Abstract
Paracoccus pantotrophus can express a periplasmic nitrate reductase (Nap) during aerobic growth. A proposed role for this enzyme is the dissipation of excess redox energy during oxidative metabolism of reduced carbon substrates. To investigate the regulation of nap expression, a transcriptional fusion between the nap promoter region of P. pantotrophus and the lacZ gene was constructed. When this fusion was used, analyses showed that transcription from the nap promoter increases as the average reduction state of the carbon atoms increases. Thus, β-galactosidase activities increase as the carbon source changes in the order succinate-acetate-butyrate. This result was obtained regardless of which of the three carbon sources was used for culture of the inoculum. If two carbon sources were presented together, the β-galactosidase activity was always the same as it was when the least-reduced carbon source was added alone. This suggests that the regulation is dependent upon metabolism of the more-reduced carbon sources rather than just their presence in the medium. Analysis of culture medium by 1H nuclear magnetic resonance showed that for aerobic growth P. pantotrophus strictly selected its carbon source in the order succinate-acetate-butyrate. This was reflected by diauxic growth kinetics on medium containing mixed carbon substrates. The regulatory mechanism underpinning such a selection is unknown but is likely to be related to the mechanism which controls the transcription of the nap operon.
Respiration in bacteria is usually associated with generation of a proton motive force and consequent ATP synthesis. However, in some instances respiration may not be coupled to transmembrane proton translocation in order to provide a mechanism for the disposal of excess reductant from cells independent of the demand for ATP. An example of this behavior is the flow of electrons from ubiquinol to the periplasmic nitrate reductase in the α-proteobacterium Paracoccus pantotrophus, a process that is not linked to the generation of a proton motive force. P. pantotrophus is capable of both aerobic and anaerobic respiration of nitrate (15). For dissimilatory nitrate reduction two different enzymes catalyze the reaction under aerobic and anaerobic conditions (1, 2). These enzymes are the periplasmic (Nap) and the membrane-bound (Nar) nitrate reductases, respectively. Nar is concerned with energy conservation during electron transfer from ubiquinol to nitrate, in the absence of oxygen as the terminal electron (e−) acceptor. In contrast, electron transfer to nitrate via Nap is not likely to conserve energy, dissipating the free energy in the ubiquinol-nitrate redox couple (4). When NADH is considered the major electron donor to the ubiquinone pool, it becomes apparent that electron transfer to oxygen via cytochrome aa3 oxidase is more highly coupled (q+/e− = 10) than electron transfer to nitrate via Nap (q+/e− = 4). A proposed role for Nap is to maintain the redox balance of the cell by providing a poorly coupled route for the oxidation of excess reducing equivalents resulting from the oxidative metabolism of highly reduced carbon substrates (13, 17). Previously, we have shown that there is differential synthesis of, and electron flux through, Nap when P. pantotrophus or Paracoccus denitrificans Pd1222 is grown aerobically with various carbon substrates. (13, 17). In addition, it was shown that Nap activity correlates with the oxidation state of the carbon substrate; lower Nap activities occurred during growth on more-oxidized carbon substrates (succinate and malate) than during growth on highly reduced carbon substrates (caproate and butyrate) (17).
Transcription initiation at the nap operon has been shown to depend on the carbon source (18), suggesting that transcription is responsive to one or more signals that reflect the reduction state of the carbon source available to the cell. There is no obvious sequence motif upstream of the transcriptional start point that gives any clue to the molecular identity of one or more transcription factors that are postulated to bind in this region. A direct test of the importance of transcriptional control, and its dependence on the redox state of the carbon source, for the production of the ubiquinol-Nap pathway in P. pantotrophus requires fusion of the promoter to the lacZ gene, which was one of the objectives of the present work. The notion that a relatively reduced carbon source (e.g., butyrate) is disadvantageous to the cell and consequently requires a reductant disposal pathway via Nap raised the question of whether this organism shows any preference in the selection of its carbon source for growth under aerobic conditions. By using nuclear magnetic resonance, we were able to show that there is indeed such a preference and that it correlates with increased nap-lacZ activity in the sense that the least-favored carbon substrate causes the greatest activation of nap expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used and their sources and genotypes are shown in Table 1. P. pantotrophus was grown on minimal medium with ammonium as the nitrogen source for growth (15). Additions of the carbon sources sodium succinate (30, 10, and 1 mM final concentrations, as appropriate), sodium acetate (50, 10, and 1 mM final concentrations, as appropriate), and/or sodium butyrate (10 mM final concentration) were made. For anaerobic growth 30 mM potassium nitrate was added as an alternative electron acceptor, and 5 mM potassium nitrate was added, where appropriate, to aerobic cultures. Escherichia coli strains were cultured on Luria-Bertani medium (16). Antibiotics were added at the following concentrations for cultures of P. pantotrophus: streptomycin, 50 μg ml−1; spectinomycin, 50 μg ml−1; gentamicin, 10 μg ml−1; and rifampin, 100 μg ml−1. Antibiotics were added to E. coli cultures at the following concentrations: ampicillin, 100 μg ml−1; streptomycin, 25 μg ml−1; gentamicin, 10 μg ml−1; and spectinomycin, 25 μg ml−1. Aerobic cultures (200 ml in 2.5-liter flasks or 20 ml in 250-ml flasks) were shaken at 200 rpm at 37°C. Anaerobic cultures were grown at 37°C in screw-top standing bottles filled to the top. Bacterial growth was monitored turbidometrically at 600 nm (optical density at 600 nm).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| P. pantotrophus | Wild type | 15 |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 relA1 | 7 |
| S17-1 | thi pro hsdR recA, integrated RP4-2 Tc::Mu Km::Tn7 | 19 |
| Plasmids | ||
| pCRSCRIPT-SK(+) | Ampr, sequencing and cloning vector | Stratagene |
| pHRP317 | Cohort vector containing Sm/Sp cassette | 12 |
| pHRP309 | lacZ fusion vector | 12 |
| pCRSCRIPT-nap | 209-bp nap promoter region fragment in pCRSCRIPT, promoter fragment with sequence −178 bp relative to the transcriptional start | This study |
| pHRP317-nap | NotI/EcoRI fragment from pCRSCRIPT-nap in pHRP317 | This study |
| pnap-lacZ | EcoICRI/SacI fragment from pHRP317-nap in pHRP309 | This study |
Genetic and molecular biology techniques.
