Abstract
Transcription initiation with CTP is an uncommon feature among Escherichia coli σ70 promoters. The fis promoter (fis P), which is subject to growth phase-dependent regulation, is among the few that predominantly initiate transcription with CTP. Mutations in this promoter that cause a switch from utilization of CTP to either ATP or GTP as the initiation nucleotide drastically alter its growth phase regulation pattern, suggesting that the choice of the primary initiating nucleotide can significantly affect its regulation. To better understand what factors influence this choice in fis P, we made use of a series of promoter mutations that altered the nucleotide or position used for initiation. Examination of these promoters indicates that start site selection is determined by a combination of factors that include preference for a nucleotide distance from the −10 region (8 > 7 > 9 ≫ 6 ≫ 10 > 11), initiation nucleotide preference (A = G ≫ CTP ≥ UTP), the DNA sequence surrounding the initiation region, the position of the −35 region, and changes in the intracellular nucleoside triphosphate pools. We describe the effects that each of these factors has on start site selection in the fis P and discuss the interplay between position and nucleotide preference in this important process.
Fis is a small nucleoid-associated protein found in a growing number of bacterial species (5, 37). It is highly abundant during the early logarithmic growth phase in Escherichia coli cells that are grown in rich medium but significantly reduced during other stages of growth or during growth in minimal medium (3, 33, 34, 47). Several processes exist that regulate fis expression at the transcription level. Integration host factor binds a site centered 116 bp upstream of the transcriptional start site to stimulate transcription in vivo three- to fourfold (39). Fis causes about sixfold repression by binding to two specific sites in the fis promoter (fis P) region that overlap the RNA polymerase (RNAP) binding region (3, 34, 39). Conditions of stringent control result in an about fivefold reduction in fis expression (34, 54). However, the growth phase-dependent regulation pattern appears to operate independently of the above three regulatory mechanisms, suggesting that additional mechanisms account for this process (39, 54).
As a member of the σ70 family of promoters, fis P exhibits a reasonable match to the −10 consensus sequence (TAatAT; matches are capitalized) and a weak match to the −35 consensus sequence (TTtcat) located 17 bp upstream (54). Other features of this promoter include an A6 tract in the spacer region that is required for maximal transcriptional activity, a GC-rich discriminator sequence between the −10 region and the start site that is required for its response to stringent control, and use of CTP as the predominant transcription initiation nucleotide (3, 54). The latter is a fairly uncommon feature among σ70 promoters (13, 14, 16, 21, 43).
Mutation analysis demonstrated that the transcription initiation region of fis P plays a critical role in its growth phase-dependent regulation (54). Replacement of the predominant C with A or G, or of the adjacent downstream T with A, resulted in high levels of fis expression throughout the logarithmic growth and early stationary phases. In each of these mutant promoters, transcription initiation was largely restricted to the respective purine at these positions. Thus, the fis regulation pattern is affected by the precise nucleoside triphosphate (NTP) chosen to initiate transcription. The position of the start site also seems to be important. Transcripts initiating with GTP 6 bp downstream of the −10 region follow a strict growth phase-dependent expression pattern, whereas transcripts initiating with GTP 8 bp downstream of the −10 region show an altered pattern of regulation. This close connection between start site selection and growth phase-dependent regulation prompted us to examine more closely the factors that guide start site selection at fis P.
Several methods have been used to assign transcription start sites in a large number of compiled promoters of the σ70 family that include primer extension analysis, RNA sequencing, S1 nuclease mapping, high-resolution sizing of RNA transcripts generated in vitro, and estimates based on genetic or mutational promoter identification (13, 14, 16, 21, 43). The most reliable results, which come from primer extension and RNA sequencing analysis, suggest that most transcriptional start sites are found within 6 to 9 bp downstream of the −10 region but are sometimes found in the range of 4 to 12 bp downstream of this region. On the basis of the number of σ70 promoters reported to initiate with A, G, C, or U, a hierarchy of initiation NTP utilization may be generally described as follows: ATP > GTP ≫ UTP > CTP. Thus, it is generally anticipated that σ70 promoters will initiate transcription with ATP or GTP 6 to 9 bp downstream of the −10 region. However, careful examination of start site selection has been limited to a relatively small number of promoters (e.g., see references 6, 18, 22, and 46) and deviations from this generalization have been observed (5, 8, 15, 23, 45, 46, 55). Thus, a thorough understanding of the processes surrounding start site selection is needed and will necessitate the inclusion of a larger set of promoters in such studies.
In this report, we describe the results of several mutations affecting transcription initiation at fis P. We find that start site selection is influenced by factors that could be broadly classified as (i) the nucleotide sequences at or near the transcriptional start site, (ii) the distance between the −10 region and the start site, (iii) the distance between the −10 and −35 regions, and (iv) the cellular NTP pools. We show that, depending on the precise promoter sequence and cellular NTP pools, transcription is able to initiate with various efficiencies in the region from 6 to 11 nucleotides downstream of the −10 region, with 8 bp downstream serving as the optimal distance. The work presented suggests that the order of initiation nucleotide preference at fis P is ATP = GTP ≫ CTP ≥ UTP and that the position preference (given as the number of nucleotides from the −10 region) is 8 > 7 > 9 ≫ 6 ≫ 10 > 11. Other factors, such as the GC richness of the region immediately upstream of the start site and the sequence in the region immediately downstream of the start site, also affect start site selection. A general model describing start site selection at fis P and its potential influence on growth phase-dependent regulation is discussed.
MATERIALS AND METHODS
Chemicals and enzymes.
Chemicals were purchased from Sigma Chemical Co., Fisher Scientific Co., Life Technologies (Gibco BRL), or Pharmacia. Radioisotopes were from Amersham Life Sciences. Enzymes were from New England Biolabs or Roche Molecular Biochemicals. Oligonucleotides were purchased from Qiagen Operon (http://www.operon.com).
