Abstract
Mutations causing rifampin resistance in vegetative cells of Bacillus subtilis 168 have thus far been mapped to a rather restricted set of alterations at either Q469 or H482 within cluster I of the rpoB gene encoding the β subunit of RNA polymerase. In this study, we demonstrated that spores of B. subtilis 168 exhibit a spectrum of spontaneous rifampin resistance mutations distinct from that of vegetative cells. In addition to the rpoB mutations Q469K, Q469R, and H482Y previously characterized in vegetative cells, we isolated a new mutation of rpoB, H482R, from vegetative cells. Additional new rifampin resistance mutations arising from spores were detected at A478N and most frequently at S487L. The S487L change is the predominant change found in rpoB mutations sequenced from rifampin-resistant clinical isolates of Mycobacterium tuberculosis. The observations are discussed in terms of the underlying differences of the DNA environment within dormant cells and vegetatively growing cells.
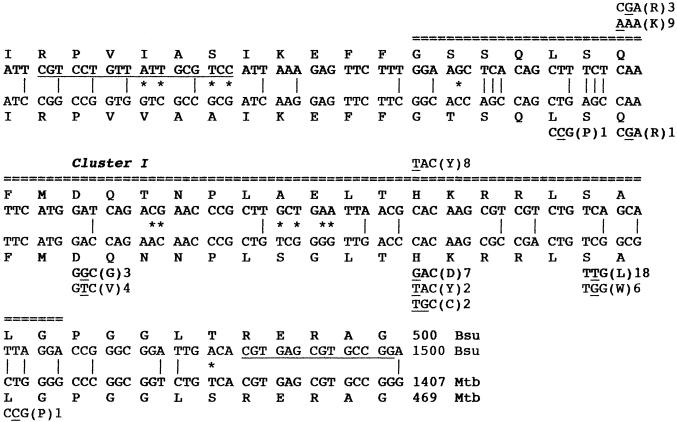
The antibiotic rifampin, a potent inhibitor of prokaryotic transcription initiation (23), has long been used to study transcription in bacteria and has also been a highly clinically effective drug, particularly in the treatment of tuberculosis. However, mycobacterial resistance to rifampin can arise from mutations in the rpoB gene encoding the β subunit of RNA polymerase; the vast majority (96%) of these mutations occur within a short 69-bp stretch within rpoB (2, 14, 24) which corresponds to rifampin resistance (Rifr) cluster I in the well-characterized Escherichia coli rpoB gene (5, 19). In Bacillus subtilis, a single Rifr mutation, called rfm2103, was historically also found to reside in rpoB (1, 15), and more recently, a larger collection of 18 Rifr mutations, both spontaneous and generated by ethyl methanesulfonate or N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis, was characterized for B. subtilis (4). To date, all of the Rifr mutations isolated in B. subtilis have been found to be single nucleotide substitutions resulting in specific amino acid changes located at only two positions, Q469R or Q469K (12 mutations) and H482Y (8 mutations), within the portion of rpoB which also corresponds to cluster I (1, 4) (Fig. 1). In contrast, the Rifr rpoB mutations found in clinical Mycobacterium tuberculosis isolates (2) exhibited a very different spectrum of nucleotide and amino acid substitutions within cluster I (Fig. 1). Out of a total of 47 Rifr mutations sequenced from clinical isolates of M. tuberculosis, the most abundant Rifr mutation occurred at amino acids (using the B. subtilis rpoB coordinates) S487L (18 isolates) and S487W (6 isolates). Additional mutations were found at L467P (one isolate), D472V (four isolates), D472G (three isolates), and L489P (one isolate) (Fig. 1). No examples were found in M. tuberculosis of the Q469K mutation, which was the predominant mutation in B. subtilis, and only one example of the Q469R mutation was found in M. tuberculosis (Fig. 1). At H482, the rpoB mutations found in M. tuberculosis isolates were H482D (seven isolates) and H482C and H482Y (two isolates each), whereas H482Y was the only Rifr rpoB mutation found in B. subtilis (Fig. 1). Notably, the single C-to-T transition resulting in S487L has been shown by several groups to be the predominant mutation in Rifr M. tuberculosis isolates, accounting for approximately 40 to 60% of all clinical isolates tested worldwide (8, 20, 22, 26), but this mutation is absent in B. subtilis.
FIG. 1.