Standard molecular biology techniques were performed as described by Sambrook et al. (16). Mobilization of plasmids from E. coli to P. pantotrophus was achieved by biparental mating by using E. coli S17-1 as a donor. Competent E. coli cells were made and transformed as described by Inoue et al. (8).
β-Galactosidase assay.
β-Galactosidase activity was assayed in triplicate as described by Miller (11). For single end point determinations of β-galactosidase activity, triplicate assays during mid-log growth were performed by using three independent cultures.
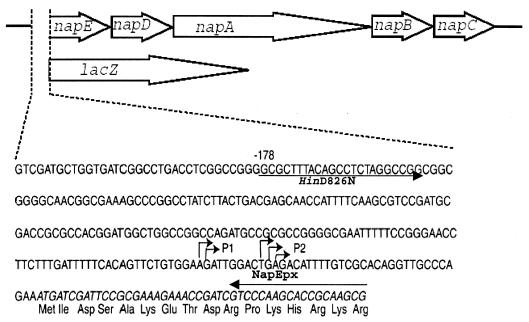
Construction of pnap-lacZ.
The primer pair consisting of HinD826N (5′-GGCGCTTTAAGCTTCTAGGCCCG-3′) and NapEpx (5′-CGCTTGCGGTGCTTGGGAC-3′) was used to PCR amplify from P. pantotrophus genomic DNA the nap promoter region from nucleotide 826 (−178 bp relative to the transcriptional start) to nucleotide 1035 of the published sequence (3), where this sequence is proximal to the napE gene. Cloning was carried out with pCRSCRIPT-SK(+), and a 263-bp EcoRV-NotI DNA fragment, derived from the amplified product, was cloned into pHRP317 to obtain pHRP317-nap. A 2.8-kb EcoICRI (Promega Corp.)-SalI fragment from pHRP317-nap was ligated into SmaI-SalI-digested pHRP309 to create pnap-lacZ. After mobilization of pnap-lacZ into P. pantotrophus by biparental mating, rifampin-, streptomycin-, gentamicin-, and spectinomycin-resistant exconjugants were selected. Plasmid DNA was isolated from liquid subcultures of positive clones, and the presence of a correct construct was confirmed by digestion with HindIII and subsequent gel electrophoresis.
Quantification of substrate utilization by proton nuclear magnetic resonance (1H-NMR) spectroscopy.
An aliquot (1 ml) of culture medium was removed and filtered through a 0.22-μm-pore-size filter to remove cellular debris. The sample was lyophilized and redissolved in 0.5 to 0.7 ml of deuterium oxide (Goss Scientific Instruments Ltd.). 3-Trimethyl-silyl-tetradeuterosodium propionate (TSP) (Goss Scientific Instruments Ltd.; 10 liters of 10 mM TSP in D2O) was added as an internal concentration and chemical shift standard.
1H-NMR spectroscopy.
All spectra were obtained at 30°C with a Varian Unity-INOVA NMR spectrometer (Varian Associates Inc., NMR Instruments, Palo Alto, Calif.) operating at a proton frequency of 400 MHz. One-pulse fully relaxed spectra were acquired with 90o pulses applied every 14 s by using a spectral width of 6000.6 Hz and 64K resolution. The decoupler was gated on the water frequency in the delay between pulses in order to suppress the residual water (HOD) peak. For a satisfactory signal/noise ratio, 256 or 512 scans were accumulated and Fourier transformed.
Metabolite identification and quantification.
Metabolite peaks found in the one-dimensional spectra were identified by their chemical shift and coupling patterns by comparison with spectra of standard metabolites at known concentrations obtained at the same pH and under the same spectroscopic conditions. Metabolite amounts were calculated from their heights in 1H-NMR spectra by reference to the internal standard TSP after baseline correction. The height of a signal in a proton spectrum is proportional to the amount of compound and the number of protons contributing to the signal.
RESULTS
Effect of oxygen and nitrate on nap-lacZ expression with succinate and butyrate as carbon sources.