Growth media and culture conditions.
Growth media were all from Difco Laboratories. Cultures were grown at 37°C in LB medium (44) or in N−C− medium (1) supplemented with 10 mM NH4Cl, 0.4% (wt/vol) glucose, 0.015 mM thiamine, 1 mM arginine, and either 1 mM uracil or 0.25 mM UMP (56). When pertinent, 100 μg of ampicillin per ml, 50 μg of kanamycin per ml, or 12 μg of tetracycline per ml was added to the growth medium. For β-galactosidase assays (28), triplicate saturated cultures of RJ1561 transformed with the various plasmids used in this work were grown in LB medium at 37°C with constant shaking for 90 min as previously described (54).
Bacterial strains and plasmids.
Transformants of strain RJ1561 [F− Δ(lac pro) thi ara str recA56 srl fis::767] (19) were used to examine start site selection in cells grown in LB. For experiments examining the effects of nucleotide concentrations on start site selection, we used RO934 (CLT42 fis::767 recA srl::Tn10), which was constructed from CLT42 (MC4100 car-94) by P1 transduction. CLT42 was kindly supplied by C. L. Turnbough, University of Alabama at Birmingham (42). First, neo-interrupted fis (fis::767) was transduced from RJ1617 (MC1000 fis::767) into CLT42. The resulting kanamycin-resistant strain was subsequently transduced with P1 phage grown in RJ1086 (MC1000 recA56 srl::Tn10). recA transductants were selected by their resistance to tetracycline and further screened by their sensitivity to UV light.
Most of the plasmids carrying the various fis P sequences used in this work were described previously (54). Essentially, they contain the wild-type or mutant fis P region from −108 to +105 inserted within the unique HindIII and EcoRI sites of the pUC18 polylinker region of pRJ800 (3), which contains a promoterless trp::laz fusion next to the EcoRI site and confers ampicillin resistance. Two additional plasmids carrying fis P mutations, pKW381 (7C→A) and pKW382 (6 to 9 GCCT→AAAA; hereafter referred to as A4), were constructed and characterized in a similar fashion. The plasmid carrying the P. vulgaris fis P region from −172 to +78 (pMB262) was described previously (5). This promoter is similar to that of E. coli in the region from −38 to +10, with three nucleotide differences in the spacer region (−30C→A, −27G→T, and −26Τ→G) and two nucleotide differences downstream of the −10 region (5C→G and 6G→C).
PCR and DNA sequencing reactions.
A standard PCR was conducted under conditions specified by the supplier (Roche Molecular Biochemicals). Site-directed mutations were generated as previously described (54), by a megaprimer PCR method (4). The mutations were confirmed by dideoxy nucleotide sequencing with alkali-denatured double-stranded DNA and Sequenase version 2.0 under conditions specified by the supplier (U.S. Biochemicals).
Primer extension analysis.
Primer extension reactions were performed as previously described (54). Saturated cultures were diluted into fresh LB medium to an optical density at 600 nm (OD600) of 0.2 and grown at 37°C for 90 min with shaking, after which a sample was collected for total-RNA extraction (7). When N−C− medium was used, saturated cultures were diluted to an OD600 of 0.1 and grown until the OD600 was approximately 0.3, at which time a sample was collected for total-RNA extraction. The reactions were performed with 10 μg of total RNA (or 500 ng of total RNA when P. vulgaris fis P was analyzed), 1 pmol of a 32P-end-labeled primer (oRO109 for E. coli fis P and oRO226 for P. vulgaris fis P) that hybridized to the fis P region from +56 to +38, and 2 U of avian myeloblastosis virus reverse transcriptase (Roche Molecular Biochemicals) in the supplied buffer. Primer-extended products were separated on 8% polyacrylamide-8 M urea gels and visualized by autoradiography. Quantitative analysis was conducted with a Storm 860 PhosphorImager and ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, Calif.).
RESULTS
Nucleotide preferences at fis P.
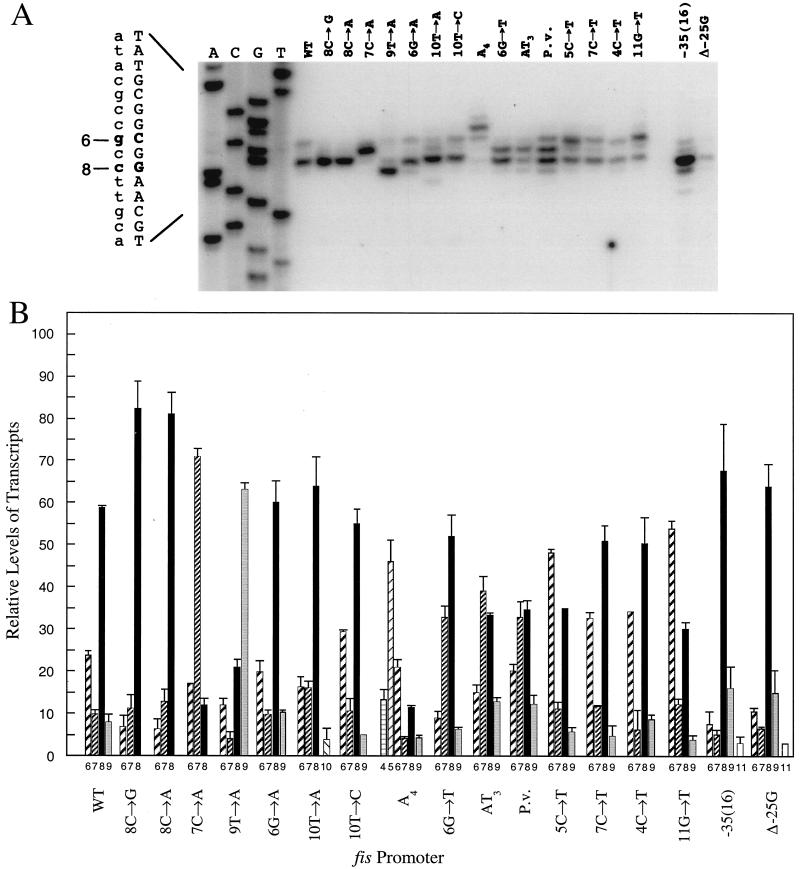
Quantitation of primer-extended products from transcripts generated by wild-type fis P indicated that transcription is able to initiate at four positions with widely different efficiencies (Fig. 1 and 2). The primary transcript initiated with CTP 8 bp downstream of the −10 region (8C) and was responsible for about 59% of the transcripts generated from this promoter. A secondary site, generating about 23% of the transcripts, occurred with GTP at position 6 (6G), while only 10% of the transcripts initiated with the C at position 7 (7C) and 8% initiated with UTP at position 9 (9T). Replacement of 8C with G (8C→G) resulted in 82% of the transcripts initiating at 8G and 11 and 7% initiating at 7C and 6G, respectively. A similar result was observed when 8C was replaced with A (8C→A), with 81% of the transcripts initiating with 8A and 13 and 6% initiating at 7C and 6G, respectively. Thus, either purine present at position 8 similarly increased the frequency of initiation from position 8 compared to that from positions 6 and 9 (Fig. 1B). More dramatic examples of nucleotide preferences were seen with fis P mutants 7C→A and 9T→A. The relative frequency of initiation at position 7 increased from 10% in the wild-type promoter to 71% in the 7C→A mutant, with only 17% at 6G and 12% at 8C. While only 8% of the total transcripts initiated at 9T in the wild-type promoter, 63% initiated at 9A in the 9T→A promoter, with 21% initiating at 8C, 4% initiating at 7C, and 12% initiating at 6G. The results obtained with all of these promoter variants clearly show that purines are more highly preferred than pyrimidines as transcription initiation nucleotides in fis P, an effect that is more noticeable at positions 7 and 9 than at position 8.