Comparison of the rifampin resistance regions of rpoB in B. subtilis and M. tuberculosis. The wild-type rpoB nucleotide and amino acid sequences of the rifampin resistance regions of rpoB in B. subtilis (Bsu) (top two lines) and M. tuberculosis (Mtb) (bottom two lines) are shown. The double dashed line indicates the region corresponding to Rifr cluster I in the E. coli rpoB gene (5, 19). Nucleotide mismatches which are silent (vertical lines) or lead to amino acid differences (asterisks) are denoted between the sequences. Mutations leading to Rifr in B. subtilis (1, 4) and M. tuberculosis (2) are denoted above and below the wild-type sequences. The number beside each mutation refers to the number of independent isolations of that mutation. Underlined nucleotides in the B. subtilis sequence denote the position of the PCR primers used to amplify the region in this study.
What might be the underlying reason for such a difference in the mutational spectrum between these two bacteria? We reasoned that clinical tuberculosis is often a reemergence of active M. tuberculosis infection after a period of dormancy in the host which can last for years (7, 13). DNA in dormant mycobacteria may therefore exist either in a different conformation or in a different cytoplasmic environment which alters its chemical reactivity, and thus, the spectrum of spontaneous mutation to Rifr. There is ample evidence supporting such a notion in the case of dormant endospores of B. subtilis and other spore-forming Bacillus spp., in which dormant spore DNA (i) is associated with α/β-type small, acid-soluble spore proteins, (ii) is packaged in an A-like conformation, and (iii) exhibits a dramatic difference in its chemical reactivity and photochemistry compared to those of DNA from vegetative cells (reviewed in references 12, 17, and 18).
We reasoned that this hypothesis could be tested directly by characterizing spontaneous Rifr mutants isolated from the same strain of B. subtilis in either the dormant spore phase or the vegetatively growing phase of its life cycle. We first plated 108 spores of B. subtilis 168 (trpC2) from a purified spore stock, which had been stored in distilled water at 4°C for 8 years, onto solid Schaeffer's sporulation medium (SSM) (16) containing 50 μg of rifampin per ml. Strain 168 cannot grow in the presence of 0.5 μg of rifampin per ml (data not shown). Six Rifr mutant colonies arose after overnight incubation at 37°C. Each colony was picked and restreaked on SSM containing rifampin, and then an isolated Rifr colony was picked and streaked again on solid Luria-Bertani (LB) medium (11). A single colony from each of the second streaks was then resuspended in 0.2 ml of TE buffer (10 mM Tris-HCl, 1 mM Na2EDTA [pH 8.0]), template chromosomal DNA was prepared by heating the cell suspension at 95°C for 40 min, debris was removed from the cell suspension by centrifugation for 1 min in a microcentrifuge (6), and the region of rpoB corresponding to cluster I was amplified by PCR using the primer pair shown in Fig. 1. Each 143-bp PCR product was prepared for sequencing (Wizard PCR Prep; Promega) and sequenced on both strands using the PCR primers as sequencing primers at the nucleotide sequencing facility at the Laboratory for Molecular Systematics and Evolution, University of Arizona. Sequencing revealed that each of the six Rifr mutants from spores contained a single missense mutation in cluster I as follows: Q469R (two of six), H482R (two of six), and S487L (two of six). In addition to the previously observed Q469R mutation (4), spores exhibited two new mutations, H482R and S487L. The S487L mutations were particularly intriguing, as they had not been isolated before in B. subtilis, but were the most prevalent Rifr mutations found in M. tuberculosis clinical isolates (2, 8, 20, 22, 26) (Fig. 1).
