes involved in the conversion of sulfate into sulfide and the incorporation of sulfide into cysteine are present in B. subtilis (14, 28). The cysH gene, which encodes PAPS sulfotransferase, is the first gene of an operon encoding a sulfate permease (CysP) and enzymes catalyzing the first steps of the sulfate assimilation pathway (18, 19). The cysJ and cysI genes, encoding the two subunits of the sulfite reductase, were recently identified (37).
Little is known about the regulation of cysteine biosynthesis in gram-positive bacteria. CmbR, a new LysR-type transcriptional activator, was shown to be important for the expression of the metC-cysK operon in Lactococcus lactis (2). In gram-positive bacteria, several genes involved in methionine or cysteine biosynthesis form a regulon controlled by a global transcriptional termination system called the S-box regulon (4). The cysH operon of B. subtilis contains an S-box motif in its leader region. However, the expression of this operon is controlled at the transcription initiation level by an unidentified repressor rather than at the level of premature termination of transcription (18).
The ssu operon is required for the assimilation of sulfur from sulfonates by B. subtilis. The expression of this operon is repressed by cysteine and sulfate and derepressed by taurine and glutathione (38). This operon is regulated at the level of transcription initiation and transcription termination by an S-box-independent mechanism (36). However, no regulator has been identified yet.
In this report, we demonstrate that the genes involved in the biosynthesis of cysteine in B. subtilis are regulated differently depending on the available sulfur source. We identified CysL (formerly YwfK), a LysR-type transcriptional activator of the cysJI operon.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains used in this work are listed in Table 1. E. coli TGI (K-12 Δ[lac pro] supE thi hsd5/F′ traD36 proA+B+ lacIq lacZΔM15) was used for the cloning experiments. E. coli cells were grown in Luria-Bertani broth. B. subtilis was grown in SP medium (31) or minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 5 mM MgCl2, 0.5% glucose, 50 mg of l-tryptophan liter−1, 22 mg of ferric ammonium citrate liter−1, 0.1% l-glutamine, 200 mM xylose) supplemented with a sulfur source (either 1 mM K2SO4, 1 mM l-methionine, 2.5 mM glutathione, 1 mM thiosulfate, 0.5 mM sulfite, 0.5 mM sulfide, or l-cysteine supplied as 1 mM l-cystine, depending on the experiment). Minimal medium plates were prepared by adding 17 g of Noble agar (Difco) liter−1 to the liquid medium. Antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; kanamycin, 5 μg ml−1; spectinomycin, 100 μg ml−1; erythromycin plus lincomycin, 1 and 25 μg ml−1, respectively. Standard procedures were used to transform E. coli (30) and B. subtilis (15). All experiments were performed in accordance with European regulations concerning the use of genetically modified organisms (level 1 containment; agreement 2735).
TABLE 1.
Bacterial strains used in this studya
| Strain | Genotype | Source |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| BSIP1168 | trpC2 cysL::spc | pDIA5619→168 |
| BSIP1195 | trpC2 cysL::spc amyE::pxyl-cysL cat | pDIA5561→BSIP1168 |
| BSIP1196 | trpC2 amyE::pΔAcysL′-lacZ cat | pDIA5559→168 |
| BSIP1197 | trpC2 cysL::spc amyE::pΔAcysL′-lacZ cat | pDIA5559→BSIP1168 |
| BSIP1206 | trpC2 ΔcysJI::aphA3 | pDIA5568→168 |
| BSIP1207 | trpC2 amyE::cysK′-lacZ cat | pDIA5566→168 |
| BSIP1210 | trpC2 cysL::spc amyE::cysK′-lacZ cat | pDIA5566→BSIP1168 |
| BSIP1219 | trpC2 amyE::pΔAcysJ′-lacZ cat | pDIA5578→168 |
| BSIP1220 | trpC2 cysL::spc amyE::pΔAcysJ′-lacZ cat | pDIA5578→BSIP1168 |
| BSIP1221 | trpC2 amyE::pΔDcysJ′-lacZ cat | pDIA5579→168 |
| BSIP1222 | trpC2 cysL::spc amyE::pΔDcysJ′-lacZ cat | pDIA5579→BSIP1168 |
| BSIP1234 | trpC2 amyE::pΔCcysL′-lacZ cat | pDIA5584→168 |
| BSIP1235 | trpC2 cysL::spc amyE::pΔCcysL′-lacZ cat | pDIA5584→BSIP1168 |
| BSIP1238 | trpC2 amyE::pΔBcysJ′-lacZ cat | pDIA5588→168 |
| BSIP1239 | trpC2 cysL::spc amyE::pΔBcysJ′-lacZ cat | pDIA5588→BSIP1168 |
| BSIP1240 | trpC2 amyE::pΔCcysJ′-lacZ cat | pDIA5589→168 |
| BSIP1241 | trpC2 cysL::spc amyE::pΔCcysJ′-lacZ cat | pDIA5589→BSIP1168 |
| BSIP1252 | trpC2 amyE::pΔBcysL′-lacZ cat | pDIA5596→168 |
| BSIP1253 | trpC2 cysL::spc amyE::pΔBcysL′-lacZ cat | pDIA5596→BSIP1168 |
| BSIP1283 | trpC2 amyE::cysH′-lacZ cat | pDIA5618→168 |
| BSIP1284 | trpC2 cysL::spc amyE::cysH′-lacZ cat | pDIA5618→BSIP1168 |
Arrows indicate construction by transformation. cat is the pC194 chloramphenicol acetyltransferase gene, aphA3 is an Enterococcus faecalis kanamycin resistance gene, and spc is a spectinomycin resistance gene from Staphylococcus aureus.
