Abstract
IS256 is a highly active insertion sequence (IS) element of multiresistant staphylococci and enterococci. Here we show that, in a Staphylococcus epidermidis clinical isolate, as well as in recombinant Staphylococcus aureus and Escherichia coli carrying a single IS256 insertion on a plasmid, IS256 excises as an extrachromosomal circular DNA molecule. First, circles were identified that contained a complete copy of IS256. In this case, the sequence connecting the left and right ends of IS256 was derived from flanking DNA sequences of the parental genetic locus. Second, circle junctions were detected in which one end of IS256 was truncated. Nucleotide sequencing of circle junctions revealed that (i) either end of IS256 can attack the opposite terminus and (ii) the circle junctions vary significantly in size. Upon deletion of the IS256 open reading frame at the 3′ end and site-directed mutageneses of the putative DDE motif, circular IS256 molecules were no longer detectable, which implicates the IS256-encoded transposase protein with the circularization of the element.
IS256 is an insertion sequence (IS) element of staphylococci and enterococci that was originally identified as a bordering component of the composite aminoglycoside resistance-mediating transposon Tn4001 (2, 8, 9). IS256, together with 32 other elements, forms a family of ISs that also includes eukaryotic relatives (3). Information about IS256 is very limited (10), despite its wide distribution. Recently, it was demonstrated that the flanking IS256 copies of Tn4001 form tandem dimers and IS circles in an Escherichia coli genetic background (17). In previous studies we have been able to show that IS256 is associated with genetic rearrangements, causing phenotypic changes in its natural host Staphylococcus epidermidis. Multiresistant S. epidermidis as well as Staphylococcus aureus are the most common causes of nosocomial infections associated with implanted biomaterials. In these clinical staphylococcal isolates IS256 proved to be highly active and caused a variety of genetic aberrations, such as reversible gene inactivations, DNA rearrangements, and large chromosomal deletions that affected the expression of virulence- and resistance-associated genes (12, 27-30). Specifically, biofilm formation, which is one of the major factors in S. epidermidis pathogenesis, is influenced by IS256. The element causes phase variation of biofilm expression in S. epidermidis by inactivation of the icaADBC operon, which encodes enzymes responsible for biofilm synthesis (28). Apparently, the icaC gene represents a hot spot for IS256 insertions. It was shown that the element creates 8- and 7-bp target site duplications during transposition (28; S. H. Cho and W. Ziebuhr, unpublished data). The spontaneous ica::IS256 insertions are reversible and, accordingly, nucleotide sequencing of biofilm-forming revertants confirmed the precise excision of the element, including the initially duplicated target sequences. In these initial studies, however, the molecular mechanisms that mediate the excision of IS256 have not been elucidated in detail. Also, we were unable to decide from Southern hybridization experiments whether IS256 transposes by a conservative or replicative strategy (I. Loessner and W. Ziebuhr, unpublished data). These observations prompted us to also consider alternative transposition mechanisms, and the data we obtained here strongly suggest that single IS256 copies can excise as extrachromosomal circular DNA molecules in S. epidermidis and S. aureus. This process depends on the IS256-encoded protein, which is predicted to be the transposase of the element.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria-Bertani (LB) broth consisting of 1% casein peptone, 0.5% yeast extract, and 0.5% sodium chloride. For DNA extraction, S. epidermidis and S. aureus were cultured in LB broth supplemented with 1% glycine. Recombinant E. coli and S. aureus were cultivated under selective antibiotic pressure with 100 μg of ampicillin and 10 μg of chloramphenicol/ml, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. epidermidis 307 | icaADBC positive; IS256 positive; strong biofilm-producer; human blood culture isolate | This study |
| S. epidermidis 307/95 | Biofilm negative, icaC::IS256 insertion mutant of S. epidermidis 307 | This study |
| S. aureus RN4220 | Restriction-deficient strain derived from S. aureus 8325-4 | 6 |
| E. coli DH5α | Host strain for construction of recombinant plasmids; lacZ negative | MBI Fermentas, St. Leon-Rot, Germany |
| E. coli GM2163 | Adenine, cytosine methylase mutant | MBI Fermentas, St. Leon-Rot, Germany |
| Plasmids | ||
| pRB472 | Gram+/gram− shuttle vector, carrying ampicillin and chloramphenicol resistance | R. Brückner (personal communication) |
| pGEM-T-Easy | AT cloning vector, carrying ampicillin resistance | Promega, Mannheim, Germany |
| pIL2 | Shuttle vector pRB472 containing the the icaC::IS256 insertion of S. epidermidis 307/95 | This study |
| pIL2Δtnp | Shuttle vector pRB472 carrying the icaC::IS256Δtnp insertion of S. epidermidis 307/95 with a mutated tnp256 gene | This study |
| pIL2-D167A | pIL2 carrying a mutated transposase gene at position D167 (D167A) | This study |
| pIL2-D233A | pIL2 carrying a mutated transposase gene at position D233 (D233A) | This study |
| pIL2-E341A | pIL2 carrying a mutated transposase gene at position E341 (E341A) | This study |
Construction of plasmids.
