Abstract
The practice of classifying organisms into hierarchical groups originated with Aristotle and was codified into nearly immutable biological law by Linnaeus. The heart of taxonomy is the biological species, which forms the foundation for higher levels of classification. Whereas species have long been established among sexual eukaryotes, achieving a meaningful species concept for prokaryotes has been an onerous task and has proven exceedingly difficult for describing viruses and bacteriophages. Moreover, the assembly of viral “species” into higher-order taxonomic groupings has been even more tenuous, since these groupings were based initially on limited numbers of morphological features and more recently on overall genomic similarities. The wealth of nucleotide sequence information that catalyzed a revolution in the taxonomy of free-living organisms necessitates a reevaluation of the concept of viral species, genera, families, and higher levels of classification. Just as microbiologists discarded dubious morphological traits in favor of more accurate molecular yardsticks of evolutionary change, virologists can gain new insight into viral evolution through the rigorous analyses afforded by the molecular phylogenetics of viral genes. For bacteriophages, such dissections of genomic sequences reveal fundamental flaws in the Linnaean paradigm that necessitate a new view of viral evolution, classification, and taxonomy.
Biological taxonomy is rooted in the Linnaean “boxes within boxes” hierarchical paradigm (80). Here, groups of organisms are defined by their shared characteristics. These groups are subdivided (boxes formed within boxes) based on greater numbers of characters shared within subgroups and on the presence of characters that distinguish between subgroups. This framework is strictly hierarchical; that is, a group at any one taxonomic level can belong to only one parental group (e.g., a species can be a member of only one genus). When devised, the Linnaean paradigm provided an orderly classification of living things, allowing natural historians to place newly found creatures into its hierarchical framework with relative ease, merely by navigating the ever more detailed sets of characteristics that defined groups within groups.
Although this concept was devised in the absence of evolutionary theory, it readily accommodated evolution as a driving force leading to such hierarchical classification of organisms. Soon after the publication of Darwin's Origin of Species (25), Haeckel (55) proposed that the more closely related forms in Linnaean classification shared more recent common ancestors than did more distantly related forms. In this way, taxonomy based on shared characteristics could be used as a framework for understanding the evolution of organisms, since it is fundamentally based on the vertical inheritance of genetic information from parent to offspring. As a caveat, if genetic information were to be transferred between distantly related forms (those residing in different “boxes”), a purely hierarchical framework would be inadequate to describe the evolution of all genes in a genome. But at the time of its conception, the potential for such cross-lineage mating was not taken seriously and thus posed no barrier to allowing a hierarchical classification system to represent the evolution of its constituent organisms (20, 24, 146, 151).
The heart of biological classification is the concept of the species, or the smallest group of organisms that can be identified robustly. These organisms—through any one of a number of mechanisms—share a common evolutionary fate. Importantly, the formation of new lineages of organisms can be equated to the formation of two species from a single ancestral stock (speciation). Although it is has been argued that virologists want and need a finely divided hierarchical taxonomy for the purpose of understanding their study organisms (138), it is clear that placing an organism into a taxonomic “box” has utility only if the box is biologically meaningful. We discuss here (i) various approaches to understanding the nature of viral species, if any; (ii) the failure of the Linnaean paradigm to represent viral and bacteriophage evolution at larger taxonomic levels; and (iii) viable mechanisms for outlining viral taxonomy in a manner that befits the organisms' complex evolutionary histories.
MATERIALS AND METHODS
Computational analyses.
The identification of shared genes employed BLAST and PSI-BLAST programs (4, 5); typically, genes with significant similarity showed BLAST E values no greater than 10−2, which became more significant as PSI-BLAST iterations were used to define protein families. In some cases, homologues were detectable only during PSI-BLAST iterative searching. Refinement of overall similarity used the Bestfit and Gap modules of the GCG program package (29).
RESULTS
Viral species: a battleground for the cladistic versus phenetic approaches.