Amylase activity was detected as previously described (33). β-Galactosidase-specific activity was measured according to the method described by Miller (22) with cell extracts obtained by lysozyme treatment. One unit of β-galactosidase is defined as the amount of enzyme that produces 1 nmol of O-nitrophenol min−1 at 28°C. Protein concentrations were determined by the method of Bradford.
DNA manipulation and plasmid construction.
Plasmid DNA from E. coli and chromosomal DNA from B. subtilis were prepared according to standard procedures. Restriction enzymes, Taq DNA polymerase, and phage T4 DNA ligase were used as recommended by the manufacturers. PCR products were purified by use of the Qiaquick kit (Qiagen). DNA sequences were determined by the dideoxy chain termination method with the Thermo Sequenase kit (Amersham Pharmacia Biotech).
Nucleotides are numbered relative to the transcriptional start sites of the different genes.
A 6,187-bp DNA fragment containing the cysL gene (formerly ywfK) was cloned into plasmid pDIA5304 to give pDIA5373 (29). A spectinomycin resistance cassette (25) was inserted into the unique XbaI site of the cysL gene within pDIA5373. The resulting plasmid, pDIA5619, was linearized with ScaI and used to transform B. subtilis strain 168. The cysL gene was disrupted with the spectinomycin resistance cassette by a double-crossover event, giving rise to strain BSIP1168.
The cysL gene was cloned into pX containing a xylose-inducible promoter (10) to complement the cysL::spc mutant. The complete coding sequence of cysL (nucleotides +30 to +980) was amplified by PCR and inserted into the SpeI and BamHI sites of pX, producing pDIA5561. This plasmid was used to transform BSIP1168, leading to the integration of the cysL gene at the amyE locus (Table 1).
To disrupt the cysJI genes, two DNA fragments containing the 5′ end of the cysJ gene (nucleotides −104 to +1091) and the 3′ end of the cysI gene (nucleotides +3043 to +3623) were amplified by PCR. These two PCR fragments and a kanamycin resistance cassette (35) were inserted into pUC18 (Roche), producing pDIA5568. This plasmid was linearized at its unique ScaI site and used to transform B. subtilis strain 168. The cysJI genes were deleted by a double-crossover event, giving rise to strain BSIP1206.
pAC6 (33) was used to construct transcriptional fusions between a series of cysJ promoter regions with various 5′ deletions and the promoterless lacZ gene. Regions ΔA (−163, +82), ΔB (−76, +82), ΔC (−70, +82), and ΔD (−52, +82) were amplified by PCR, such that 5′ EcoRI and 3′ BamHI sites were incorporated. The PCR products were inserted into pAC6, giving rise to pDIA5578 (ΔA), pDIA5588 (ΔB), pDIA5589 (ΔC), and pDIA5579 (ΔD), respectively. The different fusions were integrated at the amyE locus of B. subtilis strains 168 and BSIP1168 (Table 1).
Transcriptional fusions between a series of cysL promoter regions with 3′ deletions and the lacZ gene were also constructed and integrated at the amyE loci of strains 168 and BSIP1168. Regions ΔA (−167, +111), ΔB (−167, +19), and ΔC (−167, +31) were amplified by PCR. The fragments were inserted into pAC6, giving rise to pDIA5559 (ΔA), pDIA5596 (ΔB), and pDIA5584 (ΔC), respectively.
The same strategy was used to introduce a cysH′-lacZ fusion and a cysK′-lacZ fusion at the amyE loci of strains 168 and BSIP1168. A 431-bp SmaI-BamHI DNA fragment, corresponding to the cysH promoter region (positions −105 to +326), was inserted into pAC6, leading to pDIA5618. A 518-bp EcoRI-BamHI fragment, corresponding to the cysK promoter region (positions −169 to +349), was inserted into pAC6, producing pDIA5566.
RNA isolation and analysis.
The Trizol reagent (Gibco-BRL) was used to extract total RNA from B. subtilis 168 that had been grown in minimal medium supplemented with 1 mM thiosulfate. Reverse transcription-PCR (RT-PCR) experiments were performed with the Access RT-PCR introductory system kit (Promega). Three independent reactions with different pairs of primers corresponding to either cysJ, cysI, or cysJ and cysI, were used to detect specific transcripts. A control experiment without reverse transcriptase was used to ensure that no contaminating DNA was present.
In the primer extension experiments, oligonucleotides IV156 (cysJ) and IV159 (cysL) (for the positions of the primers see Fig. 2) were labeled with [γ-32P]ATP and hybridized with 10 μg of RNA. For the cysL primer extension experiment, RNA was extracted from a strain transformed with pDIA5643, a pHT315 derivative containing the promoter region, and the 5′ part of the cysL gene (nucleotides −167 to +448). Elongation of the DNA primer using reverse transcriptase and analysis of the products were performed as previously described (30). The same primers were used for the generation of a sequence ladder.
FIG. 2.
Promoter regions of the cysJI operon (A) and of the cysL gene (B). Vertical arrows, transcription start sites (+1) of the cysJ and cysL promoters. The −35 and −10 regions and the proposed ribosome-binding sites (RBS) are capitalized and underlined. Bent arrows, deletion end points of the different fusions with the lacZ gene (numbering is with respect to the transcriptional start site); shaded boxes, DNA regions protected by CysL in the DNase I footprint experiments. Oligonucleotides IV156 and IV159 are indicated.
Overproduction of the CysL protein in E. coli.