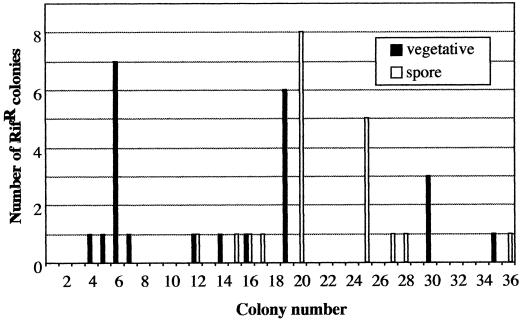
Because all six Rifr mutants discussed above were isolated from the same batch of spores, and hence cannot be assured to have arisen independently, we decided to further investigate this observation by isolating parallel sets of independent Rifr mutants from vegetative cells or spores. Strain 168 was streaked for single isolated colonies on solid LB medium, and 36 colonies were picked in duplicate and transferred onto either solid LB medium, on which B. subtilis does not sporulate efficiently, or onto solid SSM, which promotes high-efficiency sporulation. After 16 h of incubation at 37°C, the 36 colonies on LB agar were each removed completely from the plate and spread separately onto SSM plates containing 50 μg of rifampin per ml to select for Rifr mutants. Meanwhile, the set of 36 duplicate colonies on SSM were incubated at 37°C for 2 days, removed completely from the plate, heat shocked (80°C, 10 min) to select for spores, and plated onto SSM containing 50 μg of rifampin per ml to select for Rifr mutants. A total of 23 and 20 Rifr mutant colonies were obtained from the 36 separate platings of vegetative cells and spores, respectively. It was separately determined that individual colonies picked and transferred onto LB and SSM contained on average ∼5.7 × 107 and ∼9.6 × 107 cells per colony, respectively (data not shown), resulting in frequencies of spontaneous mutation to Rifr of ∼1.1 × 10−8 for vegetative cells and ∼5.8 × 10−9 for spores. The distribution of spontaneous Rifr mutants observed from the 36 pairs of colonies (Fig. 2) strongly resembled that of two independent fluctuation tests (10). Furthermore, Rifr mutant colonies arose in two different patterns among the plated colonies of vegetative cells and spores (Fig. 2), indicating that the Rifr mutations had arisen independently in the duplicate sets of colonies. One Rifr mutant arising from each separate selective plate was chosen, streak purified, PCR amplified, and sequenced as described above. As a control, chromosomal template DNA prepared in parallel from a colony of B. subtilis strain 168 grown overnight on LB agar was amplified and sequenced; its rpoB sequence in cluster I was identical to those previously published (1, 4, 9).
FIG. 2.
Distribution of Rifr mutants among 36 duplicate selective platings of B. subtilis vegetative cells and spores. See text for details.
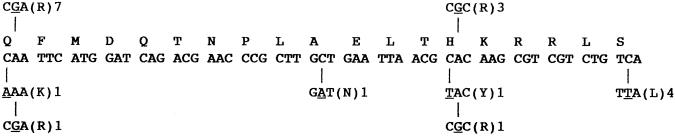
Sequence analysis of the cluster I region of rpoB in Rifr isolates obtained showed a clear difference in the spectrum of spontaneous Rifr mutations obtained from spores and vegetative cells (Fig. 3). Out of the 10 Rifr mutations sequenced from vegetative cells, 7 were found to consist of the same A-to-G transition (underlined) changing codon Q469 (CAA) to R469 (CGA), and 3 were found to consist of the same A-to-G transition changing codon H482 (CAC) to R482 (CGC) (Fig. 3). The Q469R mutation has been observed previously in B. subtilis rpoB (4), but the H482R mutation is a new mutation uncovered in this study. In sharp contrast, out of a total of nine spontaneous Rifr mutations isolated from spores, the majority of Rifr mutations (five) were found to be located in two completely new codons; most of these new mutations (four) consisted of the same C-to-T transition changing codon S487 (TCA) to L487 (TTA), and a single C-to-A transversion which changed codon A478 (GCT) to N478 (GAT) was noted (Fig. 3). In addition, single examples of the following mutations were observed: a C-to-A transversion changing codon Q469 (CAA) to K469 (AAA), an A-to-G transition changing codon Q469 (CAA) to R469 (CGA), a C-to-T transition changing codon H482 (CAC) to Y482 (TAC), and an A-to-G transition changing codon H482 (CAC) to R482 (CGC) (Fig. 3). A comprehensive comparison of all rpoB mutations isolated in B. subtilis thus far is presented in Table 1.
FIG. 3.
Spectrum of Rifr mutations in the B. subtilis rpoB gene isolated in this study. The wild-type rpoB nucleotide and amino acid sequences are shown from positions Q469 to S487. Positions of nucleotide changes leading to the indicated amino acid substitutions in vegetative cells and spores are denoted above and below the wild-type sequence, respectively, and the exact nucleotide change is underlined. Amino acid changes resulting from each mutation are in parentheses. The number of independent isolates containing each mutation is denoted to the right of the amino acid change.
TABLE 1.