FIG. 1.
Effects of mutations on start site selection at fis P. (A) Results from primer extension reactions with 1 pmol of 32P-labeled oRO109 primer and 10 μg of total RNA from RJ1561 cells transformed with fis P-containing plasmids or 1 pmol of oRO226 and 500 ng of total RNA when the P. vulgaris (P.v.) fis P was analyzed. Cells were cultured in LB medium at 37°C for 90 min and then harvested for total-RNA preparation. Primer-extended products were separated on urea-denaturing polyacrylamide gels and subjected to autoradiography. The fis promoters examined are indicated above the lanes. Dideoxy sequencing reactions (A, C, G, and T) for wild-type (WT) fis P are shown in the first four lanes on the left. They were performed with 32P-labeled oRO109, which gave comparable DNA sequencing ladders for all of the mutant promoters, allowing direct determination of start site positions. Primer oRO226 anneals to the same downstream promoter region in the P. vulgaris promoter as oRO109 in the E. coli promoter such that their resulting transcript sizes can be directly compared. The DNA sequence of the antisense strand, as read from the gel, is represented on the left by uppercase letters, and the deduced complementary sequence of the sense strand is represented by lowercase letters. The two primary start sites in the wild-type promoter (positions 8 and 6) are in boldface. (B) The percentage of transcripts from each promoter was quantified from the results of three primer extension assays on a total-RNA preparation made from each bacterial culture carrying a different fis P on a plasmid. The primer-extended signals were quantified with a PhosphorImager and ImageQuant software. Standard deviations are indicated by vertical error bars. The promoters analyzed and the mapped start site positions are indicated beneath the bars.
FIG. 2.
fis P sequences and their transcription initiation sites. Each promoter's transcriptional start sites are indicated by arrows that qualitatively depict their relative frequencies of use within each promoter, according to the results in Fig. 1. Only transcripts representing greater than 10% of the total are represented. Nucleotides in bold represent the changes from wild-type E. coli fis P. The numbers at the top indicate the distances from the −10 region (position numbers). Mutations are listed on the left. The bottom two sequences depict the transcription initiation pattern of fis P variants containing mutations outside the initiation region. The −35(16) fis P contains a perfect match to the −35 consensus sequence (TTGACA) positioned 16 bp upstream of the −10 region. The Δ−25G fis P carries a deletion of −25G that shortens the wild-type spacer region from 17 to 16 bp. P.v. is the wild-type fis P from P. vulgaris. As suggested in the text, the arrows in the A4 promoter may not represent the correct initiation sites. Relative promoter activities are shown on the right. β-Galactosidase assays were performed with RJ1561 cells carrying the respective fis promoters fused to the trp::lacZ in pRJ800. Most of the results have been previously reported (5, 54). The activity of the wild-type (WT) promoter is assigned a value of 1.00, and all other promoter activities are shown relative to this one.
Replacement of 6G with A did not result in significant changes in start site preference compared to the wild-type promoter (Fig. 1A and B). Hence, the preference for GTP appears to be equivalent to that for ATP as an initiation nucleotide at either position 6 or 8. A small preference for CTP over UTP could be detected at position 6 but not at position 7. About 9 and 15% of the transcripts initiated at 6T in the 6G→T and AT3 promoters, respectively, whereas 20% initiated at 6C in the Proteus vulgaris promoter (Fig. 1B and 2). However, no preference among pyrimidines could be reliably discerned at position 7. About 10% of the transcripts initiated at 7C in the wild-type promoter, while 12% initiated at 7T in the 7C→T mutant promoter. These differences are near the margin of experimental error (1.5%) and are thus of little or no significance. The small preference for CTP appears to be position dependent. On the basis of our results, the overall initiation NTP preference at fis P may be represented as follows: ATP = GTP ≫ CTP ≥ UTP.