The cysL gene (nucleotides +46 to +940) was amplified by PCR such that a 5′ NdeI site and a 3′ XhoI site were introduced. The amplified gene was cloned into the pET20b+ vector (Novagen), which had been digested with the same enzymes. The resulting plasmid, pET20b+cysL, was introduced into E. coli strain BL21(DE3) (Novagen), which contains pDIA17 (24), encoding the LacI repressor. Transformants were grown in Luria broth at 30°C to an optical density at 600 nm of 2. The expression of the cysL gene was induced by adding 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cells were harvested 2 h later by centrifugation, resuspended in 0.1 M potassium phosphate buffer (pH 7.3), and disrupted in a Fastprep apparatus (Bio 101). The lysate was clarified by centrifugation for 30 min at 8,000 × g and used for the gel shift and DNase I footprint experiments. The overproduction of CysL was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Comparison of a crude E. coli extract, which contains pET20b+cysL or pET20b+ alone, revealed the presence of an additional band at 34 kDa corresponding to CysL (data not shown).
Gel mobility shift assay.
DNA fragments corresponding to the promoter regions of cysJ (nucleotides −163 to +82), cysL (nucleotides −167 to +111), cysH (nucleotides −105 to +326), and cysK (positions −169 to +349) were amplified by PCR using specific primers 5′ end labeled with [γ-32P]ATP. Unincorporated oligonucleotides were removed with the QIAquick-spin PCR purification kit (Qiagen). Protein-DNA complexes were formed in 10-μl volumes by incubating the 32P-end-labeled DNA fragments with different amounts of crude E. coli extract containing the CysL protein in binding buffer (20 mM HEPES [pH 7.6], 4 mM MgCl2, 100 mM KCl, 0.5 mM dithiothreitol, 0.2 mM EDTA, 6% glycerol) in the presence of 5 ng of poly (dI-dC) μl−1 and 0.1 μg of bovine serum albumin μl−1. After 30 min at 25°C, 5 μl of loading buffer (0.125% [wt/vol] bromophenol blue, 0.125% [wt/vol] xylene cyanol, 10 mM Tris-HCl, 1 mM EDTA, 5% glycerol) was added to the reaction mixture. Samples were separated on a 6% polyacrylamide native gel in Tris-borate-EDTA buffer at 4°C at 10 V cm−1 for 1 h.
DNase I footprinting.
The labeled DNA fragments corresponding to the cysJ and the cysL promoter regions were used to analyze the CysL protected regions in DNase I footprinting reactions. Briefly, constant amounts of the 5′-end-labeled 245-bp top strand DNA fragment of the cysJ promoter region (nucleotides −163 to +82) or the 278-bp bottom strand DNA fragment of the cysL promoter region (nucleotides −167 to +111) were incubated without protein or with various concentrations of the crude E. coli extract containing the CysL protein. The binding conditions were similar to those used for gel mobility shift assays. DNase I (0.05 U μl−1; Roche) was added to the reaction mixtures. After 30 s at 25°C, the DNase I was inactivated by adding 40 μl of phenol and 190 μl of stop solution (0.4 M sodium acetate, 2.5 mM EDTA, 50 μg of poly[dI-dC] ml−1, 10 μg of glycogen ml−1). For each reaction, DNA was then subjected to ethanol precipitation followed by electrophoresis in a 7 M urea-polyacrylamide gel containing 6% acrylamide in Tris-borate-EDTA buffer. The sequence ladder (G+A) was produced as previously described (20).
RESULTS
CysL (YwfK) is a regulator of sulfur metabolism in B. subtilis.
Three LysR-type activators are involved in the regulation of sulfur metabolism in E. coli: CysB, Cbl, and MetR (8, 11, 21). Nine of the 14 LysR-type regulators of unknown function in B. subtilis (14) were tested for their possible role in the regulation of sulfur metabolism. The genes encoding eight of these regulators (yofA, ytlI, yusT, yvbU, ywqM, yclA, yxjO, and yybE) were inactivated as part of European and Japanese projects for the functional characterization of the B. subtilis genome (http://locus.jouy.inra.fr/cgibin/genmic/madbase/progs/madbase.operl and http://bacillus.genome.ad.jp). We disrupted the ywfK gene by inserting a spectinomycin resistance cassette. The resulting mutants were screened on minimal medium plates in the presence of sulfate, cystine, or methionine as the sulfur source. Interestingly, the B. subtilis ywfK mutant (BSIP1168) could not grow in the presence of sulfate or sulfite, but grew similarly to the wild-type strain in the presence of sulfide, thiosulfate, cystine, or methionine (Table 2). A single copy of the ywfK gene, expressed under the control of the xylA promoter, was introduced into the amyE locus of the BSIP1168 strain. The presence of ywfK in trans restored the growth of the ywfK::spc mutant on all of the sulfur sources tested (Table 2).
TABLE 2.
Phenotypes of the B. subtilis cysL and cysJI mutants grown in the presence of various sulfur sourcesa
| Sulfur source | Generation time (min) for strainb
|
|||
|---|---|---|---|---|
| 168 | BSIP1168 | BSIP1195 | BSIP1206 | |
| Sulfate | 55 | NGc | 55 | NG |
| Sulfite | 70 | NG | 70 | NG |
| Sulfide | 55 | 55 | NDd | 70 |
| Cystine | 55 | 55 | 55 | 60 |
| Methionine | 50 | 55 | 55 | ND |
| Thiosulfate | 55 | 60 | 55 | ND |
All strains were grown in minimal medium. This medium was supplemented with a sulfur source at 1 mM except for sulfite and sulfide (0.5 mM). For strain BSIP1195, 5 mM xylose was added to the medium to induce transcription from the xylA promoter.
For strain genotypes, see Table 1.
NG, no growth.
ND, not determined.