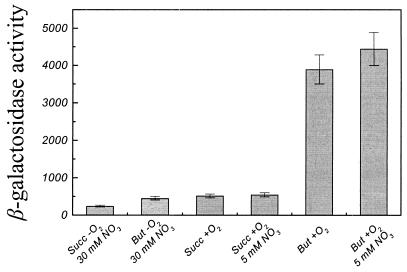
In order to quantitatively investigate expression from the nap promoter, a nap-lacZ transcriptional fusion was constructed (Table 1 and Fig. 1) and conjugatively transferred into P. pantotrophus. Deletion analysis has previously shown that this region of DNA is essential for maximal Nap enzyme activity (18). Controls consisting of wild-type P. pantotrophus and P. pantotrophus/pHRP309 were assayed for β-galactosidase activity in triplicate by using preparations from three independent cultures during the mid-log phase of growth (optical density at 600 nm, ∼0.7). β-Galactosidase activities were low for both controls under all aerobic or denitrifying conditions with succinate and butyrate carbon substrates, and a maximum value of 100 U of β-galactosidase activity was recorded for P. pantotrophus/pHRP309. For P. pantotrophus/pnap-lacZ β-galactosidase activity was initially assayed during the mid-logarithmic phase of growth on a relatively oxidized carbon substrate, succinate, and a relatively reduced carbon substrate, butyrate, in the presence and absence of oxygen. Under anaerobic conditions expression from the nap promoter remained low regardless of the carbon substrate used for growth, and both substrates resulted in less than 500 ± 50 U of β-galactosidase activity (Fig. 2). Aerobic growth on succinate also yielded a relatively low level of expression from the nap promoter (500 ± 50 U of β-galactosidase activity). The level of expression increased approximately 9- to 10-fold (4,000 ± 400 U of β-galactosidase activity) during aerobic growth on butyrate (Fig. 2). Addition of 5 and 50 mM nitrate to aerobic cultures utilizing butyrate resulted in moderate increases in expression from the nap promoter to approximately 4,500 ± 450 and 5,000 ± 500 U of β-galactosidase activity, respectively. No detectable effect of nitrate was found in aerobic succinate cultures, demonstrating that in succinate- or butyrate-containing medium, nitrate has little effect on transcription from the nap promoter even at a high concentration.
FIG. 1.
P. pantotrophus nap operon and nap promoter-lacZ fusion. The nap promoter sequence is expanded, and two transcription initiation sites are indicated. The translated napE sequence is italicized, and the amino acid sequence is indicated at the bottom. The primer sites are labeled HinD826N and NapEpx, and the arrows indicate the 5′-to-3′ direction of the primers. There are a small number of cloning site bases between the nap promoter sequence and the ATG codon for lacZ.
FIG. 2.
β-Galactosidase activity in P. pantotrophus pnap-lacZ cultures is dependent upon the carbon substrate and terminal electron acceptor. β-Galactosidase assays were determined for cells taken during mid-log-phase growth, and activity was calculated as described by Miller (11). Succ, succinate; But, butyrate.
Because of the effects of succinate and butyrate on transcription from the nap promoter under aerobic conditions, the effects of other substrates were investigated under aerobic conditions. Table 2 shows data obtained from triplicate β-galactosidase assays of three independent P. pantotrophus/pnap-lacZ cultures grown on a range of carbon substrates. A trend of increasing expression levels from the nap promoter as the average oxidation state of the carbon atoms in the carbon substrate decreased is apparent. The most reduced substrates, butyrate and caproate, gave β-galactosidase activities that were 9- to 10-fold higher than those obtained with the relatively oxidized substrates succinate and malate. Substrates such as acetate and glycerol, which have an intermediate average oxidation state, resulted in intermediate levels of expression from the nap promoter, which were approximately threefold higher than the levels of expression obtained with succinate.
TABLE 2.
β-Galactosidase activities in P. pantotrophus/pnap-lacZ grown aerobically on increasingly reduced carbon substrates
| Carbon substrate (final concn[mM]) | Avg oxidation no. of carbons | β-Galactosidase activity (U) |
|---|---|---|
| Malate (30) | +1.50 | 700 |
| Pyruvate (30) | +0.66 | 800 |
| Succinate (30) | +0.50 | 520 |
| Acetate (50) | 0.00 | 1,400 |
| Glycerol (30) | −0.66 | 1,700 |
| Butyrate (10) | −1.00 | 4,100 |
| Caproate (7) | −1.33 | 4,000 |
Growth kinetics and nap-lacZ expression with succinate, acetate, or butyrate as the carbon source.
To monitor the regulation of expression from the nap promoter during growth, β-galactosidase assays were carried out at different times during aerobic batch culture growth experiments. Cultures were supplied with succinate, acetate, and butyrate as substrates, which during the mid-log phase of growth gave low, medium, and high levels of expression from the nap promoter, respectively. Experiments were carried out with inocula precultured on each of the three carbon substrates, in order to analyze the effect of a switch in substrate on expression from the nap promoter at the start of growth.
In all experiments different growth rates were observed for cultures utilizing the three carbon substrates. When a succinate-cultured inoculum was used, succinate cultures had a short lag phase and a relatively high rate of growth (doubling time, 80 min); this resulted in the stationary phase being reached approximately 6 to 7 h after inoculation (Fig. 3A). With acetate, growth was slower than growth on succinate (doubling time, 90 min), and the stationary phase was reached approximately 11.5 to 12 h after inoculation. Cultures utilizing butyrate displayed a longer lag phase and a lower growth rate (doubling time, 185 min) and reached the stationary phase approximately 15 to 16 h after inoculation.