Position preferences at fis P.
Despite the general preference for initiation with purines, initiation at 8C was much more efficient than initiation at 6G or 6A (Fig. 1 and 2). This suggests that position 8 is more highly favored than position 6 for transcription initiation. To better evaluate the position preference, we examined mutant promoters with an A substitution at each of positions 6 through 10. Initiation with ATP accounted for 20% of the total fis P transcripts at position 6 (6G→A), 71% of those at position 7 (7C→A), 81% of those at position 8 (8C→ A), 63% of those at position 9 (9T→A), and 4% of those at position 10 (10T→A). Thus, the position preference at fis P may be represented as follows: 8 > 7 > 9 ≫ 6 ≫ 10.
Naturally occurring G nucleotides are found at positions 3 and 11 (Fig. 2), but we have never observed initiation from position 3 and only in two distinctive mutant promoters [−35(16) and Δ−25G] were we able to detect a very small proportion of transcripts initiating at 11G (Fig. 1B). This suggests that positions 3 and 11 are not ordinarily used as start sites. In P. vulgaris fis P, which contains a G at position 5, no appreciable levels of transcript initiating at 5G could be observed. Thus, transcription initiation appears to be mostly confined to positions 6 through 9, with positions 8 and 7 serving as the most highly preferred. Positions 10 and 11 serve as very weak sites detected only in a few mutant promoters. Outside the region between positions 6 and 11, transcription initiation was not detected.
Effect of T in the fis P initiation region.
In several promoters, a T nucleotide was placed at a position upstream of the primary start site in place of a C or G in each case. Each of these substitutions resulted in an increase in the frequency of initiation at a nearby downstream position. For example, replacement of 5C with a T resulted in a dramatic shift in the predominant initiation site from 8C to 6G, which accounted for 48% of the transcripts in the 5C→T promoter (Fig. 1 and 2). In the 4C→T mutant, the proportion of transcripts initiating at 6G (34%) also increased compared to that in the wild-type promoter (23%), although not as much as that in the 5C→T mutant. A stretch of T nucleotides at positions 4, 5, and 6 (AT3 promoter) also caused a notable shift in the predominant initiation site, in this case from 8C to 7C, and the 6G→T mutation caused an increase in initiation at 7C compared to the wild-type promoter. In the 7C→T promoter, a modest increase in the proportion of the transcripts was observed at a neighboring upstream position. About 33% of its transcripts initiated at 6G, compared to 23% in the wild-type promoter. These results suggest that the presence of a T nucleotide in the initiation region can significantly affect start site selection.
There was one mutation downstream of the primary start site (11G→T) that also affected the relative transcript levels from this promoter. In this case, the predominant transcript initiated at 6G rather than at 8C (Fig. 1).
Effect of core promoter structure of fis P.
Mutations reducing the distance between the −35 and −10 regions also modified the efficiency of initiation sites. A triple mutation (−33G, −31C, and −30A) created a perfect match to the consensus −35 region positioned 16 bp upstream of the −10 region [−35(16) fis P], thus shortening the spacer region by 1 bp (54). This mutant promoter resulted in higher frequencies of initiation at 8C, 9T, and 11G and a reduced frequency of initiation at 6G compared to the wild-type promoter (Fig. 1). A similar effect on start site selection was caused by a deletion of 1 bp in the spacer region of the wild-type promoter (Δ−25G). The −35 (16) promoter results in a greater-than-threefold increase in transcription, while the Δ−25G promoter results in about a threefold decrease relative to the wild-type promoter (Fig. 2). Thus, the similar effects of these promoters on position preference were independent of their effects on transcriptional activity and were most likely due to their shortened spacer lengths.
Effect of relative NTP concentrations on fis P.
We had previously observed that cells grown in M9 salts with 0.2% glucose showed a modest increase in the proportion of wild-type fis P transcripts initiating at 6G compared to the same cells grown in LB (54). In addition, transcripts initiating at 10A in the 10T→A promoter were more prominent in cells growing in M9-glucose than in cells growing in LB. Although several explanations for these effects might be conceived, we explored the possibility that changes in cellular NTP pools under different growth conditions might affect start site selection in fis P.
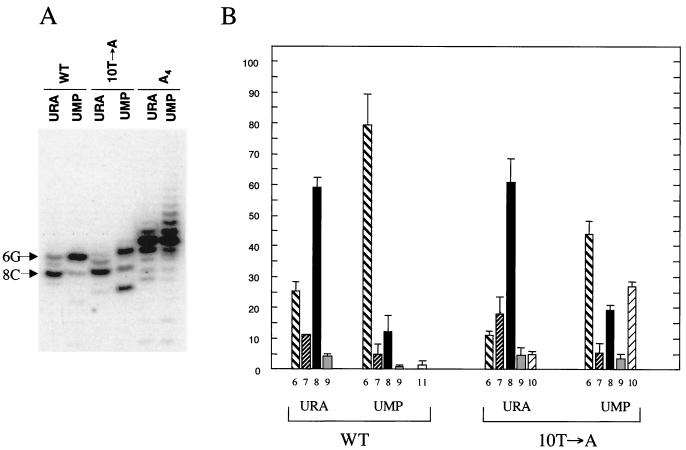
To examine the effects of various NTP concentrations on start site selection, we transformed strain RO934 with plasmids carrying either the wild-type or mutant fis P. This pyrimidine auxotroph can be cultured in growth medium supplemented with UMP to create conditions of pyrimidine limitation and purine excess or with uracil to approach normal intracellular NTP concentrations (38). The proportions of transcripts initiating at 8C (59%) and 6G (26%) in wild-type fis P when RO934 is grown in uracil (Fig. 3) are comparable to those observed when RJ1561 is grown in LB (Fig. 1). However, growth of RO934 in UMP resulted in a notable switch in start site preference from 8C (only 12%) to 6G (80%) (Fig. 3). A very small proportion of transcripts initiating at 11G (2%) could also be detected in the latter. In the 10T→A promoter, the percentage of transcripts initiating at 10A was improved from 5% in cells grown in uracil to 27% in cells grown in UMP (Fig. 3B). Additional effects on this promoter from growth in UMP include increased initiation at position 6 (from 11 to 47%) and decreased initiation at position 8 (from 61 to 19%). These results strongly suggest that start site selection at fis P can be tailored to the availability of intracellular NTPs.