The phenotypes of a B. subtilis ywfK mutant and of an E. coli cysB mutant are very similar, except that the cysB mutant is unable to grow in the presence of thiosulfate (11), whereas the B. subtilis ywfK mutant can grow on this sulfur source. Each of the LysR-type transcriptional regulators contains an N-terminal DNA-binding domain with a helix-turn-helix (HTH) motif and two regions probably involved in coinducer recognition and/or response (32). A sequence comparison revealed that the YwfK polypeptide has only 28% identity to CysB of E. coli. Only 3 of the 18 residues known to be important for the activity of CysB are conserved in YwfK (16). Accordingly, the YwfK polypeptide was not able to restore the growth of a cysB mutant (CB64, cysB93) in the presence of sulfate (data not shown), indicating that YwfK cannot substitute for CysB. The fact that the ywfK gene could not complement the cysB mutant led us to rename the YwfK regulator CysL.
Regulation of the expression of the cysJI operon.
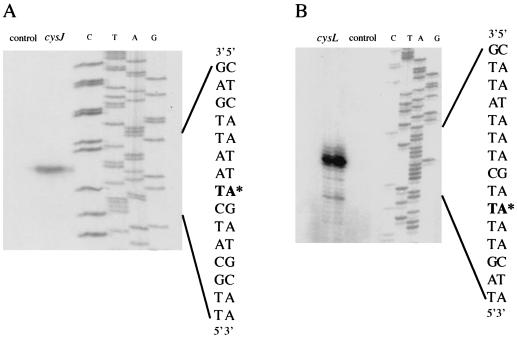
The cysJ and cysI genes of B. subtilis were recently shown to encode the two subunits of the sulfite reductase (37), which reduces sulfite to sulfide (Fig. 1). The cysJ and cysI genes are adjacent in the B. subtilis chromosome, suggesting that these two genes are organized in an operon. To confirm this hypothesis, RT-PCR experiments were performed with total RNA isolated from B. subtilis 168 grown in the presence of thiosulfate. Three DNA fragments were amplified by using primer pairs specific to either cysJ, cysI, or cysJ and cysI (data not shown). A single DNA fragment corresponding to a cysJI common transcript was detected, confirming that cysJ and cysI form an operon. The translation initiation codon of cysJ is a TTG preceded by a consensus ribosome-binding site. The 5′ end of the cysJI transcript was identified by primer extension analysis using total RNA extracted from the wild-type strain. Transcription was initiated at a single A located 44-bp upstream of the translational start point (Fig. 3A). This initiation start site is slightly different from the one identified previously, which is located 46 bp upstream of the translation initiation codon of cysJ (37). The deduced −35 (TTTACT) and −10 (TAAAGT) boxes of the promoter are quite similar to the consensus sequences for σA-dependent promoters.
FIG. 3.
Mapping of the transcription start sites of the cysJI operon (A) and cysL gene (B) by primer extension. Primer extension experiments were performed using oligonucleotides IV156 (A) and IV159 (B), which hybridized with the cysJ and cysL genes, respectively. The labeled oligonucleotides were loaded as a control. Sequencing reactions (lanes C, T, A, and G) were performed with the same oligonucleotides and pDIA5578 (cysJ) or pDIA5582 (cysL) as the template. ∗, 5′ end of mRNA.
The phenotype of the cysL mutant corresponds to the phenotype expected for a mutant inactivated in the cysJI genes. The ΔcysJI::aphA3 mutant (BSIP1206) was unable to grow when sulfate or sulfite was the sole sulfur source (Table 2). To study the regulation of the expression of the cysJI operon in response to sulfur availability and the role of CysL in this regulation, a cysJ′-lacZ transcriptional fusion containing the promoter region from positions −163 to +82 was integrated at the amyE locus of B. subtilis 168 (strain BSIP1219) and of the cysL mutant (strain BSIP1220). In strain BSIP1219, the level of β-galactosidase activity was high in the presence of sulfate or sulfite but was 10-fold lower when cystine was the sulfur source (Table 3). An intermediate level of expression was observed in the presence of thiosulfate, and a low level of expression was detected in the presence of glutathione, methionine, or cystine (Table 3). These results confirm that the transcription of the cysJI operon is sulfur source dependent. To investigate the role of CysL in the regulation of the cysJI operon, we then compared the level of expression of this fusion in the wild-type and cysL backgrounds. The expression of a cysJ′-lacZ fusion was 11-, 4-, and 2.5-fold lower in the cysL mutant than in the wild-type strain for cells grown in the presence of thiosulfate, cystine, or methionine as the sole sulfur source (Table 3). The CysL regulator is therefore involved in the transcriptional activation of the cysJI operon.
TABLE 3.
Expression of cysJ′-lacZ, cysH′-lacZ, and cysK′-lacZ fusions in the presence of different sulfur sources
| Sulfur source | β-Galactosidase activity (U mg of protein−1)a in the indicated strain for:
|
|||||
|---|---|---|---|---|---|---|
|
cysJ′-lacZ
|
cysH′-lacZ
|
cysK′-lacZ
|
||||
| BSIP1219 (cysL+) | BSIP1220 (cysL::spc) | BSIP1283 (cysL+) | BSIP1284 (cysL::spc) | BSIP1207 (cysL+) | BSIP1210 (cysL::spc) | |
| Sulfate | 190 | NG | 600 | NG | 120 | NG |
| Sulfite | 260 | NG | 470 | NG | 90 | NG |
| Thiosulfate | 75 | 7 | 760 | 1,590 | 120 | 910 |
| Cystine | 20 | 5 | 400 | 360 | 110 | 130 |
| Glutathione | 40 | 50 | 1,035 | 1,360 | 680 | 640 |
| Methionine | 40 | 15 | 625 | 660 | ND | ND |
Cells were grown in minimal medium in the presence of the indicated sulfur source (1 mM) except for sulfite (0.5 mM) and glutathione (2.5 mM). The β-galactosidase activities were obtained from cultures in mid-exponential growth. The values represent means of at least three independent experiments. Standard deviations were less than 20% of the means. NG, no growth; ND, not determined.