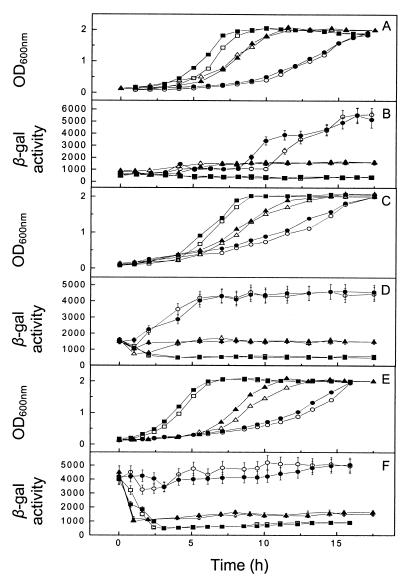
FIG. 3.
Correlation of β-galactosidase activity with switching carbon substrate at the time of inoculation in P. pantotrophus/pnap-lacZ cultures. The carbon sources used for preculture of the inocula were 30 mM succinate (A and B), 50 mM acetate (C and D), and 10 mM butyrate (E and F). The culture medium contained 30 mM succinate (▪), 30 mM succinate and 5 mM nitrate (□), 50 mM acetate (▴), 50 mM acetate and 5 mM nitrate (▵), 10 mM butyrate (•), or 10 mM butyrate and 5 mM nitrate (○). Growth was measured by determining the optical density at 600 nm (OD600nm). β-Galactosidase (β-gal) activity was calculated as described by Miller (11).
The expression levels from the nap promoter remained low (500 ± 50 U of β-galactosidase activity) throughout growth for cells utilizing succinate (Fig. 3B). Cultures utilizing acetate or butyrate as the substrate showed an increase in β-galactosidase activity during the lag phase. With acetate as the carbon source, β-galactosidase activities of 1,400 ± 140 U were observed after the early log phase of growth. The β-galactosidase activities in butyrate-containing cultures reached 4,000 ± 400 U during early logarithmic growth and increased to 5,000 ± 500 U as a culture approached the late stationary phase.
When similar experiments were carried out with an inoculum precultured on acetate or butyrate, the growth kinetics were essentially identical to those observed when an inoculum precultured on succinate was used (Fig. 3C and E). However, slight variations in the lag times were evident when carbon substrate switches occurred. Intermediate levels of expression from the nap promoter were observed for an inoculum precultured on acetate; β-galactosidase activities of 1,500 ± 150 U were recorded at zero time (Fig. 3D). Transcriptional activity from pnap-lacZ was then down- and up-regulated for cultures whose carbon source was switched from acetate to succinate and butyrate, respectively. This occurred within 3 h of inoculation (Fig. 3D). When no change in carbon source occurred with the acetate culture, expression remained constant at 1,500 ± 150 U of β-galactosidase activity (Fig. 3D).
When the inoculum was precultured on butyrate, cultures utilizing butyrate as the carbon source exhibited constant levels of β-galactosidase activity (4,000 ± 400 U) throughout growth (Fig. 3F). For cultures whose carbon source was switched from butyrate in the preculture inoculum to acetate, and for cultures whose carbon source was switched from butyrate in the preculture inoculum to succinate, expression from the nap promoter was down-regulated to 1,400 ± 140 and 500 ± 50 U of β-galactosidase activity, respectively, within 3 h of inoculation (Fig. 3F). Addition of 5 mM nitrate to these cultures marginally slowed growth in most cases, but expression from the nap promoter was essentially unaffected. The exception was the transition from the lag phase to the logarithmic phase of growth in cultures utilizing butyrate which were derived from a succinate-grown inoculum (Fig. 3B). In these cultures, the levels of expression from the nap promoter were slightly higher when there was no nitrate present.
Clearly, expression from the nap promoter is dependent on the carbon substrate under aerobic conditions, and addition of nitrate has little effect. In addition, the constant levels of expression in cultures in which there was no change of carbon substrate indicate that regulation of expression from the nap promoter is independent of the growth phase. However, expression levels are strictly controlled in response to the carbon substrate.
Carbon substrate utilization during growth of P. pantotrophus on mixed carbon substrates.
To investigate whether there is simultaneous or hierarchical use of substrates when two substrates are present, and to monitor the corresponding effect on expression from the nap promoter, mixed-carbon-substrate batch cultures were analyzed. Disappearance of the carbon substrate was monitored by 1H-NMR spectroscopy and was considered a measure of carbon substrate metabolism. Expression from the nap promoter was again measured by assaying β-galactosidase activity. The carbon substrates succinate, acetate, and butyrate were used in pairwise mixtures at various concentrations, and experiments were repeated with inocula grown on each of the substrates in a mixture. Three pairwise combinations of carbon substrates were prepared, and the concentration of one of the substrates in each was varied. In succinate-butyrate mixtures, the succinate concentration was 30, 10, or 1 mM, and the butyrate concentration was 10 mM. In succinate-acetate mixtures, the succinate concentration was 30, 10, or 1 mM, and the acetate concentration was 50 mM. Acetate-butyrate mixtures contained 50, 10, or 1 mM acetate along with 10 mM butyrate.