Summary of mutational changes in B. subtilis and M. tuberculosis rpoB genes leading to Rifra
| Codon positionb | Codon change | Amino acid change | No. (% of total) isolated in:
|
||
|---|---|---|---|---|---|
| B. subtilis vegetative cells | B. subtilis spores | M. tuberculosis clinical isolates | |||
| 467 | CTG to CCG | L to P | 0 | 0 | 1 (2) |
| 469 | CAA to AAA | Q to K | 9 (30) | 1 (11) | 0 |
| 469 | CAA to CGA | Q to R | 10 (33) | 1 (11) | 1 (2) |
| 472 | GAC to GGC | D to G | 0 | 0 | 3 (7) |
| 472 | GAC to GTC | D to V | 0 | 0 | 4 (9) |
| 478 | GCT to GAT | A to N | 0 | 1 (11) | 0 |
| 482 | CAC to TAC | H to Y | 8 (27) | 1 (11) | 2 (4) |
| 482 | CAC to CGC | H to R | 3 (10) | 1 (11) | 0 |
| 482 | CAC to TGC | H to C | 0 | 0 | 2 (4) |
| 482 | CAC to GAC | H to D | 0 | 0 | 7 (16) |
| 487 | TCR to TTRc | S to L | 0 | 4 (44) | 18 (40) |
| 487 | TCR to TGR | S to W | 0 | 0 | 6 (13) |
| 489 | CTG to CCG | L to P | 0 | 0 | 1 (2) |
Data are summarized from references 1, 2, and 4 and this study.
Codon positions using B. subtilis coordinates.
R, purine (A or G).
Thus, the spectrum of independent spontaneous Rifr mutations differed considerably in vegetative cells and spores of B. subtilis which were derived from a common set of 36 colonies. This observation is very likely a reflection of the different conformations and cytoplasmic environments of DNA in actively growing cells and dormant spores (12). The major class of spontaneous Rifr mutations observed in B. subtilis spores consisted of a unique C-to-T transition resulting in S487L (four of nine, or 44% of isolates); this identical nucleotide and amino acid substitution is also the major class of mutations found in clinical Rifr M. tuberculosis isolates and is recovered at approximately the same frequency (2, 8, 20, 22, 26) (Table 1).
To verify that the new rpoB mutations identified in B. subtilis by sequencing were indeed sufficient to cause a Rifr phenotype, the following experiments were performed. First, by standard techniques (3), chromosomal DNA (500 ng) prepared from mutants containing the new mutations Q469R, H482R, A478N, and S487L was introduced into competent cells of strain 168 simultaneously with 50 ng of DNA from plasmid pWN162, which carries the wild-type allele of the trpC2 mutation (25). Tryptophan-prototrophic (Trp+) transformants were selected on Spizizen’s minimal medium (SMM) (21) and replica plated onto SMM containing rifampin (50 μg/ml) to select for Trp+ Rifr congressants. DNA was prepared from individual colonies of congressants and PCR amplified with the primers indicated in Fig. 1, and the resulting 143-bp PCR products were sequenced. In all cases, the identical nucleotide substitution from the donor strain was detected in the resulting Rifr congressant (data not shown).
However, because only a 143-bp PCR product spanning cluster I was sequenced, it was formally possible that additional nucleotide changes responsible for Rifr and residing outside the cluster I region could be present in the original mutants, hence transferred to the congressants. Because the 143-bp PCR product from Rifr mutants was too small to be efficiently transferred by transformation (data not shown), we designed two primers, 5′-GCGAAAAGCTTGCTTGATTC-3′ and 5′-CCAACAAGAAGATCTCCGTC-3′, which amplified a 1,781-bp region of rpoB from a HindIII site at nucleotide 756 to a BglII site at nucleotide 2517 (underlined portions of the primers) and which included codons 252 to 840, including cluster I. Using the above primers, we amplified chromosomal DNA from the Rifr mutant carrying the S487L and A478N mutations. Sequencing of the 1,781-bp PCR products confirmed that the only nucleotide changes present consisted of the S487 (TCA) to L487 (TTA) mutation and the A478 (GCT) to N478 (GAT) mutation, respectively (data not shown). We introduced 500 ng of the 1,781-bp PCR products and 50 ng of plasmid pWN162 into competent B. subtilis 168 cells by transformation as described above. In the experiment using S487L as the donor DNA, from a total of 960 Trp+ transformants, we obtained four Trp+ Rifr congressants, one of which was shown by PCR amplification of the 1,781-bp segment of rpoB from chromosomal DNA and sequencing to contain only the S487L mutation (data not shown). In the experiment using A478N as the donor DNA, from a total of 1,640 Trp+ transformants, we obtained six Trp+ Rifr congressants, one of which was shown by PCR amplification of the 1,781-bp segment of rpoB from chromosomal DNA and sequencing to contain only the A478N mutation (data not shown). Thus, the single C-to-T transition resulting in S487L or the single C-to-A transversion resulting in A478N on the 1,781-bp rpoB fragment was sufficient to cause the Rifr phenotype in B. subtilis.