Role of the CysL regulator in the expression of the cysH operon and of the cysK gene.
To determine whether CysL is necessary for the regulation of cysH and cysK (Fig. 1), cysH′-lacZ and cysK′-lacZ transcriptional fusions were integrated at the amyE loci of a wild-type strain and of a cysL mutant (Table 1). β-Galactosidase activity was measured after growth of the resulting strains in minimal medium in the presence of different sulfur sources (Table 3). The levels of expression of each fusion in the presence of cystine, sulfate, sulfite, or thiosulfate were similar. In contrast, the levels of expression of the cysH′-lacZ and cysK′-lacZ fusions were 2.5- to 6-fold higher in the presence of glutathione than in the presence of cystine. In a cysL mutant, the expression of the cysH′-lacZ and the cysK′-lacZ fusions, after growth in the presence of thiosulfate, were 2- and 7.5-fold higher than that for the wild-type strain, respectively (Table 3). For these fusions, there was no difference in expression between the wild-type and cysL strains after growth in the presence of cystine or glutathione. Therefore, the transcription of the cysH operon and that of the cysK gene are increased in a cysL background in the presence of thiosulfate.
Regulation of the expression of the cysL gene.
The translation initiation codon of cysL is a TTG preceded by a consensus ribosome binding site (Fig. 2B). The 5′ end of the cysL transcript was identified by primer extension using total RNA extracted from a strain containing a plasmid carrying the promoter region and the 5′ part of the cysL gene (nucleotides −167 to +448). Transcription was initiated at a single A located 46 bp upstream of the translational start point (Fig. 3B). The deduced −35 (TTTACC) and −10 (TACAAT) boxes (Fig. 2B) of the promoter are quite similar to the consensus sequences for σA-dependent promoters.
To study the regulation of expression of the cysL gene, the cysL promoter region (positions −167 to +111) was fused to the lacZ gene to create a transcriptional cysL′-lacZ fusion. A single copy of this fusion was integrated at the amyE loci of the wild-type strain and of the cysL mutant, resulting in strains BSIP1196 and BSIP1197, respectively. The β-galactosidase activity was measured for these strains after growth in minimal medium in the presence of various sulfur sources. The expression of the cysL gene was low and was not modulated by the sulfur source (data not shown). However, the expression of a cysL′-lacZ fusion was twofold higher in a cysL mutant (8 U mg of protein−1) than in a wild-type strain (4 U mg of protein−1). This result suggests that the CysL protein negatively regulates its own transcription, as observed for other LysR-type transcriptional regulators (9, 32).
Binding of the CysL regulator to the promoter regions of genes involved in cysteine biosynthesis.
We used electrophoretic mobility shift assays to investigate the capacity of CysL for binding to the DNA of the promoter regions of the cysJ, cysL, cysH, and cysK genes. The B. subtilis CysL protein was overproduced in E. coli, and the cell-free crude extract was used in the DNA-binding experiments.
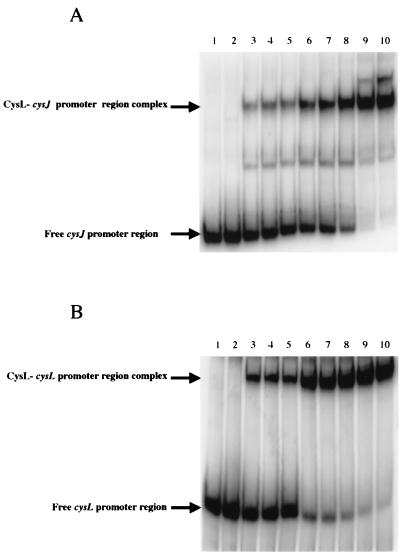
A 245-bp radiolabeled DNA fragment corresponding to the cysJ promoter region (positions −163 to +82) was incubated with increasing amounts of the CysL protein. Native polyacrylamide gel electrophoresis revealed that the band corresponding to the 245-bp labeled fragment was shifted when the CysL protein was present (Fig. 4A). Indeed, the DNA fragment containing the cysJ promoter region was fully retarded by 50 ng of the E. coli extract containing CysL (Fig. 4A, lane 9). This shift was not observed with 200 ng of the E. coli extract without CysL (Fig. 4A, lane 2). The addition of an excess of an unlabeled DNA fragment corresponding to the cysJ promoter region prevented CysL binding to the same labeled DNA fragment. In contrast, an excess of a cold noncompetitor DNA fragment (the serA promoter region) did not prevent CysL binding to the cysJ promoter region (data not shown). These results demonstrate that CysL binds specifically to the cysJ promoter region.
FIG. 4.
Binding of the CysL regulator to a cysJ or cysL promoter fragment in a gel mobility shift assay. A 245-bp 5′-labeled fragment containing the cysJ promoter region (positions −163 to +82) (A) or a 278-bp fragment containing the cysL promoter (nucleotides −167 to + 111) (B) was incubated with various amounts of a cell-free crude E. coli extract either containing overproduced CysL or not. Lane 1, free DNA probe; lane 2, negative control with a crude E. coli extract without CysL (200 ng); lanes 3 to 10, increasing amounts of the E. coli extract overproducing CysL (10, 12, 15, 18, 20, 30, 50, and 100 ng, respectively).