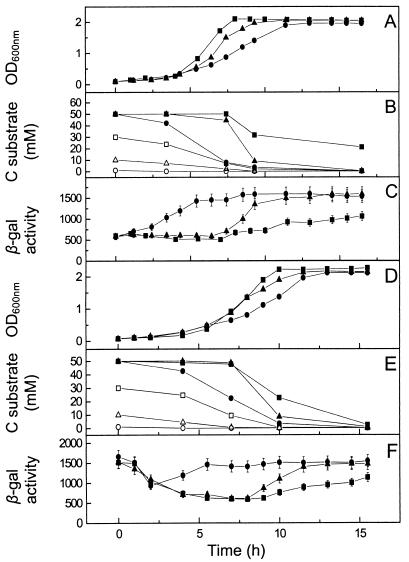
Media containing various initial concentrations of succinate and a fixed concentration of butyrate were inoculated by using cultures grown on succinate (Fig. 4A to C) and butyrate (Fig. 4D to F). Plots of the growth curves (Fig. 4A and D) show that with 30 mM succinate and 10 mM butyrate the growth rates (doubling time, 90 min) were comparable to those for cultures containing succinate as the single carbon substrate (doubling time, 80 and 90 min), suggesting that the cells were exclusively utilizing succinate. The suggestion that there was exclusive substrate use was further supported by the β-galactosidase activity data (Fig. 4C and F), which reflected the levels observed for cultures growing exclusively on succinate (Fig. 3B). Either cultures had a constant β-galactosidase activity of 500 ± 50 U (cultures inoculated from a succinate preculture) until late stationary phase (Fig. 4C), or the level of activity started at 4,000 ± 400 U and dipped in the lag phase to 500 ± 50 U of β-galactosidase activity (cultures inoculated from a butyrate preculture) (Fig. 4F). It should be noted that in the late stationary phase the β-galactosidase activities increased towards those seen in butyrate cultures. This suggests that butyrate was being utilized at this late stage to provide maintenance energy. Confirmation was obtained by using 1H-NMR spectroscopy, which clearly showed the hierarchical and exclusive disappearance of succinate over butyrate (Fig. 4B and E).
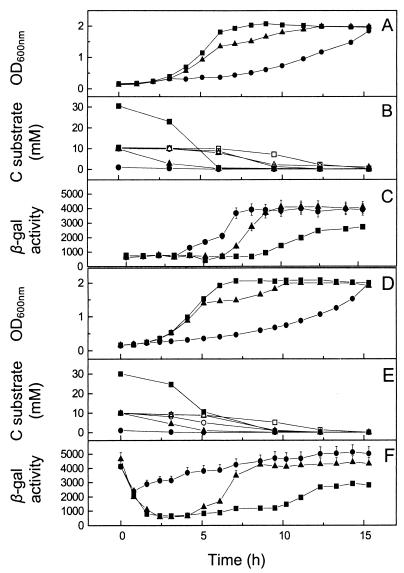
FIG. 4.
Hierarchical use of succinate over butyrate in mixed-carbon-substrate cultures of P. pantotrophus/pnap-lacZ and correlation with nap promoter activity. The carbon sources used for preculture of the inocula were 30 mM succinate (A, B, and C) and 10 mM butyrate (D, E, and F). The culture medium contained 30 mM succinate and 10 mM butyrate (squares), 10 mM succinate and 10 mM butyrate (triangles), or 1 mM succinate and 10 mM butyrate (circles). In panels B and E the solid symbols indicate the succinate concentration and the open symbols indicate the butyrate concentration. The carbon substrate concentration was measured by 1H-NMR spectroscopy. β-Galactosidase (β-gal) activity was calculated as described by Miller (11). Growth was measured by determining the optical density at 600 nm (OD600nm).
In the presence of 1 mM succinate and 10 mM butyrate, the growth rates were lower (doubling time, 220 min) and were comparable to those in cultures exclusively utilizing butyrate for growth (doubling times, 180 and 220 min), suggesting that butyrate was the substrate being used (Fig. 4A and D). As in the other experiments, measurements of β-galactosidase activities supported these observations (Fig. 4C and F); there was either a prolonged period of high activity (Fig. 4F) or an 8- to 10-fold increase in β-galactosidase activity postinoculation (Fig. 4C) depending on the substrate used for culturing the inocula. Again, 1H-NMR spectroscopy confirmed the observations described above (Fig. 4B and E).
Cultures grown with 10 mM succinate and 10 mM butyrate produced diauxic growth curves, and the initial growth rates (up to 6 h) (Fig. 4A and D) were similar to those of cultures growing exclusively on succinate (doubling time, 100 min), regardless of the history of the inoculum (compare Fig. 3A and 4A and D). After 6 h (mid-log to late log phase) the growth rate decreased significantly to a doubling time of 250 min, suggesting that the culture had switched in the late log phase from utilizing succinate to utilizing butyrate (Fig. 4A and D). β-Galactosidase assay data reflected this, with increased expression from the nap promoter observed with the cultures utilizing butyrate as the carbon substrate. Interestingly, the level of expression from the nap promoter in cultures inoculated with a butyrate preculture dipped sharply (Fig. 4C) and stayed low while succinate was being utilized, increasing again only once butyrate was being utilized. This demonstrates that there is tight regulation of nap promoter activity in relation to the carbon substrate used for growth.