FIG. 3.
Effect of altered NTP pools on start site selection at fis P. (A) Results of primer extension reactions with 10 μg of total RNA from RO934 transformed with plasmids containing either the wild-type (WT), 10T→A, or A4 fis P. Cells were cultured in N−C− medium containing either uracil (URA) or 5′-UMP. Signals resulting from initiation at 6G or 8C in the wild-type promoter are indicated on the left. (B) The percentage of each transcript was determined for the wild-type and 10T→A fis promoters from two primer extension assays on a total-RNA preparation made from each bacterial culture carrying one of the fis promoters on a plasmid. Quantification was done as described in the legend to Fig. 1. The fis P's analyzed and the mapped start site positions are indicated beneath the bars.
The A4 promoter, in which the four nucleotides at positions 6 through 9 were replaced with A, gave a predominant transcript with a length that corresponded to initiation at 5C when RJ1561 was grown in LB (Fig. 1A and 2) or when RO934 was grown in uracil (Fig. 3A, A4 URA). This was surprising, given the choices for initiation with ATP at the more highly preferred positions 8, 7, and 9. We reasoned that if this promoter were actually relying on CTP to initiate transcription from 5C, then growth under pyrimidine-limiting/purine excess conditions would likely bring about a shift toward initiation with ATP from a more highly preferred position in the region between positions 6 and 9. Instead, however, we continued to observe the same predominant signal with a length corresponding to that of a transcript initiating at 5C that was now accompanied by a ladder of larger transcripts varying in size by one-base increments (Fig. 3A). This ladder of bands is strongly suggestive of reiterative transcription, a process whereby nucleotides are repetitively added to the 3′ end of a nascent transcript (12, 49, 52, 58). The reiterative process is most likely enhanced by the large ATP pools found in cells cultured in UMP, which may facilitate incorporation of ATP molecules into the nascent mRNA (38).
DISCUSSION
Nucleotide preference at fis P.
The preference for purines over pyrimidines as initiation nucleotides was demonstrated at fis P by comparing the effects that different NTPs have when placed at positions 6 through 10. At each of these positions, purines were preferred over pyrimidines. Positions 6 and 10 seem too poor to allow predominant initiation with a purine, while position 8 seems good enough to favor adequate use of CTP such that the preference for purines at this position becomes less conspicuous than that at 7 or 9. The preference for purines as initiation NTPs has been widely observed in other promoters (13, 14, 18, 22) and may result from the combined effects of higher concentrations of purines than pyrimidines in vivo (31, 36, 38, 50, 53) and the greater affinity that RNAP exhibits for purines than pyrimidines at its initiation nucleotide binding site (2, 30, 41, 57). A slight preference was noted in fis P for CTP over UTP at position 6 but not at the more favorable position 7, consistent with the notion that NTP discrimination may be better detected at less favored initiation positions. On the basis of our observations, we describe the NTP preference at fis P as follows: ATP = GTP ≫ CTP ≥ UTP. A somewhat different assessment has been made in the case of the pyrC promoter, in which UTP is seemingly preferred over CTP and ATP is preferred over GTP as an initiation nucleotide (22).
Position preference at fis P.
Predominant initiation with ATP was observed only when the A nucleotide was placed at position 7, 8, or 9, strongly suggesting that these are the most highly preferred positions. Initiation from position 8 with either ATP or GTP gave the greatest proportion of transcripts measured in any of the promoters analyzed (81 or 82%), followed by position 7 (71%) and then position 9 (63%). G or A at position 6 normally served as a secondary initiation site, resulting in 23 and 20% of the total transcripts, respectively, and giving way to a greater preference for CTP at position 8. Initiation with ATP from position 10 (10Τ→A) was very inefficient, and initiation from 11G was rarely observed. Thus, we summarize our observations regarding position preference as follows: 8 > 7 > 9 ≫ 6 ≫ 10 > 11. In other promoters, such as those of pyrC and lacUV5 (18, 22), position 7 is the most strongly preferred, followed by positions 8, 6, 9, and 10. Thus, a unique hierarchy of position preference does not apply to all σ70 promoters. Clearly, sequences surrounding the initiation site, and even upstream of the −10 region, are able to influence the position preference.
It is possible that the relative proportions of some of the fis P transcripts may be influenced by differences in RNA decay rates that might result from differences in their 5′ termini. Effects of 5′ termini on RNA decay rates generally depend on whether they participate in a stable secondary structure or remain unpaired (10, 20, 22, 24); the nucleotide composition of the 5′ terminus itself appears to have little or no effect (9, 48, 51). RNA secondary-structure predictions (with M-fold software) suggest that the 5′ termini of fis P transcripts initiating at positions 7, 8, and 9 remain unpaired, irrespective of the initiating nucleotide. The 5′ termini of transcripts initiating with 6G can potentially participate in a moderately stable secondary structure. However, our work showed that replacement of 6G with A, which impairs this putative structure, does not affect the proportion of transcripts observed. Thus, we believe it is unlikely that the various fis P transcripts will exhibit structural differences at their 5′ termini that could result in substantial changes in degradation rates.
Influence of upstream T nucleotides on start site selection.