As CysL regulates its own transcription, a 278-bp DNA fragment containing the cysL promoter region (positions −167 to +111) was used in similar binding studies (Fig. 4B). A gel shift was observed only with the E. coli extract overproducing CysL, and 50 ng of the CysL crude extract caused complete retardation of the DNA fragment containing the cysL promoter region (Fig. 4B, lane 9). The specificity of this interaction was verified under the same experimental conditions as those described for the binding of CysL to the cysJ promoter region. The CysL regulator bound specifically to its own promoter in a dose-dependent manner.
Gel mobility shift assays were also performed to determine if CysL can directly interact with the cysH and cysK promoter regions. The labeled DNA fragments containing the cysH and cysK promoter regions (nucleotides −105 to +326 and −169 to +349, respectively) were tested with increasing amounts of the E. coli crude extract containing CysL (10 ng to 1 μg). A limited amount of the DNA fragment corresponding to the cysK promoter region was retarded in the presence of 1 μg of an E. coli crude extract containing CysL (data not shown). The affinity of CysL for the cysK promoter region was about 20-fold lower than its affinity for the cysJ or cysL promoter regions. No detectable binding of CysL to the cysH promoter region was observed in the conditions tested (data not shown).
Characterization of a cis-acting target in the promoter regions of the cysJ and the cysL genes.
To identify the DNA sequences involved in the sulfur-dependent regulation of the cysJI operon, cysJ promoter regions containing various 5′ deletions were fused to the promoterless lacZ gene (Fig. 2A). A single copy of these fusions was introduced at the amyE locus of B. subtilis 168. β-Galactosidase activities in the different strains grown in minimal medium in the presence of sulfate, thiosulfate, or cystine as a sulfur source were measured (Table 4). The ΔA (−163, +82) and ΔB (−76, +82) cysJ′-lacZ fusions were expressed 10- and 4-fold more highly in the presence of sulfate or thiosulfate than in the presence of cystine. The ΔC (−70, +82) and ΔD (−52, +82) cysJ′-lacZ fusions, which were expressed at lower levels, were weakly or not regulated in these conditions. This indicates that the DNA fragment located between positions −76 and −70 of the cysJ promoter is important for the sulfur source-dependent regulation of the cysJI operon. To identify cis-acting DNA sequences required for activation of the cysJI operon by the CysL regulator, the same fusions were introduced into a cysL background. β-Galactosidase activities of the wild-type and cysL::spc strains after growth in the presence of thiosulfate were compared (Table 4). The ΔA (−163, +82) and ΔB (−76, +82) cysJ′-lacZ fusions were expressed 10-fold more highly in the wild-type strain than in the cysL mutant. No activation by CysL was observed with the ΔC (−70, +82) and the ΔD (−52, +82) cysJ′-lacZ fusions. Thus, the same DNA fragment (positions −76 to −70) is important for both the CysL-dependent and the sulfur source-dependent regulation of the cysJI operon.
TABLE 4.
Effect of the sulfur source and of the CysL regulator on the expression of different cysJ′-lacZ transcriptional fusionsa
| Transcriptional fusion of the cysJ promoter at the amyE locus | β-Galactosidase activity (U mg of protein−1) in the presence of:
|
|||
|---|---|---|---|---|
| Sulfateb (cysL+) | Cystineb (cysL+) | Thiosulfate for strain with genotype:
|
||
| cysL+ | cysL::spc | |||
| ΔA (−163, +82) cysJ′-lacZ | 190 | 20 | 75 | 7 |
| ΔB (−76, +82) cysJ′-lacZ | 200 | 20 | 90 | 9 |
| ΔC (−70, +82) cysJ′-lacZ | 20 | 4 | 7 | 6 |
| ΔD (−52, +82) cysJ′-lacZ | 3 | 2 | 3 | 6 |
β-Galactosidase activities were determined in extracts prepared from exponentially growing cells (optical density at 600 nm, 0.8 to 1). The values represent means of at least three independent experiments. Standard deviations were less than 15% of the means. The cysL+ and cysL::spc strains used in this study are described in Table 1.
Relevant strain genotype is in parentheses.
To identify the cis-acting DNA target required for the autoregulation of cysL expression, the cysL promoter region with different 3′ deletions was fused to the lacZ gene. The fusions were introduced into strains 168 and BSIP1168, and the β-galactosidase activity was measured after growth of the strains with thiosulfate as a sulfur source. The ΔA (−167, +111) and ΔC (−167, +31) cysL′-lacZ fusions were twofold more highly expressed in the cysL mutant (8 and 22 U mg of protein−1, respectively) than in the wild-type strain (4 and 9 U mg of protein−1, respectively). No regulation by CysL was observed with the ΔB (−167, +19) cysL′-lacZ fusion. These results indicate that the DNA fragment located between nucleotides +19 and +31 of the cysL promoter is important for negative autoregulation by CysL.
DNase I protection assays of the cysI and cysL promoter regions by CysL.