A 2,413-bp DNA fragment containing a part of icaC with the inserted IS256 was amplified from S. epidermidis 307/95 by PCR. Primer 1 (5′-ATA AAC TTG AAT TAG TGT ATT-3′) and primer 2 (5′-ATA TAT AAA ACT CTC TTA ACA-3′) bind at positions 3131 and 4099 of the published ica sequence (accession number U43366). Reaction conditions for PCR were as follows: 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 1 min for 42°C, and 1.5 min for 70°C. The PCR product was inserted into plasmid pGEM-T-Easy (Promega, Mannheim, Germany) by AT cloning, yielding plasmid pIL1. For construction of pIL2, the icaC::IS256 containing fragment was removed by PstI and SphI digestion from pIL1 and inserted into the PstI/SphI-digested pRB472 shuttle vector, resulting in plasmid pIL2. To construct plasmid pIL2Δtnp, pIL1 was transformed into the adenine-cytosine methylase-deficient host E. coli GM2163 (MBI Fermentas, St. Leon-Rot, Germany). After reisolation, the vector was restricted with ClaI, which cleaves the icaC::IS256 insert twice in a distance of 20 bp. The 20-bp fragment was eliminated, the linearized vector was isolated, purified, and religated, resulting in pIL1Δtnp. For construction of pIL2Δtnp, the PstI/SphI insert of pIL1Δtnp was ligated into pRB472. Insertion and the correct sequence of cloned PCR fragments and the 20-bp ClaI deletion were confirmed by nucleotide sequencing. After propagation in E. coli DH5α, the shuttle vectors pIL2 and pIL2Δtnp were transformed into S. aureus RN4220 by electroporation (20).
Site-directed mutagenesis of the DDE motif of the IS256 transposase.
Site-directed mutagenesis of the transposase gene was performed by recombination PCR as described previously (26). Residues D167, D233, and E341 of the IS256 copy on the pIL1 vector were replaced by an alanine residue, respectively, by using the following mutagenic oligonucleotides: mutation D167A (primer 1, 5′-AAA AAT TAT CCT TAC TTA ATG ACC gct GTA CTC TAT-3′; primer 2, 5′-TTC TCG TAC TTT TAT ATA GAG TAC agc GGT CAT TAA-3′), mutation D233A (primer 3, 5′-CAA GGT ACG GAA CTC GTT ATT TCT gct GCG CAC AAA-3′; primer 4, 5′-GGC AGA GAC TAA TCC TTT GTG CGC agc AGA AAT AAC-3′, and mutation E341A (primer 5, 5′-CGA CTA AAG AGT ACC AAT CTA ATT gca CGA CTG AAT-3′; primer 6, 5′-TCT GCG TAC TTC TTG ATT CAG TCG tgc AAT TAG ATT-3′) (replaced triplets are marked by lowercase letters). PCR amplifications of the whole vectors with the mutated transposase genes were carried out with 1 ng of pIL1 as the template DNA. PCR conditions were 1 min at 94°C, 1 min at 45°C, and 7 min at 72°C for 35 cycles. To remove any residual template DNA, the PCRs were treated with DpnI, which restricts methylated DNA and leaves the PCR amplicons intact. The 5.3-kb linear PCR products were then transformed directly into competent E. coli DH5α cells and plated onto LB agar containing 100 μg of ampicillin/ml. The resulting clones were picked and analyzed for the presence of vector DNA. Introduction of the correct mutations were verified by nucleotide sequence analyses of the complete transposase genes in pIL1-D167A, pIL1-D233A, and pIL1-E341A. For propagation in S. aureus, the PstI/SphI-restricted inserts were cloned again in the pRB472 shuttle vector resulting in in pIL2-D167A, pIL2-D233A, and pIL2-E341A and transformed into S. aureus 4220 by electroporation (20).