Our results therefore lend support to the hypothesis that DNA in dormant M. tuberculosis cells may exist either in a conformation or cellular environment analogous to that seen in dormant bacterial endospores. This structural analogy between DNA in Bacillus spores and dormant mycobacteria could be extended to imply that reactivation of latent M. tuberculosis infection may share features in common with spore germination, which in turn could yield important new insights into the treatment and prevention of tuberculosis.
Acknowledgments
We thank Helen Jost and Stephen Billington for critical reading of the manuscript.
This work was supported in part by a grant from the Arizona Agricultural Experiment Station (USDA Hatch) to W.L.N. and a Research Training Grant from the Department of Ecology and Evolutionary Biology to H.M.
REFERENCES
- 1.Boor, K. J., M. L. Duncan, and C. W. Price. 1995. Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase. J. Biol. Chem. 270:20329-20336. [DOI] [PubMed] [Google Scholar]
- 2.Garcia, L., M. Alonso-Sanz, M. J. Rebollo, J. C. Tercero, and F. Chaves. 2001. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis isolates in Spain and their rapid detection by PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 39:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harwood, C. R., and S. M. Cutting. (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Sussex, England.
- 4.Ingham, C. J., and P. A. Furneaux. 2000. Mutations in the β subunit of the Bacillus subtilis RNA polymerase that confer both rifampicin resistance and hypersensitivity to NusG. Microbiology 146:3041-3049. [DOI] [PubMed] [Google Scholar]
- 5.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 6.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kell, D. B., and M. Young. 2000. Bacterial dormancy and culturability: the role of autocrine growth factors. Curr. Opin. Microbiol. 3:238-243. [DOI] [PubMed] [Google Scholar]
- 8.Kim, B.-J., K.-H. Lee, B.-N. Park, S.-J. Kim, E.-M. Park, Y.-G. Park, G.-H. Bai, S.-J. Kim, and Y. H. Kook. 2001. Detection of rifampin-resistant Mycobacterium tuberculosis in sputa by nested PCR-linked single-strand conformation polymorphism and DNA sequencing. J. Clin. Microbiol. 39:2610-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 10.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, J. 1972. Experiments in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of bacterial spores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plorde, J. J. 1994. Mycobacteria, p. 401-416. In K. J. Ryan (ed.), Sherris medical microbiology, 3rd ed. Appleton & Lang, East Norwalk, Conn.
- 14.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 15.Rothstein, D. M., C. L. Keeler, and A. L. Sonenshein. 1976. Bacillus subtilis RNA polymerase mutants temperature-sensitive for sporulation, p. 601-616. In R. L. Losick and M. Chamberlin (ed.), RNA polymerase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 18.Setlow, P. 2001. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 38:97-104. [DOI] [PubMed] [Google Scholar]
- 19.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited: new rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 20.Sintchenko, V., W. K. Chew, P. J. Jelfs, and G. L. Gilbert. 1999. Mutations in rpoB gene and rifabutin susceptibility of multidrug-resistant Mycobacterium tuberculosis strains isolated in Australia. Pathology 31:257-260. [DOI] [PubMed] [Google Scholar]
- 21.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi, H., H. Aramaki, Y. Nikaido, Y. Mizuguchi, M. Nakamura, T. Koga, and S. Yoshida. 1996. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 144:103-108. [DOI] [PubMed] [Google Scholar]
- 23.Wehrli, W., F. Knusel, K. Schmid, and M. Staehelin. 1968. Interaction of rifamycin with bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA 61:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams, D. L., L. Spring, M. Salfinger, T.-P. Gillis, and D. H. Persing. 1998. Evaluation of polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampicin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin. Infect. Dis. 26:446-450. [DOI] [PubMed] [Google Scholar]
- 25.Xue, Y., and W. L. Nicholson. 1996. The two major spore DNA repair pathways, nucleotide excision repair and spore photoproduct lyase, are sufficient for the resistance of Bacillus subtilis spores to artificial UV-C and UV-B but not to solar radiation. Appl. Environ. Microbiol. 62:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, B., H. Koga, H. Ohno, K. Ogawa, M. Fukuda, Y. Hirakata, S. Maesaki, K. Tomono, T. Tashiro, and S. Kohno. 1998. Relationship between antimycobacterial activities of rifampicin, rifabutin, and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621-628. [DOI] [PubMed] [Google Scholar]