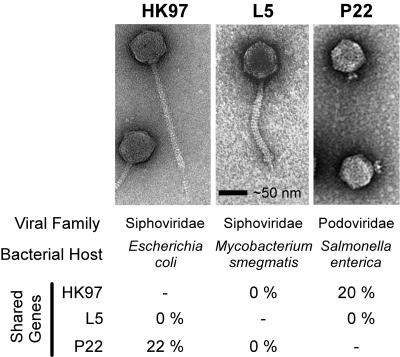
The recognition of viral and bacteriophage taxa, including the difficult task of delineating species, has been the subject of much debate (10, 37, 48, 58, 73, 90, 91, 118, 132, 135, 136, 138). Viral taxonomy (134) has its roots firmly in a taxonomic species concept, discussed below, wherein one knows a member of a species when one sees it. Many features can be employed to discriminate among viral forms, including host range, pathogenic nature, method of transmission, biophysical or antigenic properties of the virion itself, overall morphology, and DNA sequence relatedness. Yet relationships among bacteriophages that are inferred from one set of features may not be congruent with relationships inferred from a disparate set of features. For example, virion morphology is utilized to group double-stranded DNA (dsDNA)-tailed bacteriophages into three groups: the Siphoviridae (bearing long flexible tails), the Myoviridae (bearing contractile tails), and the Podoviridae (bearing short tail stubs). As seen in Fig. 1, these families can include organisms with little or no sequence similarity (e.g., HK97 and L5) and exclude more closely related phages based solely on their tail morphology (e.g., HK97 is a close relative of phage P22).
FIG. 1.
Conflicts between morphological classification and genetic relatedness in dsDNA-tailed bacteriophages. Although bacteriophages HK97 (70) and L5 (59) share very similar morphologies, including a long flexible tail, there are no genes or encoded proteins that are detectable as close homologues. In contrast, bacteriophage P22 (132) has substantial numbers of genes in common with HK97; genetic differences lead to different tail morphologies, which led to classification into two distinct families. Proteins shared between HK97 and P22 exhibit BLASTP (4, 5) E values between 10−16 and 10−101; BLASTP E values for comparisons with L5 proteins failed to reveal matches with significance values lower than 100.
Moreover, some existing taxonomic groups may not reflect common ancestry at all, even when considering their defining characteristics; for example, the Myoviridae include phages Mu and T4, whose contractile tails are encoded by sets of genes that are not recognizably similar in sequence or organization, likely making the Myoviridae a polyphyletic group (meaning that all of the descendants of the common ancestor of all phages classified as Myoviridae may not belong to this group). The Podoviridae are distinguished not by a set of defining features but rather the lack thereof (no long tail), a long-standing problem in taxonomy (e.g., the absence of a nucleus led to the still-professed idea that “prokaryotes” form a cohesive group [20], even though Archaea are relatives of Eukarya [50, 68]). In addition, proteins comprising the diminutive tail assemblies are not recognizably similar among some Podoviridae (e.g., between P22 and T7) and exhibit markedly different protein folds (124, 125), also indicating likely polyphyly of this group.
Since it has long been recognized that no single feature can be usefully employed to describe viral species, the concept of polythetic groupings—whereby firm boundaries are abandoned in favor of fuzzy ones—has become more accepted (133, 135). In addition, the concept of a quasispecies (37), whereby heterogeneous populations (presumably resulting from replication errors) are lumped together, has been often applied, especially to animal and plant viruses such as human immunodeficiency virus, hepatitis viruses, coronaviruses, arenavirues, tobamoviruses, and bromoviruses. However, this approach to rescuing the Linnaean paradigm involves a somewhat dangerous application of phenetic methodologies: that is, identification of groups by overall similarity, whereby one delineates boundaries by deciding how different the organisms must be before they must belong to distinct groups. First, caveats could be raised on how accurately the diversity of these populations can be measured, since differences between sequences are artifactually introduced due to the error-prone nature of the PCR used in these analyses (118). Second, as seen above (Fig. 1) and as detailed below, measures of overall similarity can lead to spurious conclusions regarding the relationships among taxa, especially when one considers chimeric genomes and the multiple ancestries of their constituent genes.