Preparation of extrachromosomal DNA from E. coli, S. aureus, and S. epidermidis for circle-specific PCR.
Extrachromosomal template DNA was prepared from a 1.5-ml overnight culture incubated at 37°C, followed by incubation at 4°C for 2 h (15) according to the alkaline lysis method (19). For S. epidermidis, possible chromosomal DNA contaminations were eliminated by treatment of the cytoplasmic DNA fraction with 200 U of exonuclease III (Amersham Pharmacia, Freiburg, Germany) and 20 U of exonuclease VII (Amersham Pharmacia, Freiburg, Germany) for 18 h at 37°C. For IS256 circle detection in E. coli, S. aureus, and S. epidermidis, 1 μl of the final volume of 50 μl was used as the template DNA for circle-specific PCR.
PCR assay to detect IS256 circles.
IS256 circle junctions were detected by using a set of PCR primers directed in opposite orientations (with 3′ extended ends directed outwardly). Outward primer 1 (5′-CTC ATA ATA GCC ATT TCG TTG-3′) and outward primer 2 (5′-GCT TGC GCA TCA TTG GAT G-3′) bind at positions 279 and 1029 of the published IS256 sequence (accession number M18086). The reaction conditions for the PCR were as follows: 95°C for 2 min, followed by 45 cycles of 95°C for 30 s, 54°C for 30 s, and 70°C for 40 s. PCR products were ligated into the plasmid pGEM-T-Easy by AT cloning according to the manufacturer's protocol. The sequence of the insert was determined by nucleotide sequencing. Exonuclease-treated samples were checked for the presence of genomic DNA by PCR with icaA-specific primers that amplify a 814-bp fragment of the icaA locus (28).
Native agarose gel electrophoresis and Southern blotting for detection of IS256 circular forms.
A total of 2 μg of extrachromosomal DNA from S. aureus pIL2 and S. aureus pIL2Δtnp were restricted with ClaI and loaded on a 0.7% Tris-phosphate-EDTA (TPE)-agarose gel. Southern hybridization with an enhanced chemiluminescence-labeled IS256-specific probe was performed according to instructions of the manufacturer (Amersham Pharmacia, Freiburg, Germany).
RESULTS
Identification of IS256 circles in a S. epidermidis clinical isolate.
For our studies we used the biofilm-forming S. epidermidis clinical isolate S. epidermidis 307. In addition to a single Tn4001 insertion, the strain carries multiple free IS256 copies in its chromosome which are active and produce spontaneous, biofilm-negative ica::IS256 insertion mutants. For our studies the icaC::IS256 insertion mutant S. epidermidis 307/95 was selected and analyzed for its capacity of precise IS256 excisions as described previously (28). Repeated passages resulted in biofilm-positive revertants that had lost the IS256 insertion and nucleotide sequence analyses revealed the precise excision of the element from the icaC gene, including the duplicated 8-bp target site (data not shown).
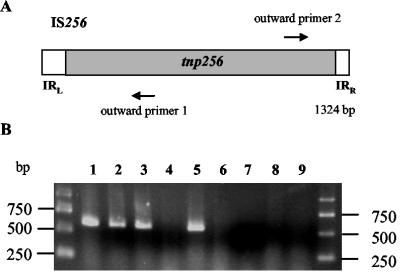
In a first set of experiments we wanted to answer the question whether IS256 can exist as circular DNA molecule in S. epidermidis 307 and its spontaneous icaC::IS256 insertion mutant S. epidermidis 307/95. For this purpose, we isolated extrachromosomal DNA from both strains and used it as template DNA in PCRs with IS256-specific outward-directed primers (Fig. 1A). In this experiment, a specific 600-bp PCR product can only be generated if IS256 forms an extrachromosomal DNA circle or if two adjacent IS256 copies are present on the chromosome. To exclude the latter possibility, we removed remaining traces of chromosomal DNA from the extrachromosomal DNA fraction by treatment with exonuclease III and exonuclease VII. The efficiency of this procedure was confirmed by doing a PCR control amplification of a chromosomally encoded gene (i.e., icaA). No PCR products were amplified in these control reactions, indicating that the DNA preparations were free of chromosomal DNA (data not shown). In contrast, when the same DNA fractions were used as templates for the IS256-specific PCR, products of ca. 600 bp in size were obtained (Fig. 1B, lanes 1 and 2).