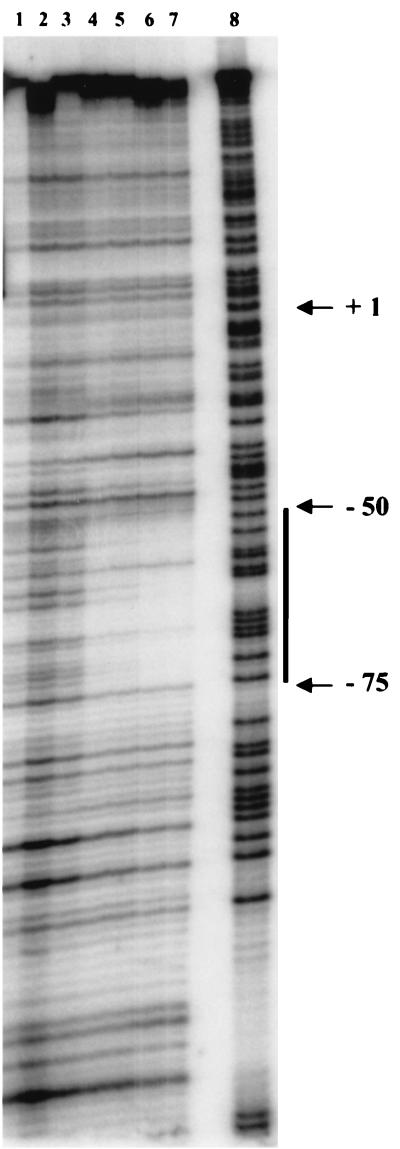
DNase I footprint experiments were carried out to determine the precise location of the CysL binding site(s) within the cysJ and cysL promoter regions. Comparison of the sequence patterns produced in the absence of CysL and in the presence of saturating concentrations of CysL located protected regions consisting of 25 nucleotides on the top strand of the cysJ promoter region (Fig. 5) and 29 nucleotides on the bottom strand of the cysL promoter region (data not shown). The binding sites extended from positions −75 to −50 and from positions +4 to +33 relative to the transcriptional start sites of cysJ and cysL, respectively.
FIG. 5.
DNase I footprint of the CysL regulator on the cysJ promoter region. The 245-bp PCR fragment representing the coding strand of the cysJ promoter region (positions −163 to +82) was 5′ end labeled, and incubated in separate reaction mixtures without protein (lane 1), in the presence of 200 ng of a crude protein extract without CysL (lane 2), or in the presence of 10 (lane 3), 50 (lane 4), 100 (lane 5), 200 (lane 6), or 300 ng (lane 7) of the crude E. coli extract overproducing CysL and subjected to DNase I digestion. Lane 8, G+A sequencing ladder (20). Vertical bar, position of the protected region. Numbers indicate the distances from the transcription initiation site of the cysJI operon.
These results are consistent with the roles of the CysL protein as activator of the expression of the cysJI operon and repressor of the expression of the cysL gene. These experiments also confirm the location of the cis-acting target sequences identified by using deletions of the cysJ and of the cysL promoter regions.
DISCUSSION
The regulation of methionine and cysteine biosynthesis genes in E. coli involves two LysR-type transcriptional activators (CysB and MetR) and a repressor (MetJ) (11). In this study, we identified a new member of the LysR regulator family, CysL (YwfK), which is involved in the regulation of expression of the cysJI operon, encoding both subunits of the sulfite reductase of B. subtilis. The CysL protein is the first regulator of at least one step of the sulfur-containing amino acid biosynthesis pathway to be identified in B. subtilis. Proteins with a high level of similarity to CysL have been found in Bacillus stearothermophilus (60% identity), Bacillus anthracis (45% identity), Clostridium difficile (44% identity), Clostridium acetobutylicum (40% identity), and Listeria monocytogenes (45% identity). No CysL-like protein could be detected in L. lactis, Bacillus halodurans, or Staphylococcus aureus. CmbR, a newly characterized LysR-type transcriptional activator of L. lactis, controls the expression of the metC-cysK operon (2). The CysL and CmbR proteins show only 25% identity, suggesting that these two regulators are not functionally equivalent. Moreover, the genes responsible for sulfate uptake and reduction are absent from the L. lactis genome. Sulfur metabolism and its regulation are probably different in B. subtilis and L. lactis.
Most of the genes encoding members of the LysR family are adjacent to and transcribed divergently from the genes they regulate (32). This is not the case for the B. subtilis cysL gene. A few other LysR-type transcriptional regulator genes, such as E. coli cysB and oxyR, are also located some distance from their target genes on the chromosome. Interestingly, a B. stearothermophilus gene encoding a protein with high similarity to CysL was found to be located adjacent to a cysJI-like operon (http://www.genome.ou.edu). The CysL-like activator of B. stearothermophilus probably controls the expression of this operon.
In enteric bacteria, the cysJ and cysI genes are cotranscribed with the cysH gene (17, 27). In contrast, in B. subtilis and B. stearothermophilus, cysJ and cysI form an operon, whereas cysH belongs to another operon encoding the first steps of the sulfate assimilation pathway (18). The location of cysH and cysJI in two independent transcriptional units suggests that the synthesis of PAPS sulfotransferase could be regulated differently from the synthesis of sulfite reductase in B. subtilis. Indeed, the cysJI operon was transcribed at a high level when the cells were grown in the presence of sulfate or sulfite. Transcription was 10-fold lower when cells were grown in the presence of cystine. The levels of expression of the cysH and cysK (encoding the cysteine synthase) genes were not regulated by these sulfur sources (this work; 1, 18, 37). The transcription of the cysJI operon is positively controlled by the CysL activator, whereas the transcription of the cysH and cysK genes is higher in a cysL mutant. These results indicate that the transcription of the cysJI operon in response to sulfur availability is regulated differently from that of the cysH and cysK genes. The expression of the cysJI operon can either be repressed by cysteine or induced by an intermediate compound from the sulfate assimilation pathway, via the CysL regulator. As in E. coli and S. enterica serovar Typhimurium (11, 13, 17), the cysK and cysH genes of B. subtilis were expressed at the highest levels in sulfur-limiting conditions (in the presence of glutathione or during cystine limitation) (this work; 1, 18). A yet-unidentified repressor, CysR, was recently proposed to prevent the transcription of cysH in these conditions (18). The activity of this CysR regulator is probably controlled by the intracellular concentration of O-acetylserine, which may induce the expression of the cys genes. The CysL regulator is not this CysR repressor. Indeed, CysL is not involved in the regulation of the cysK and cysH genes in sulfur-limiting conditions (Table 3) (I. Guillouard, unpublished results). This suggests that the cysteine biosynthetic pathway is controlled at several levels in B. subtilis and that this control involves at least two regulators, CysR and CysL. The precise roles of these regulators deserve further investigation.