More importantly, a focus on how different organisms must be before they are placed in different groups does not consider the biological forces which lead to similarity among organisms that comprise those groups. Even among purely clonal organisms, groups may retain genetic coherence by periodic selection (e.g., among influenza viruses [14, 15, 42]) or by disruptive frequency-dependent selection (79). Alternatively, groups may owe their distinctiveness to independent origins (e.g., dsDNA-tailed phages versus single-stranded DNA [ssDNA] filamentous phages) or to ecological constraints that diminish the frequency of successful gene exchange among groups (e.g., constraints on DNA packaging size in λ-like phages may reduce the likelihood of their acquiring accessory genes found in the larger T4-like phage genomes). Lastly, gene exchange may act to unify organisms based on shared gene pools (discussed below). We propose that the identification of mechanisms that lead to cohesion among members of viral groups at any taxonomic level is necessary to allow viral taxonomy to reflect meaningful biological relationships.
General species concepts and why they are inadequate for viruses and bacteriophages.
The taxonomic species concept identifies groups of organisms solely based on their overall similarity and is the foundation of current bacteriophage taxonomy (134). This model can be effective when large numbers of morphological features are available for the delineation of biologically relevant groups and has been applied with considerable success—e.g., to plants and animals—in the absence of molecular or genetic data. As detailed above, this approach is inadequate for bacteriophages since morphological differences can arise from very few genetic differences and since morphological similarity can be maintained by organisms that retain little or no genetic similarity or be converged upon by different routes. As a result, this species concept vastly oversimplifies the genetic complexity of viruses and bacteriophages.
Aside from the concept for viral species currently in practice, there have been numerous alternative species concepts that have been devised, primarily for the classification of eukaryotes. The most widely applied genetically based species concept is the biological species concept, wherein Ernst Mayr proposed that species could be defined as organisms that share a common gene pool (86-88). Here, members of a species frequently recombine their genetic material (in many eukaryotes, it is obligatory for reproduction), thereby conferring genetic “cohesion” to the group. Although delineation of these boundaries is relatively straightforward among diploid, freely recombining eukaryotes (but not without its difficulties in some cases), it is less obvious how to draw the boundaries among groups of rarely recombining organisms that exchange only portions of their genomes, such as bacteria or bacteriophages. Similarly, the recognition species concept proposes that members of a species share a common mate recognition system (105), but it is not clear what such a system would be for viruses. Since recombination occurs within a host cell, one could use the host range as a surrogate measure for mate recognition, but disparate phages clearly recognize the same suite of hosts. In both of these models, the frequency of gene exchange lies at the heart of describing a species, thereby hampering their effectiveness in describing viral lineages but, as described below, it may provide great utility in delineating groups of phages at higher levels of taxonomic inclusiveness.
Viral taxonomy has traditionally eschewed use of the biological species concept, or any other framework that involves gene exchange, because bacteriophages and other viruses are viewed as primarily clonal organisms, reproducing almost exclusively without sexual exchange (134). Yet there is ample evidence for gene exchange by both homologous recombination (99) and illegitimate means (63, 123) among bacteriophages. Therefore, not only should gene exchange be considered in constructing a taxonomic framework, it effectively invalidates any taxonomy that a priori assumes that gene exchange is nonexistent. This impasse has led to vigorous debates on the validity of existing viral taxonomic groups (see, for example, references 48 and 138).
Alternative models for species descriptions avoid direct discussion of gene exchange. The ecological species concept (139) defines a species as a group of organisms sharing a common ecological niche. Here, if two distinct groups of organisms were attempting to occupy precisely the same niche at the same time, one would competitively eliminate the other by purely stochastic means. Alternatively, if the two ecologically identical species were separated in time or space (allopatry), they may both persist; if so, genetic and ecological differences would arise, thereby allowing coexistence should they ever return to sympatry. For viral forms, it is difficult to delineate an ecological niche, so this model seems impractical. Moreover, the cosmopolitan distribution of bacteriophages would preclude the allopatric separation of lineages. However, as with the biological species concept, ecological differences may be useful in delineating viral groupings at higher taxonomic levels where phages may be constrained from exchanging genes (e.g., between eukaryotic viruses and bacteriophages, which rely upon radically different transcription and translation apparati in their respective hosts).