FIG. 1.
(A) IS256 and positions of outward-reading primers used for circle-specific PCRs. White boxes mark the left and right arms of the element containing the left (IRL) and right (IRR) inverted repeats. The gray rectangle represents the putative transposase gene (tnp256) of IS256. (B) Agarose gel electrophoresis of PCR fragments amplified with circle-specific outward-reading primers. Templates were obtained from extrachromosomal DNA preparations of S. epidermidis 307 (lane 1), S. epidermidis 307/95 (lane 2), E. coli(pIL2) (lane 3), E. coli(pIL2Δtnp) (lane 4), S. aureus(pIL2) (lane 5), S. aureus(pIL2Δtnp) (lane 6), S. aureus(pIL2-D167A) (lane 7), S. aureus(pIL2-D233A) (lane 8), and S. aureus(pIL2-E341A) (lane 9).
Detection of IS256 circles in recombinant S. aureus and E. coli.
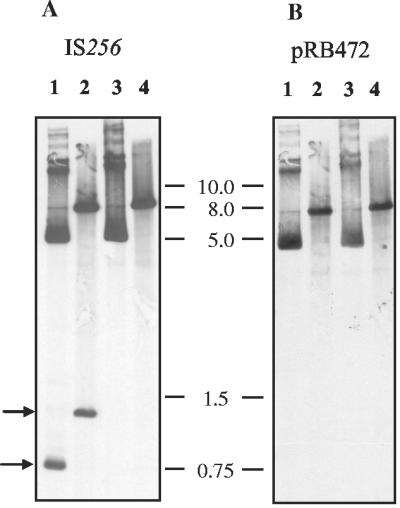
To confirm the existence of IS256 circles, we decided to study the circularization process in a more controlled manner by using recombinant plasmids in IS256-free S. aureus and E. coli genetic backgrounds. To this end, the natural chromosomal icaC::IS256 insertion from S. epidermidis 307/95 was cloned into the E. coli-Staphylococcus shuttle vector pRB472, and the resulting plasmid pIL2 was propagated in the recombination deficient hosts E. coli DH5-α and S. aureus RN4220, respectively. For visualization of putative IS256-circles by agarose gel electrophoresis, extrachromosomal DNA was prepared from S. aureus(pIL2) and S. aureus(pIL2Δtnp), carrying a mutated transposase gene (see below). IS256-specific Southern hybridization was performed with undigested and ClaI-restricted plasmid DNA of pIL2 and pIL2Δtnp, respectively (Fig. 2A). Figure 2A shows the appearance of an additional IS256-specific fragment of ca. 0.75 kb in size in the plasmid fraction obtained from S. aureus cells harboring the pIL2 plasmid (Fig. 2A, lane 1). The signal was not detectable in cells carrying the pIL2Δtnp vector with a mutated transposase gene (Fig. 2A, lane 3). Since IS256 contains two immediately adjacent ClaI restriction sites, use of this enzyme should result in linearization of a putative IS256 circle. As also indicated in Fig. 2A, ClaI restriction of the pIL2 plasmid fraction resulted in an IS256-specific fragment of ca. 1.3 kb, which exactly matches the size of an linearized IS256 circle (Fig. 2A, lane 2). In contrast, the signal was absent in the ClaI-restricted plasmid fraction of S. aureus(pIL2Δtnp) (Fig. 2A, lane 4). Rehybridization of the blot with the pRB472 plasmid indicated that the 0.75- and 1.3-kb fragments in S. aureus(pIL2) did not contain DNA of the vector backbone (Fig. 2B, lanes 1 and 2). We concluded from these experiments that the 0.75-kb fragment in S. aureus(pIL2) represents IS256 circles that differ in mobility from their linear form.
FIG. 2.