We observed that the presence of T nucleotides upstream of the start site enhanced initiation preference at neighboring positions. Examples of these effects were seen with promoters 5C→T, 6G→T, and AT3 (Fig. 1). A reduction in the GC richness upstream of the initiation site could facilitate or help stabilize DNA melting in this region during open-complex formation. This could, in turn, increase the opportunity for initiation at a site neighboring the T nucleotide(s). Introduction of A nucleotides would have a similar effect on melting, but the strong preference for ATP tends to favor its use rather than that of a neighboring pyrimidine. A similar effect of T nucleotides upstream of the initiation site could also be observed in the lacUV5 promoter (18). While the 7A→C mutation in the lacUV5 promoter resulted in predominant initiation at 8A (or +2A, according to the authors' numbering), the double mutation of 5G→T and 7A→C caused a sizeable shift toward 6G (or −1G) as the predominant initiation site.
Effect of downstream sequences on start site preference.
Sequences downstream of the start site can also affect the proportion of transcripts initiating at various start sites, most likely for reasons different from those of mutations upstream of the start site. For instance, 11G→T caused a notable shift in predominant initiation from 8C to 6G (Fig. 1A and B). We suspect that this downstream change might somehow reduce the efficiency of elongation from 8C such that initiation from 6G prevails. The mutation changed the initial tetrameric transcript starting at position 8 from CUUG to CUUU. The stretch of three U nucleotides immediately following initiation may result in weak base pairing between the nascent transcript and the DNA template strand (26) such that extension of the nascent transcript may become inefficient. Nonproductive events, such as abortive initiation (25), RNAP backtracking (35), and reiterative transcription (12, 49, 58), are among the processes that could result in less efficient elongation from 8C. A stretch of four A nucleotides in the fis P initiation region (A4) gave rise to a ladder of transcripts under conditions of purine excess, suggesting that reiterative transcription may occur at fis P, provided there is a stretch of three or more A or U nucleotides in the initiation region. In the 11G→T promoter, initiation at 6G may be more efficiently elongated than that at 8C because of the stability provided by the GCC trinucleotide in the nascent transcript (35).
Influence of promoter structure on start site selection.
Two different kinds of promoter mutations [−35(16) and Δ−25G] altered the promoter structure, shortening the spacer region by 1 bp. In both cases, a similar increase in initiation preference was observed at 8C, 9T, and 11G while a decrease occurred at 6G. We attribute the shifts in initiation preference to the shortened spacer regions, which is the only effect the two mutations have in common. We have not determined if the distance between the −35 and −10 regions or the distance between the −35 region and the initiation region is the most critical factor. Nevertheless, this suggests that interactions between the RNAP and the −35 region play an important role in establishing start site position preference. Other examples of mutations in the −35 and spacer regions, or the length of the spacer region, affecting start site selection have been observed (6, 17, 29), but the mechanisms behind these effects remain elusive.
Intracellular NTP concentrations.
In cells grown under conditions of limited pyrimidines and excess purines, wild-type fis P exhibits a substantial shift in start site preference from 8C to 6G and the 10T→A mutant exhibits a shift from 8C to 6G and 10A. Thus, while position preference is a primary determinant in start site selection, changes in NTP availability can affect the range of positions that may be efficiently used.
Alteration of intracellular NTP concentrations has been shown to affect start site selection at several other promoters of genes involved in pyrimidine metabolism (22, 40, 46, 49). For example, the pyrC promoters of Salmonella enterica serovar Typhimurium and E. coli initiate with CTP when pyrimidine levels are sufficient but switch to initiation with GTP 2 bases downstream when pyrimidine levels are limiting (22, 46). The data from this and other, similar, work indicate that the switching process appears to require the following three conditions: (i) a poor initiation nucleotide located at the optimal position, (ii) a good initiation nucleotide a short distance from the optimal position, and (iii) limiting concentrations of the nucleotide needed for initiation at the optimal position. All three of these conditions are met by fis P.
It has been suggested that formation of the first dinucleotide bond is a critical step in the initiation process and that sufficient levels of the first two nucleotides are required (11, 27, 32). For example, low UTP concentrations at the E. coli codBA and upp promoters result in a shift in initiation from A-U to G-A, where UTP is no longer the second nucleotide added (40, 49). Thus, if NTP levels are not sufficient for formation of the first dinucleotide bond, alternative start sites may be used. Growth of a car strain with UMP significantly reduces the UTP pool but has a relatively smaller effect on the CTP pools (38, 53). Such conditions might hinder initiation with C-U at position 8 more severely than with G-C at position 6.
Model for start site selection.
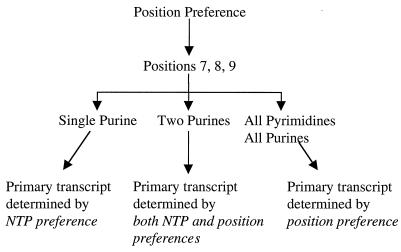
How does RNAP weigh the nucleotide preference against the position preference to select its primary initiation site? Upon open-complex formation at wild-type fis P, position 8 is most likely the nearest one to the RNAP initiation site such that the greatest preference is given to this position. If a purine is present at this position, then initiation takes place with such high efficiency that little opportunity is available for probing of alternate sites. If, instead, a pyrimidine is present at this position, the relatively lower concentration of pyrimidines in vivo, together with their lower affinity for the RNAP initiation site, may cause the holoenzyme to hesitate at the initiation step. This offers an opportunity for RNAP to probe adjacent positions 7 and 9. If a purine is present at either of these positions, then rapid initiation would ensue, superseding the use of a pyrimidine at position 8. Otherwise, initiation with pyrimidine at position 8 remains the predominant choice. Thus, the dominating factor in start site selection at fis P is the preference for the region encompassing positions 7 through 9 in the order 8 > 7 > 9 (Fig. 4). NTP preference serves mainly to influence selection of the primary start site among these three positions. If this region consists of all purines or all pyrimidines, such that NTP preference cannot be used as a discriminating factor, then preference for position 8 will prevail. If one purine is available in this region, it will be chosen as the primary start site. If two or three purines are available in this region, then we predict that they will be used with a relative efficiency that is guided by their position preference.