The evolutionary species concept (147) dictates that species comprise organisms that share a common evolutionary fate. For bacteriophages, measuring what that fate has been would appear to entail a description of their shared genetic ancestry and reduces, at the functional level, to the biological species concept. Lastly, the cohesion species concept (129) stipulates that members of a species retain similarity through the action of cohesion mechanisms, which are not rigorously defined and offer no foundation for describing viral species. Yet the idea that a taxonomic group should be defined by some cohesion mechanism that retains similarity among its members provides a meaningful biological foundation to any taxonomic system. In sum, while there appears to be no adequate model to delineate a viral species, requiring compromise or extension of one or more of these ideas, these concepts offer strong conceptual frameworks for devising taxonomic systems that maximize their underlying biological utility.
Bacterial species concepts and reconciling lateral gene transfer.
The issues detailed above have arisen in the description of bacterial species, and the lessons learned there offer insights into the problems of viral species and taxonomy. The earliest models for bacterial species recognized the fundamentally asexual nature of bacteria, a feature shared with bacteriophages and other viruses, wherein recombination between individuals is not tied to reproduction. Here, advantageous mutations could arise in populations of organisms, and individuals carrying this new information would sweep the population. In this way, periodic selection for successively fitter forms leads to distinct groups of closely related organisms representing the descendents of organisms initiating these events (78). The dissemination of advantageous alleles by homologous recombination—which occurs at a much higher frequency than initially suspected (39, 40, 54, 85, 119)—led to the idea that bacterial species could be defined by their shared gene pools, essentially invoking the biological species concept (35). Phylogenies drawn from different genes among different “species” would be congruent, whereas the same phylogenies constructed from conspecific individuals would not be congruent, reflecting homologous exchange among members of this group. A barrier to gene exchange via homologous recombination is mediated by bacterial mismatch correction systems, which prevent successful integration of DNA strands that are too dissimilar (82, 83, 113, 142, 143, 156).
However, illegitimate recombination allows for the introduction of genes from very distantly related taxa, leading to incongruent phylogenies among genes found in distantly related organisms (33, 34, 77, 101). This horizontal gene transfer both muddles the application of the biological species concept to bacterial lineages (since it appears that all Bacteria and Archaea, and many Eukarya, share the same gene pool), and it obfuscates higher-ordered taxonomic relationships (74). The transmission of genetic material across large phylogenetic distances is incongruent with the strictly hierarchical “boxes-within-boxes” Linnaean paradigm. Although the concepts of bacterial “species” have been vigorously debated (23, 74, 75, 145), all would agree that a species' members share significant numbers of genes by common ancestry and almost always participate in allelic exchange by homologous recombination. Groups above the species level are recognized by a common set of genes transmitted primarily through vertical inheritance (the “core genome”) which are more recalcitrant to lateral gene transfer, possibly due to their ubiquity or to their high degree of integration with other components of cellular machinery (69). It has also been suggested that horizontal gene transfer itself could serve as a cohesion mechanism, if the likelihood of transfer decreased with overall phylogenetic distance (49). In addition, shared ecology serves to unify the gene content of organisms (e.g., niche-specific genes among methanogens or cyanobacteria), leading to phylogenetic cohesion at these higher taxonomic levels. As discussed below, these same cohesion mechanisms may provide coherence to viral taxonomic groups as well.
Gene exchange among bacteriophage genomes: the failure of hierarchy.
The underlying processes that give rise to the complex and diverse viral population can be illustrated by consideration of the tailed bacteriophages, likely the most abundant of all virus types; in fact it is likely that there are more individual tailed phage particles in the biosphere than of all other organisms combined (13). These viruses are also remarkably diverse and thus serve well to illustrate the Procrustean difficulties of trying to fit a real world viral population into a hierarchical Linnaean taxonomy. Here, we discuss how gene exchange can serve as a mechanism for cohesion among diverse phages, even as it serves to disrupt strict hierarchical relationships at small scales.
Pathways: homologous recombination.