(A) Detection of IS256 circular forms by IS256-specific Southern hybridization of extrachromosomal DNA obtained from S. aureus(pIL2) (lanes 1 and 2) and S. aureus(pIL2Δtnp). (lanes 3 and 4). Lanes: 1, undigested plasmid fraction of pIL2; 2, ClaI-restricted plasmid fraction of pIL2; 3, undigested plasmid fraction of pIL2Δtnp; 4, ClaI-restricted plasmid fraction of pIL2Δtnp. The arrows mark the circular (lower) and linearized (upper) forms of the IS256 circles.(B) Rehybridization of the same blot with the vector backbone pRB472 but without the icaC:: IS256 insert.
Next, plasmid fractions of E. coli(pIL2) and S. aureus(pIL2) were used as templates in circle-specific PCRs. Figure 1B (lanes 3 and 5) shows that circle-specific PCR products were obtained both from E. coli(pIL2) (lane 3) and S. aureus(pIL2) (lane 5). The combined data indicate that IS256 circularization proceeds in S. epidermidis wild-type strains, as well as in a recombinant S. aureus and E. coli genetic background, suggesting that the reaction is independent from species-specific host factors.
Circle formation depends on the function of the putative IS256 transposase protein.
IS256 is predicted to contain a single open reading frame encoding a protein of 390 amino acid residues (2). The amino acid sequence shows strong homologies to bacterial transposases and contains a typical conserved “D,D,(107)E” motif (residues D167, D233, and E341) that is supposed to be involved in the formation of the catalytic center of the enzyme (4). To investigate whether IS256 circle formation is mediated by the putative IS256 transposase protein or by host-cell mediated processes, the plasmid pIL2Δtnp was constructed which carries a stop codon at position 927 of the transposase gene tnp256 (GenBank accession no. M18086). Thus, the IS256 copy on pIL2Δtnp encodes a protein lacking the C-terminal 82 amino acid residues, including the glutamate residue of the putative DDE motif. The vector was propagated in E. coli and S. aureus, respectively, and extrachromosomal DNA preparations from E. coli(pIL2Δtnp) and S. aureus(pIL2Δtnp) were used as templates in IS256 circle-specific PCRs. As indicated in Fig. 1B (lanes 4 and 6), no circle-specific PCR fragments were detected both in E. coli and in S. aureus. The results of this experiment suggest that circular forms of IS256 do not exist in E. coli(pIL2Δtnp) and S. aureus(pIL2Δtnp), and it is tempting to speculate that this is due to the deletion of the putative transposase protein at the C terminus. To investigate whether or not the predicted DDE motif of the putative transposase protein is involved in the circle-forming activity of the enzyme, the charged amino acid residues D167, D233, and E341 of the IS256 copy on pIL2 were replaced by alanine. The resulting plasmids pIL2-D167A, pIL2-D233A, and pIL2-E341A were propagated in S. aureus, and extrachromosomal DNA preparations were used again in circle-specific PCRs. As indicated in Fig. 1B (lanes 7, 8, and 9), no circle-specific PCR fragments were detectable in S. aureus strains carrying site-specific mutated transposase genes. The data give evidence that the transposase of the element is crucial for the generation of IS circles and that the predicted DDE motif is essentially involved in the function of the enzyme.
Analyses of circle junctions.