1H-NMR spectroscopy (Fig. 4B and E) confirmed that utilization of the carbon substrates occurred in the order succinate-butyrate and that butyrate was not used until the succinate concentration fell below 1 mM (Fig. 4B and E). This selection of carbon substrates was also independent of the carbon substrate used to preculture the inoculum.
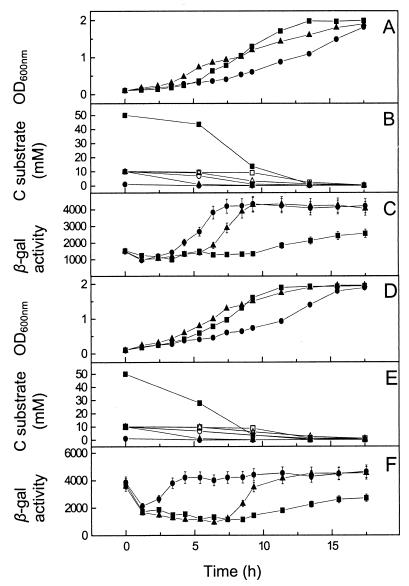
The hierarchical order for succinate and acetate utilization was also investigated by using inocula precultured with acetate and succinate (Fig. 5). The data for cultures grown on 30 mM succinate plus 50 mM acetate show a relatively high growth rate (doubling time, 80 min), suggesting that succinate is the preferred substrate. Indeed, 1H-NMR spectroscopy confirmed that succinate was used preferentially (Fig. 5B and E). As previously observed with a succinate-butyrate mixed-substrate culture, these studies revealed limited usage of acetate once the cells were in the stationary phase and the succinate concentration was below 1 mM in a succinate-acetate mixed-substrate culture (Fig. 5B and E). β-Galactosidase activities reflected these patterns of carbon substrate utilization; the activities remained low until the stationary phase, and then they increased towards, but did not attain, levels expected during growth on acetate. This suggests that the cultures started to utilize acetate for maintenance once the succinate concentration became low during the stationary phase.
FIG. 5.
Hierarchical use of succinate over acetate in mixed-carbon-substrate cultures of P. pantotrophus/pnap-lacZ and correlation with nap promoter activity. The carbon sources used for preculture of the inocula were 30 mM succinate (A, B, and C) and 50 mM acetate (D, E, and F). The culture medium contained 30 mM succinate and 50 mM acetate (squares), 10 mM succinate and 50 mM acetate (triangles), or 1 mM succinate and 50 mM acetate (circles). In panels B and E the solid symbols indicate the acetate concentration and the open symbols indicate the succinate concentration. The carbon substrate concentration was measured by 1H-NMR spectroscopy. β-Galactosidase (β-gal) activity was calculated as described by Miller (11). Growth was measured by determining the optical density at 600 nm (OD600nm).
In the presence of 10 mM succinate and 50 mM acetate the initial growth rate was high (Fig. 5A and D), suggesting that succinate was utilized initially. The substrate utilized then appeared to change to acetate, as suggested by a lower rate of growth. 1H-NMR spectroscopy again confirmed the exclusive utilization of succinate until its concentration fell below 1 mM, whereupon acetate was utilized (Fig. 5B and E).
When 1 mM succinate was present, the growth rate was relatively low throughout (Fig. 5A and D). The succinate present in the medium was utilized at the start of growth, as revealed by 1H-NMR spectroscopy, while acetate was initially utilized slowly, even taking into account the low culture density. Expression from the nap promoter reflected the substrate used for growth or maintenance, and the levels of β-galactosidase activity were 500 ± 50 U during periods of succinate utilization and around 1,400 ± 140 U when acetate was being utilized (Fig. 5C and F). Changes in substrate utilization caused changes in regulation of expression from the nap promoter to occur within 3 to 4 h of the substrate change.
The data presented above clearly demonstrate that acetate and butyrate are less favored as growth substrates than succinate. Carbon substrate mixtures containing acetate and butyrate were also investigated. Cells cultured in the presence of 50 mM acetate and 10 mM butyrate (Fig. 6A and D) grew at a rate comparable to that of cultures growing solely on acetate, suggesting that acetate was used exclusively in the presence of butyrate. 1H-NMR spectroscopy showed that there was clear exclusive disappearance of acetate from the medium in the presence of butyrate. When the starting concentration of acetate was 50 mM, butyrate did not start to disappear from the medium until stationary phase, suggesting that butyrate was used only for maintenance in stationary-phase cultures (Fig. 6B and E). The β-galactosidase activity remained at levels expected for cultures using acetate until butyrate started to disappear from the medium, when the activity started to increase (Fig. 6C and F). Compared to the growth rates of cultures growing on butyrate alone, cultures growing on 10 mM acetate and 10 mM butyrate showed higher initial rates of growth until the mid-exponential to late exponential phase, and this was followed by a premature decrease in the growth rate before the stationary phase, suggesting a switch of substrate utilization from acetate to butyrate. Cultures containing 10 mM acetate and 10 mM butyrate exhausted the acetate by the mid-exponential to late exponential phase, as shown by 1H-NMR spectroscopy (Fig. 6B and E). The β-galactosidase activities were at the corresponding levels for the substrates being utilized (Fig. 6C and F). Cultures grown with 1 mM acetate and 10 mM butyrate did not display an acetate growth rate; however, 1H-NMR spectroscopy revealed that acetate was utilized first, with butyrate providing the carbon substrate for exponential-phase growth and stationary-phase maintenance. Changing the substrate in the preculture used as the inoculum from acetate to butyrate had no effect on the order of utilization. As noted above, changes in substrate utilization caused changes in expression from the nap promoter within 2 to 3 h.