Like most members of the LysR family, the CysL protein can also inhibit the transcription of its structural gene. The expression of the cysL gene is independent of the sulfur source added to the medium. A genetic approach showed that the expression of four different genes or operons (cysH, cysJ, cysK, and cysL) is modified in a cysL background. The CysL protein can specifically bind in vitro to the cysJ and cysL promoter regions in the absence of any added cofactor, as observed for several LysR-type regulators (32). CysL may also interact with the cysK promoter region, but with a much lower affinity. The specificity and the physiological significance of this low binding deserve further experiments. The mobility shift assays did not show that CysL can bind to the cysH promoter region. The derepression of cysH expression in a cysL mutant in the presence of thiosulfate could be due to the depletion of sulfite reductase synthesis, which may modify the concentrations of several metabolites of the cysteine biosynthesis pathway.
The DNA sequence of the cysJ promoter region found to be protected in the DNase I footprint experiment is located between positions −76 and −50. This is consistent with the genetic data indicating that the −76 to −70 DNA region is important for sulfur- and CysL-dependent regulation. Similar experiments with the cysL promoter region showed that the DNA sequence extending from nucleotides +19 to +31 is necessary for cysL negative autoregulation. These locations correspond to the positions of the recognition sites of other LysR-type regulators (32). Many LysR-type-regulated promoters contain characteristic sequence TN11A at the core of a larger motif with dyad symmetry (3, 32). Sequence ATTA-N7-TAAT, consistent with this consensus motif, was found upstream of the B. subtilis and B. stearothermophilus cysJI operons. However, this motif was not found in the cysK promoter region and in the protected DNA sequence of the cysL promoter region. The target sequences of the OxyR and CysB regulators, which control a large set of genes, are not highly conserved (6, 34). This could be also the case for CysL. Additional experiments are necessary to determine the precise DNA sequences required for recognition by CysL.
Acknowledgments
We thank E. Presecan-Siedel for constructing the cysL mutant, M. S. Gelfand, A. A. Mironov, and D. A. Rodionov for helpful discussions, and G. Rapoport, M. Débarbouillé, and I. Moszer for careful reading of the manuscript. We are grateful to V. Polonais and C. Mulot for their help during this work.
This research was supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Centre National de la Recherche Scientifique (URA 2171), the Institut Pasteur, the Université Paris 7, and the European Biotech Program (contract QLG2 CT9901455).
REFERENCES
- 1.Coppee, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goethals, K., M. Van Montagu, and M. Holsters. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 5.Hryniewicz, M., A. Sirko, A. Palucha, A. Bock, and D. Hulanicka. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 172:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hryniewicz, M. M., and N. M. Kredich. 1995. Hydroxyl radical footprints and half-site arrangements of binding sites for the CysB transcriptional activator of Salmonella typhimurium. J. Bacteriol. 177:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hryniewicz, M. M., and N. M. Kredich. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 9.Jagura-Burdzy, G., and D. Hulanicka. 1981. Use of gene fusions to study expression of cysB, the regulatory gene of the cysteine regulon. J. Bacteriol. 147:744-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 12.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 13.Kredich, N. M. 1971. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. I. Effects of growth of varying sulfur sources and O-acetyl-L-serine on gene expression. J. Biol. Chem. 246:3474-3484. [PubMed] [Google Scholar]
- 14.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 15.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 17.Loughlin, R. E. 1975. Polarity of the cysJIH operon of Salmonella typhimurium. J. Gen. Microbiol. 86:275-282. [DOI] [PubMed] [Google Scholar]
- 18.Mansilla, M. C., D. Albanesi, and D. de Mendoza 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182:5885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxon, M. E., B. Redfield, X. Y. Cai, R. Shoeman, K. Fujita, W. Fisher, G. Stauffer, H. Weissbach, and N. Brot. 1989. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. USA 86:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Monroe, R. S., J. Ostrowski, M. M. Hryniewicz, and N. M. Kredich. 1990. In vitro interactions of CysB protein with the cysK and cysJIH promoter regions of Salmonella typhimurium. J. Bacteriol. 172:6919-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munier, H., A. M. Gilles, P. Glaser, E. Krin, A. Danchin, R. Sarfati, and O. Barzu. 1991. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 196:469-474. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 26.Ostrowski, J., G. Jagura-Burdzy, and N. M. Kredich. 1987. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 262:5999-6005. [PubMed] [Google Scholar]
- 27.Ostrowski, J., J. Y. Wu, D. C. Rueger, B. E. Miller, L. M. Siegel, and N. M. Kredich. 1989. Characterization of the cysJIH regions of Salmonella typhimurium and Escherichia coli. B. DNA sequences of cysI and cysH and a model for the siroheme-Fe4S4 active center of sulfite reductase hemoprotein based on amino acid homology with spinach nitrite reductase. J. Biol. Chem. 264:15726-15737. [PubMed] [Google Scholar]
- 28.Pasternak, C. A., R. J. Ellis, M. C. Jones-Mortimer, and C. E. Crichton. 1965. The control of sulphate reduction in bacteria. Biochem. J. 96:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Presecan-Siedel, E., A. Galinier, R. Longin, J. Deutscher, A. Danchin, P. Glaser, and I. Martin-Verstraete. 1999. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol. 181:6889-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 33.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 34.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 35.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 36.van der Ploeg, J. R., M. Barone, and T. Leisinger 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 37.van der Ploeg, J. R., M. Barone, and T. Leisinger 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 38.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]