FIG. 4.
Determination of primary transcripts. Position preference initially guides RNAP to positions 8, 7, and 9, in that order. Depending on the nucleotides present at these positions, either NTP preference, position preference, or both will aid in determining which site(s) is chosen for initiation.
Once the predominant start site is selected in the region including positions 7 through 9, then the remaining two positions in this region may serve as secondary start sites. In fis P, position 6 is also used as a secondary start site and the frequency with which it is chosen is determined by NTP preference and availability. Because selection of one site necessarily excludes that of alternate sites within the same initiation event, the efficiency with which such secondary initiation events occur will be inversely related to the efficiency of initiation at the primary site. Structural constraints may hinder the probing of positions as they become farther from position 8. However, as we have seen, the precise range of positions used for initiation can be influenced by factors such as NTP availability (Fig. 3), promoter structure (Fig. 1), and, as has been suggested, the strength of the RNAP-promoter interactions around the initiation region (6).
Start site selection and growth phase-dependent regulation.
We had previously observed that initiation with 8A, 8G, and 9A profoundly altered the growth phase-dependent regulation pattern (54). The work presented here indicates that such transcription initiation conditions are among the most favored at fis P. Given that the high NTP preference and optimal (or nearly optimal) position preference requirements are both met in these cases, efficient transcription initiation ensues. We envision that, because of the relatively large intracellular pools of purines and their relatively lower Km as initiation NTPs (compared to pyrimidines), efficient transcription initiation with purines at optimal positions in fis P will be sustained throughout the logarithmic growth phase. However, if the intracellular pools of the initiating NTP were to become limiting, then the initiation process could become affected by changes in NTP pools during logarithmic growth, a condition that may lead to the growth phase-dependent expression pattern of fis P. One way in which initiating NTP levels may become limiting is if pyrimidines serve as the predominant initiating nucleotide. Another way is to initiate with a purine from a poor position.
fis P employs a poor initiating NTP (CTP) at an optimal position and a good initiating NTP (GTP) at a poor position (position 6). Both transcripts obey a growth phase-dependent expression pattern. We have recently observed that replacing 8C with a T (T. S. Pratt and R. Osuna, unpublished results) or 6G with an A had almost no effect on the growth phase-dependent regulation pattern (data not shown). However, as could be predicted from our observations, both mutants 7C→A and A4 showed altered growth phase-dependent regulation patterns comparable to those of 8C→A and 8C→G (data not shown). Thus, the growth phase-dependent regulation pattern normally exhibited by fis P requires that initiation efficiency be compromised in some way. We suggest that this may be achieved by use of a poor initiating NTP, use of a poor starting position, or both.
Acknowledgments
We are grateful to Carey L. Atkins for the construction of several of the fis P mutants used in this work. We also thank Timothy S. Pratt for useful comments.
This work was supported with funds from Public Health Service grant GM52051.
REFERENCES
- 1.Alper, M. D., and B. N. Ames. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J. Bacteriol. 133:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, D. D., D. A. Goldthwait, and C. W. Wu. 1969. Studies with the ribonucleic acid polymerase. II. Kinetic aspects of initiation and polymerization. Biochemistry 8:246-256. [DOI] [PubMed] [Google Scholar]
- 3.Ball, C. A., R. Osuna, K. C. Ferguson, and R. C. Johnson. 1992. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 174:8043-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barik, S., and M. Galinski. 1991. “Megaprimer” method of PCR: increased template concentration improves yield. BioTechniques 10:489-490. [PubMed] [Google Scholar]
- 5.Beach, M. B., and R. Osuna. 1998. Identification and characterization of the fis operon in enteric bacteria. J. Bacteriol. 180:5932-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpousis, A. J., J. E. Stefano, and J. D. Gralla. 1982. 5′ nucleotide heterogeneity and altered initiation of transcription at mutant lac promoters. J. Mol. Biol. 157:619-633. [DOI] [PubMed] [Google Scholar]
- 7.Case, C. C., S. M. Roels, J. E. Gonzalez, E. Simons, and R. Simons. 1988. Analysis of the promoters and transcripts involved in IS10 anti-sense transcriptional RNA control. Gene 72:219-236. [DOI] [PubMed] [Google Scholar]
- 8.de Boer, H., and M. Nomura. 1979. In vivo transcription of rRNA operons in Escherichia coli initiates with purine nucleoside triphosphates at the first promoter and with CTP at the second promoter. J. Biol. Chem. 254:5609-5612. [PubMed] [Google Scholar]
- 9.Ehretsmann, C. P., A. J. Carpousis, and H. M. Krisch. 1992. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 6:149-159. [DOI] [PubMed] [Google Scholar]
- 10.Feng, Y., and S. N. Cohen. 2000. Unpaired terminal nucleotides and 5′ monophosphorylation govern 3′ polyadenylation by Escherichia coli poly(A) polymerase I. Proc. Natl. Acad. Sci. USA 97:6415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gourse, R. L. 1988. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 16:9789-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, X., and C. L. Turnbough, Jr. 1998. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heck, J. D., and G. W. Hatfield. 1988. Valyl-tRNA synthetase gene of Escherichia coli K12. Molecular genetic characterization. J. Biol. Chem. 263:857-867. [PubMed] [Google Scholar]