For investigation of circle junctions, IS256 circle-specific PCR fragments obtained from S. epidermidis, S. aureus, and E. coli were cloned into E. coli(pGEM-T-Easy), and from each PCR product four different clones were analyzed by nucleotide sequencing. The experiments revealed that the fragments consisted of the left and right ends of IS256 connected by short DNA stretches of various lengths and nucleotide sequences (Fig. 3B). Thus, one circle junction obtained from E. coli(pIL2) contained the left and right arms of IS256 abutted by a 19-bp nucleotide stretch (Fig. 3B, circle junction 1). These 19 bp corresponded perfectly to the nucleotide sequence of the icaC gene immediately upstream of the IS256 insertion site in pIL2, indicating that the IS256 insertion on the plasmid represents the source for the detected IS256 circle (Fig. 3A). Nucleotide sequence analysis of four clones obtained from S. aureus(pIL2) revealed two different circle junctions (Fig. 3, circles 2 and 3). They contained 5- and 6-bp nucleotide stretches, respectively, matching the icaC gene downstream of the IS256 insertion site in S. aureus pIL2 (Fig. 3A and B, circles 2 and 3). These results suggest that each end of the element can be subject of a strand transfer reaction. Also, the PCR products of S. epidermidis 307/95 represented a mixture of IS256-specific DNA. The nucleotide sequences of the circle junctions varied (Fig. 3B, circles 4 and 6), which is due to the different insertion sites of the element in the genome of S. epidermidis 307/95. Interestingly, one circular intermediate, detected in this spontaneous icaC::IS256 insertion mutant, contained a 6-bp circle junction which corresponded perfectly to the nucleotide sequence of the icaC gene immediately adjacent to the IS256 insertion site in S. epidermidis 307/95 (Fig. 3B, circle junction 5). This result suggests also that the chromosomal IS256 copy inserted in the icaC gene of S. epidermidis 307/95 undergoes circle formation. In addition to circle junctions consisting of both IS256 ends and adjacent DNA, fragments were analyzed from S. epidermidis and E. coli, which contained only one entire terminus linked to a truncated part of the opposite end. Truncations could affect either end and varied in size from 8 to 253 bp (Fig. 3C). It is tempting to speculate that these fragments reflect small IS256 circles that might be formed when the strand transfer reaction of a released IS end is directed to the nucleotide sequence of the element itself and not to the adjacent DNA. Together with the data on the variable length of the nucleotide sequence stretches in the complete circles, these results suggest a relatively low specificity of the strand transfer reaction during the circle-forming process.
FIG. 3.
Nucleotide sequence analyses of IS256 circle junctions detected in S. epidermidis, S. aureus, and E. coli. (A) Schematic representation of the IS256 insertion in the icaC gene in pIL2. The 8-bp target site duplications are marked by boldface letters. Dotted arrows indicate the putative strand transfer reactions of transposon ends into adjacent DNA. (B and C) Schematic illustration of IS256 circle junctions and nucleotide sequence analyses of complete (B) and truncated (C) circle junctions detected in S. epidermidis 307, S. epidermidis 307/95, E. coli(pIL2), and S. aureus(pIL2). IRL, inverted repeat (left); IRR, inverted repeat (right).
DISCUSSION
IS elements have adopted different mechanisms to move from one insertion site to another (for a recent review, see reference 4). Transposition can proceed by a conservative cut-and-paste mechanism, in which the element is excised from a donor molecule and inserted into a new target site. Another strategy involves semiconservative replication of the IS by the formation of a cointegrate. Increasing experimental evidence obtained during the past several years demonstrates that some bacterial IS elements use an alternative transposition mechanism which is characterized by circular IS intermediates. In general, the outcome of transposition reactions depends on the liberation of an element from the donor DNA by action of the transposase. Cut-and-paste transposition requires the complete excision of the IS from a donor DNA molecule and results, therefore, in double-strand cleavages at both transposon ends. In contrast, elements that transpose by cointegrate formation are only released from adjacent DNA at their 3′ end and remain linked to the donor backbone with the 5′ terminus. In both reactions, free 3′ OH groups are generated at the transposon ends that can attack a phosphodiester bond at a new insertion site. Circle-mediated transposition proceeds by a similar sequel of chemical reactions, but the most intriguing feature of this mechanism is an intramolecular strand transfer reaction of one released transposon terminus to its own opposite end. A well-studied element transposing by this pathway is IS911, a member of the IS3 family. Here, circularization is generated by introduction of a single-strand break at one transposon end and transfer of the released 3′OH group to the opposite end of the same strand (16). This single transposon strand is then resolved by an unknown mechanism, resulting in a circularized copy of the element in which both transposon ends are abutted. IS circularization was also observed in several other elements, such as IS1, IS2, IS492, Tn7, Tn10, Tn916, IS30, and others (1, 5, 7, 13, 15, 21, 24). Recently, it was shown in E. coli that a Tn4001 copy on a plasmid results in tandem IS dimers and circles which arose from the flanking IS256 at the ends of the transposon (17). The data presented in this study indicate that IS256 can to excise as an extrachromosomal DNA circle in its natural hosts S. epidermidis and S. aureus. Use of a recombinant S. aureus plasmid carrying a single IS256 insertion shows that IS256 circularization occurs also independently from Tn4001. Moreover, the results give evidence that the putative IS256-encoded transposase is essential for the circularization reaction. Specifically, the site-directed mutagenesis of the predicted DDE motif shows that these amino acid residues are indeed critically involved in the function of the protein. At the present stage of experimental work we cannot decide how IS256 donor strand cleavage and strand transfer reactions proceed, but transposases and recombinases with the DDE signature are known for their activity to execute single-strand cleavages at transposon termini (4), and it is tempting to speculate that this is also true for IS256. In addition, the possible involvement of host factors in the circularization process is an interesting question that needs to be addressed in future experiments.