FIG. 6.
Hierarchical use of acetate over butyrate in mixed-carbon-substrate cultures of P. pantotrophus/pnap-lacZ and correlation with nap promoter activity. The carbon sources used for preculture of the inocula were 50 mM acetate (A, B, and C) and 10 mM butyrate (D, E, and F). The culture medium contained 50 mM acetate and 10 mM butyrate (squares), 10 mM acetate and 10 mM butyrate (triangles), or 1 mM acetate and 10 mM butyrate (circles). In panels B and E the solid symbols indicate the acetate concentration and the open symbols indicate the butyrate concentration. The carbon substrate concentration was measured by 1H-NMR spectroscopy. β-Galactosidase (β-gal) activity was calculated as described by Miller (11). Growth was measured by determining the optical density at 600 nm (OD600nm).
DISCUSSION
In this paper an analysis of growth kinetics, carbon substrate utilization, and nap promoter activity in P. pantotrophus with a range of carbon substrates is described. Our data show the strict hierarchical preference for succinate over acetate, which in turn is preferred over butyrate as the carbon source for growth. This hierarchical preference shows a strong correlation with the relative expression levels of the nap promoter when it is fused to the lacZ gene. Thus, it is likely that a signal or signals that control carbon source selection are also involved in regulating nap expression. The molecular identities of these signals will have to await further investigation. Here we have analyzed the possible reasons for the hierarchical selection of carbon sources, in the context that it is already known that the amounts of the Nap protein in butyrate-grown cells are higher than those in succinate-grown cells (13). Thus, for the first time we demonstrate that there is a correlation between transcription from the nap promoter and the amount of holoenzyme produced.
The data in Fig. 3A, C, and E show the low growth rates for cultures growing on butyrate. This is significant because it suggests that cells growing on butyrate are stressed compared to cells growing on succinate. From an energetic standpoint this is logical as butyrate is more reduced than the average cell material. This poses a problem for the cell as the excess reducing equivalents, which could be generated in the cytoplasm from its oxidative metabolism, must be discarded.
Why should acetate be selected in preference to butyrate? At first sight the greater average reduction state of each carbon in butyrate might suggest that this compound would be the preferred substrate; more electrons would be available for transfer to a respiratory chain electron acceptor (e.g., oxygen), with a concomitant greater yield of ATP per carbon atom supplied to the cell. The fact that acetate is preferred indicates that use of butyrate presents the cell with a surfeit of reductant and thus potentially a surfeit of ATP. An excess of ATP should result in a very low concentration of ADP, and since respiration is coupled to phosphorylation of ADP, a constraint will be placed on the respiratory rate. The excess reductant can be disposed of only if there is a pathway for respiratory electron flow that is uncoupled from ATP synthesis. Such a pathway is provided by the ubiquinol-Nap nitrate reductase. As we have proposed before (17), this explains why the activity of the latter pathway is elevated in butyrate-grown cells. Thus, nitrate is reduced to nitrite via this pathway, when it is added to aerobic cultures growing on butyrate. The extent to which this pathway has to be utilized during growth on butyrate is not known, although this study shows that addition of 5 mM nitrate has no effect on growth with butyrate. In the absence of nitrate the cells have other strategies for the dissipation of excess reductant (14), and it is likely these operate both in the presence and in the absence of nitrate.
Based on the metabolism of acetate and the metabolism of butyrate, it is probable that the two compounds are taken up by a similar transport system and then phosphorylated at the expense of ATP to give an acyl-phosphate. Reaction with coenzyme A (CoA) in each case would give acyl-CoA. In the case of butyrate one round of β-oxidation would then generate two molecules of acetyl-CoA, from which point the metabolism would be identical for the two carbon sources. This would involve partition of the acetyl groups between oxidation via the tricarboxylic acid cycle and oxidation via the glyoxylate cycle for formation of malate, which would then go on to be incorporated into cell material. Hence, we conclude that the need to use some acetyl groups derived from acetate as the sole carbon source cannot place a severe demand for ATP on the cell; otherwise it would be logical for butyrate to be selected in preference to acetate.
The selection of succinate over either of the monocarboxylic acids suggests that the ATP requirement of the cells can be met as a result of the succinate dehydrogenase reaction and the oxidation of the NADH formed in the conversion of malate to oxaloacetate. It is also conceivable that succinate is converted via oxaloacatate to pyruvate and then to acetyl-CoA for oxidation. The nature of the signal or signals controlling the hierarchy for succinate, acetate, and butyrate utilization is not known. The evidence from this work suggests that regulation is dependent on the metabolism of a carbon substrate rather than its presence in the medium, but in the case of acetate and butyrate the only obvious intermediate metabolite specific to either pathway is butyryl-CoA.