- 16.Hershberg, R., G. Bejerano, A. Santos-Zavaleta, and H. Margalit. 2001. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 29:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquet, M. A., R. Ehrlich, and C. Reiss. 1989. In vivo gene expression directed by synthetic promoter constructions restricted to the −10 and −35 consensus hexamers of E. coli. Nucleic Acids Res. 17:2933-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong, W., and C. Kang. 1994. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 22:4667-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. C., C. A. Ball, D. Pfeffer, and M. I. Simon. 1988. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc. Natl. Acad. Sci. USA 85:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kushner, S. R. 1996. mRNA decay, p. 849-860. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 21.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, J., and C. L. Turnbough, Jr. 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 176:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machida, C., Y. Machida, and E. Ohtsubo. 1984. Both inverted repeat sequences located at the ends of IS1 provide promoter functions. J. Mol. Biol. 177:247-267. [DOI] [PubMed] [Google Scholar]
- 24.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 25.Martin, C. T., D. K. Muller, and J. E. Coleman. 1988. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry 27:3966-3974. [DOI] [PubMed] [Google Scholar]
- 26.Martin, F. H., and I. Tinoco, Jr. 1980. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 8:2295-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClure, W. R., C. L. Cech, and D. E. Johnston. 1978. A steady state assay for the RNA polymerase initiation reaction. J. Biol. Chem. 253:8941-8948. [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Mulligan, M. E., J. Brosius, and W. R. McClure. 1985. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J. Biol. Chem. 260:3529-3538. [PubMed] [Google Scholar]
- 30.Naryshkina, T., A. Mustaev, S. A. Darst, and K. Severinov. 2001. The beta′ subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J. Biol. Chem. 276:13308-13313. [DOI] [PubMed] [Google Scholar]
- 31.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 445-473. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 1st ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 32.Nierman, W. C., and M. J. Chamberlin. 1979. Studies of RNA chain initiation by Escherichia coli RNA polymerase bound to T7 DNA. Direct analysis of the kinetics and extent of RNA chain initiation at T7 promoter A1. J. Biol. Chem. 254:7921-7926. [PubMed] [Google Scholar]
- 33.Nilsson, L., H. Verbeek, E. Vijgenboom, C. van Drunen, A. Vanet, and L. Bosch. 1992. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J. Bacteriol. 174:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ninnemann, O., C. Koch, and R. Kahmann. 1992. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J. 11:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33-41. [DOI] [PubMed] [Google Scholar]
- 36.O'Donovan, G. A. 1970. Nucleotide pool changes in mutants of Escherichia coli. Biochim. Biophys. Acta 209:589-591. [DOI] [PubMed] [Google Scholar]
- 37.Osuna, R., D. Lienau, K. T. Hughes, and R. C. Johnson. 1995. Sequence, regulation, and functions of fis in Salmonella typhimurium. J. Bacteriol. 177:2021-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulsen, P., and K. F. Jensen. 1987. Effect of UTP and GTP pools on attenuation at the pyrE gene of Escherichia coli. Mol. Gen. Genet. 208:152-158. [DOI] [PubMed] [Google Scholar]
- 39.Pratt, T. S., T. Steiner, L. S. Feldman, K. A. Walker, and R. Osuna. 1997. Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J. Bacteriol. 179:6367-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi, F., and C. L. Turnbough, Jr. 1995. Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 254:552-565. [DOI] [PubMed] [Google Scholar]
- 41.Record, M. T., Jr., W. S. Reznikoff, M. L. Craig, K. L. McQuade, and P. J. Schlax. 1996. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation, p. 792-821. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 42.Roland, K. L., F. E. Powell, and C. L. Turnbough, Jr. 1985. Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 163:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg, M., and D. Court. 1979. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 13:319-353. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Shinagawa, H., K. Makino, M. Amemura, S. Kimura, H. Iwasaki, and A. Nakata. 1988. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J. Bacteriol. 170:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen, K. I., K. E. Baker, R. A. Kelln, and J. Neuhard. 1993. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J. Bacteriol. 175:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. F., L. Moitoso de Vargas, C. Koch, R. Kahmann, and A. Landy. 1987. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell 50:901-908. [DOI] [PubMed] [Google Scholar]
- 48.Toivonen, J. E., and D. P. Nierlich. 1974. Biological decay of the 5′-triphosphate termini of the RNA of E. coli. Nature 252:74-76. [DOI] [PubMed] [Google Scholar]
- 49.Tu, A. H., and C. L. Turnbough, Jr. 1997. Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J. Bacteriol. 179:6665-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel, U., S. Pedersen, and K. F. Jensen. 1991. An unusual correlation between ppGpp pool size and rate of ribosome synthesis during partial pyrimidine starvation of Escherichia coli. J. Bacteriol. 173:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Gabain, A., J. G. Belasco, J. L. Schotte, and A. C. Y. Chang. 1983. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc. Natl. Acad. Sci. USA 80:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, L. A., R. B. Weiss, R. Driscoll, D. S. Dunn, and R. F. Gesteland. 1990. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 18:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker, K. A. 2001. Ph.D. dissertation. State University of New York, Albany.
- 54.Walker, K. A., C. L. Atkins, and R. Osuna. 1999. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J. Bacteriol. 181:1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe, W., G. Sampei, A. Aiba, and K. Mizobuchi. 1989. Identification and sequence analysis of Escherichia coli purE and purK genes encoding 5′-phosphoribosyl-5-amino-4-imidazole carboxylase for de novo purine biosynthesis. J. Bacteriol. 171:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson, H. R., C. D. Archer, J. K. Liu, and C. L. Turnbough, Jr. 1992. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J. Bacteriol. 174:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, C. W., and D. A. Goldthwait. 1969. Studies of nucleotide binding to the ribonucleic acid polymerase by equilibrium dialysis. Biochemistry 8:4458-4464. [DOI] [PubMed] [Google Scholar]
- 58.Xiong, X. F., and W. S. Reznikoff. 1993. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J. Mol. Biol. 231:569-580. [DOI] [PubMed] [Google Scholar]