A surprising result of this study were the various sizes of the circle junctions in the IS circles, suggesting that in IS256 the circle formation-associated strand transfer reaction occurs with low specificity and results therefore in a range of different circle species which differ in size and nucleotide sequence. This finding is in contrast to other circle-forming elements, in which circle junctions mostly include a constant number of neighboring DNA molecules. In this respect it is an interesting question whether or not circle formation can cause the excision of an element from a donor DNA molecule. If this is the case, the mechanism could mediate a precise excision of the element when the strand transfer reaction into the flanking sequence includes the 8- or 7-bp duplicated target sequence. We do not yet have enough evidence to substantiate this hypothesis unambiguously. However, precise excisions of IS256 from the icaC gene were proven previously in S. epidermidis in the course of biofilm phase variation (28), and also the icaC::IS256 insertion in S. epidermidis 307/95 has the capacity for exact excisions and restoration of the wild-type sequence. More experimental work is needed to elucidate whether the excision reaction is associated with the circle-forming process or mediated by a transposase-independent recombination event between the duplicated target sites.
The data of this study also indicate that circularization of the single IS256 copy on the pIL2 plasmid is not oriented. In fact, circle junction analyses revealed that each end of the element can attack the opposite terminus, respectively. This behavior differs from that of the IS256 copies in Tn4001, where the left end of the element constantly attacks the right end (17). Moreover, no IS256 dimers were observed on the pIL2 plasmid, because linearization of the vector with a restriction enzyme that cuts in the multiple cloning site (i.e., PstI) clearly resulted in a single vector fragment (data not shown). Formation of tandem IS256 repeats were detected, however, in Tn4001 when the transposon was propagated on an E. coli plasmid (17). One explanation for this difference might be that the dimerization reaction detected in Tn4001 is related to the number of IS copies on the replicon. Tn4001 carries two IS256 elements at the ends of the transposon that might undergo a similar inter-IS recombination event as described for IS911 (25). In contrast, the single IS256 insertion on pIL2 only has the capacity for a circularization reaction, which might be due to an intra-IS, transposase-mediated recombination process between the IS256 termini.
In several IS elements it was shown that head-to-tail junctions, which can arise by circle formation or tandem IS duplication, are highly active in transposition (14, 22, 23, 25). Although we only give experimental evidence for the formation of extrachromosomal IS256 circles in the present study, it is conceivable that these structures represent similar intermediates in transposition since they have been associated with other circle-forming elements. In this connection, the possible role of IS256 in the flexibility of the staphylococcal genome is an interesting issue. Multiresistant S. epidermidis and S. aureus carry multiple IS256 copies on their chromosomes, and phenotypic changes in these isolates (e.g., heterogenous gene expression of virulence and resistance traits) are often accompanied by variant IS256 hybridization patterns (27-29). With respect to the IS256 circle-forming process observed here, it is conceivable that incompletely removed target site duplications or partially truncated IS copies might generate a great variety of mutations and deletions. Moreover, IS256 does not contain transcription termination signals, and the nucleotide sequence is predicted to contain various promoter structures that have the capacity to drive neighboring gene expression (11, 17, 18, 28). It is therefore tempting to speculate that the presence and activity of IS256 in the staphylococcal genome might act as a driving force in the generation of heterogeneous gene expression and the microevolution of nosocomial staphylococci. However, more experimental work is needed to substantiate this assumption.
Acknowledgments
We thank Hennes Kränzler for excellent technical assistance and Reinhold Brückner, Department for Microbiology, University of Kaiserslautern, Kaiserslautern, Germany, for providing plasmid pRB472.