A major metabolic difference between growth on succinate and growth on either butyrate or acetate is that the glyoxylate cycle is required for the latter two growth substrates. Nothing is known about the factors controlling the regulation of expression of the enzymes of the glyoxylate cycle in Paracoccus species, but it is known that that in E. coli the ArcAB and Crp (6), IclR (10, 22), and AtoC/FadR (9, 20) proteins control transcription of genes coding for tricarboxylic acid cycle, glyoxylate cycle, and β-oxidation enzymes, respectively. However, knowledge concerning some of the signals and effectors acting on these regulators is limited. For glyoxylate bypass control, the prime candidates might be considered to be acetate or a high acetyl-CoA/CoA ratio; the latter can be expected during growth on acetate and butyrate. Future work will be devoted to answering how the butyrate-specific pathway of metabolism is induced and how the presence of acetate prevents the expression of this pathway in Paracoccus species.
During revision of the present paper two studies of nap regulation in other organisms appeared. In Rhodobacter sphaeroides DSM158 it was found that nap-lacZ transcriptional fusions resulted in no change in β-galactosidase activity irrespective of (i) the presence or absence of nitrate under either aerobic or anaerobic growth conditions and (ii) the nature of the carbon source, which ranged from butyrate to malate. For reasons that are not understood yet, this behavior differs from the situation in P. pantotrophus, although increased flux through the Nap system attributed to enzyme activation was seen when R. sphaeroides was grown phototrophically on a highly reduced carbon source, such as butyrate (5). The regulation of nap expression in E. coli presents a complete contrast, as reemphasized by another recent paper. Stewart et al. (21) showed that NapA in E. coli is differentially expressed in response to nitrate and supported the work of others in proposing that Nap is designed to function in E. coli when low nitrate concentrations limit the bioenergetic efficiency of nitrate respiration using the membrane-bound reductase (Nar) in E. coli.
Acknowledgments
We thank H. J. Sears, G. P. Butland, and E. H. J. Gordon for helpful discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council (grants C0866 and P15877 to S.J.F. and D.J.R.) and by a BBSRC PMS committee studentship to M.J.K.E. K.K.B. was supported by the Medical Research Council.
REFERENCES
- 1.Bell, L. C., D. J. Richardson, and S. J. Ferguson. 1990. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 265:85-87. [DOI] [PubMed] [Google Scholar]
- 2.Bell, L. C., M. D. Page, B. C. Berks, D. J. Richardson, and S. J. Ferguson. 1993. Insertion of transposon Tn5 into a structural gene of the membrane-bound nitrate reductase of Thiosphaera pantotropha results in anaerobic overexpression of periplasmic nitrate reductase activity. J. Gen. Microbiol. 139:3205-3214. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., D. J. Richardson, A. Reilly, A. C. Willis, and S. J. Ferguson. 1995. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem J. 309:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks, B. C., M. D. Page, D. J. Richardson, A. Reilly, A. Cavill, F. Outen, and S. J. Ferguson. 1995. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol. Microbiol. 15:319-331. [DOI] [PubMed] [Google Scholar]
- 5.Gavira, M., M. D. Roldan, F. Castillo, and C. Moreno-Vivian. 2002. Regulation of nap gene expression periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J. Bacteriol. 184:1693-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guest, J. R., and G. C. Russell. 1992. Complexes and complexities of the citric acid cycle in Escherichia coli. Curr. Top. Cell. Regul. 33:231-247. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-563. [DOI] [PubMed] [Google Scholar]
- 8.Inoue, H., H. Nojima, and H. Okayame. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins, L. S., and W. D. Nunn. 1987. Regulation of the ato operon by the atoC gene in Escherichia coli. J. Bacteriol. 169:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maloy, S. R., and W. D. Nunn. 1982. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J. Bacteriol. 149:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 13.Richardson, D. J., and S. J. Ferguson. 1992. The influence of the carbon substrate on the activity of the periplasmic nitrate reductase in aerobically grown Thiosphaera pantotropha. Arch. Microbiol. 157:535-537. [Google Scholar]
- 14.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146:551-571. [DOI] [PubMed] [Google Scholar]
- 15.Robertson, L. A., and J. G. Kuenen. 1983. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulphur bacterium. J. Gen. Microbiol. 129:2847-2855. [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Sears, H. J., S. Spiro, and D. J. Richardson. 1997. Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd 1222. Microbiology 143:3767-3774. [DOI] [PubMed] [Google Scholar]
- 18.Sears, H. J., G. Sawers, B. C. Berks, S. J. Ferguson, and D. J. Richardson. 2000. Control of periplasmic nitrate reductase gene expression (napEDABC) from Paracoccus pantotrophus in response to oxygen and carbon substrates. Microbiology 146:2977-2985. [DOI] [PubMed] [Google Scholar]
- 19.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 20.Simons, R. W., P. A. Egan, H. T. Chute, and W. D. Nunn. 1980. Regulation of fatty acid degradation in Escherichia coli: isolation and characterization of strains bearing insertion and temperature-sensitive mutations in gene fadR. J. Bacteriol. 142:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart, V., Y. R. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunnarborg, A., D. Klumpp, T. Chung, and D. C. LaPorte. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]