This work was supported by the Sonderforschungsbereich 479 of the University of Würzburg and grant ZI 655/1-1 of the Deutsche Forschungsgemeinschaft to W.Z.
REFERENCES
- 1.Biery, M. C., M. Lopata, and N. L. Craig. 2000. A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol. 297:25-37. [DOI] [PubMed] [Google Scholar]
- 2.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 3.Eisen, J. A., M. I. Benito, and V. Walbot. 1994. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 5.Kiss, J., and F. Olasz. 1999. Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Mol. Microbiol. 34:37-52. [DOI] [PubMed] [Google Scholar]
- 6.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 7.Lewis, L. A., and N. D. Grindley. 1997. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol. Microbiol. 25:517-529. [DOI] [PubMed] [Google Scholar]
- 8.Lyon, B. R., M. T. Gillespie, and R. A. Skurray. 1987. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 133(Pt. 11):3031-3038. [DOI] [PubMed] [Google Scholar]
- 9.Lyon, B. R., J. W. May, and R. A. Skurray. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193:554-556. [DOI] [PubMed] [Google Scholar]
- 10.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mempel, M., H. Feucht, W. Ziebuhr, M. Endres, R. Laufs, and L. Grüter. 1994. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 38:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morisato, D., and N. Kleckner. 1987. Tn10 transposition and circle formation in vitro. Cell 51:101-111. [DOI] [PubMed] [Google Scholar]
- 14.Olasz, F., R. Stalder, and W. Arber. 1993. Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. Mol. Gen. Genet. 239:177-187. [DOI] [PubMed] [Google Scholar]
- 15.Perkins-Balding, D., G. Duval-Valentin, and A. C. Glasgow. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 9:2846-2858. [DOI] [PubMed] [Google Scholar]
- 17.Prudhomme, M., C. Turlan, J. P. Claverys, and M. Chandler. 2002. Diversity of Tn4001 transposition products: the flanking IS256 elements can form tandem dimers and IS circles. J. Bacteriol. 184:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouch, D. A., M. E. Byrne, Y. C. Kong, and R. A. Skurray. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J. Gen. Microbiol. 133(Pt. 11):3039-3052. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 20.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 21.Scott, J. R., P. A. Kirchman, and M. G. Caparon. 1988. An intermediate in transposition of the conjugative transposon Tn916. Proc. Natl. Acad. Sci. USA 85:4809-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ton-Hoang, B., P. Polard, and M. Chandler. 1998. Efficient transposition of IS911 circles in vitro. EMBO J. 17:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ton-Hoang, B., P. Polard, L. Haren, C. Turlan, and M. Chandler. 1999. IS911 transposon circles give rise to linear forms that can undergo integration in vitro. Mol. Microbiol. 32:617-627. [DOI] [PubMed] [Google Scholar]
- 24.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turlan, C., B. Ton-Hoang, and M. Chandler. 2000. The role of tandem IS dimers in IS911 transposition. Mol. Microbiol. 35:1312-1325. [DOI] [PubMed] [Google Scholar]
- 26.Yao, Z., D. H. Jones, and C. Grose. 1992. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1:205-207. [DOI] [PubMed] [Google Scholar]
- 27.Ziebuhr, W., K. Dietrich, M. Trautmann, and M. Wilhelm. 2000. Chromosomal rearrangements affecting biofilm production and antibiotic resistance in a Staphylococcus epidermidis strain causing shunt-associated ventriculitis. Int. J. Med. Microbiol. 290:115-120. [DOI] [PubMed] [Google Scholar]
- 28.Ziebuhr, W., V. Krimmer, S. Rachid, I. Loessner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]
- 29.Ziebuhr, W., I. Loessner, V. Krimmer, and J. Hacker. 2001. Methods to detect and analyze phenotypic variation in biofilm-forming staphylococci. Methods Enzymol. 336:195-205. [DOI] [PubMed] [Google Scholar]
- 30.Ziebuhr, W., I. Loessner, S. Rachid, K. Dietrich, F. Götz, and J. Hacker. 2000. Modulation of the polysaccharide intercellular adhesin (PIA) expression in biofilm forming Staphylococcus epidermidis: analysis of genetic mechanisms. Adv. Exp. Med. Biol. 485:151-157. [DOI] [PubMed] [Google Scholar]