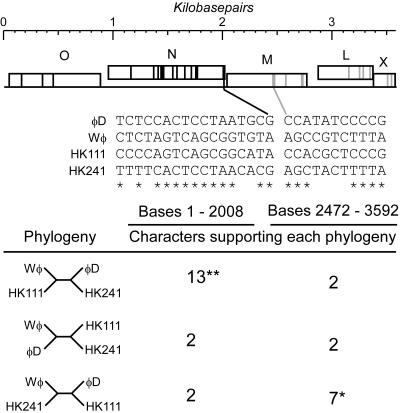
Plausibly, phage populations could be largely clonal if they are unable to participate in recombination events that can assort mutations that arise through DNA replication inaccuracies. However, this does not seem likely since phage and prophage partners should have ample opportunity to exchange genetic information via homologous recombination events by using host- or phage-encoded recombination machinery. These opportunities should be considerably greater than those observed in bacteria since recombining phage genomes may successfully evade host mismatch repair systems. Moreover, phages are expected to be versatile in exploiting a broad range of ecological situations, frequently infecting a variety of host bacterial species residing in different environments. Variant alleles providing a benefit in any of these environments may spread quickly throughout the viral population via homologous recombination. A specific example of this is shown in Fig. 2, wherein the analysis of phylogenetically informative sites within the head gene region of a group of P2-like phages—augmenting a previous analysis demonstrating a significant absence of homoplasy within portions of these genomes (99)—provides evidence for homologous recombination events in the ancestry of these phages.
FIG. 2.
Evidence for gene exchange via homologous recombination among P2-related bacteriophages Wφ, φD, HK111, and HK241, which was initially detected by significant lack of homoplasy among certain gene sequences (99). Here phylogenetically informative sites (listed at the center of the figure) were extracted from sequences encoding the structural genes from four bacteriophages sufficiently closely related that multiple substitutions have not likely occluded phylogenetic relationships. A recombination event is evident between bases 2008 and 2472 of the aligned sequences and is denoted by the gap in the alignment of informative sites. Nucleotide positions supporting the significantly most parsimonious phylogeny on either side of the recombination join point are noted with asterisks; different phylogenies are robustly supported by the 5′ and 3′ portions of the sequence. ✽, P < 0.05 by Felsenstein's S test (41); ✽✽, P < 0.05 by both Felsenstein's S test and C test (41).
Homologous recombination is also evident in genomes of eukaryotic and archaeal viruses. For example, the attenuated polio vaccine virus has recombined several times with natural isolates to yield virulent forms (53, 72, 81, 84). Both homologous and illegitimate recombination events have also been seen in the Potyviridae (102), the Herpesviridae (46, 100), the Adenoviridae (98), the Retroviridae (27, 38, 93), archaeal viruses SIRV1 and SIRV2 (107), and both positive- and negative-stranded RNA viruses (2, 110).
Homologous recombination between phage and prophage genomes could be mediated by host- or phage-encoded recombination enzymes. For example, it is not uncommon for phages to encode homologues of RecA, e.g., coliphage T4 and mycobacteriophages Bxz1 and Cjw1(155; unpublished results); single-stranded binding proteins, e.g., T4, PVL, and Cjw1, among others (31, 71; unpublished results); and Holliday junction resolving enzymes, e.g., T4, L5, TM4, and HK97 (45, 59, 70, 111). Although these enzymes may play a primary role in DNA replication or in DNA packaging, their actions will also result in the assortment of mutations arising through replication errors. The potency of these recombination systems should not be taken lightly, particularly since they may require little more than 20 bp for efficient recombination (26, 95, 96).
If phage populations lack clonality, then the lack of sequence similarity between phages such as L5 and HK97 (Fig. 1) could indicate that they have originated completely independently. Alternatively, they may have a shared ancestry but enjoyed long-term genetic isolation from each other. A resolution to this is provided by the observation that some phage and prophage genomes carry one or more genes with homology to their counterparts in L5, whereas others are related to homologues in HK97. An example is that of φC31, in which the products of the terminase, portal, protease, and capsid genes have obvious sequence similarity to their functional counterparts in HK97, whereas the products of genes 9a, 16, and 20 are related to the products of mycobacteriophages TM4 gp70, L5 gp48, and D29 gp36.1 (63). Therefore, one cannot conclude that morphological differences result from different ancestry, nor can a lack of sequence similarity be construed as indicating independent ancestry.
Pathways: illegitimate recombination and genetic mosaicism.
The existence of frequent genetic exchanges among members of the tailed phages was first recognized in the “lambdoid” phages (such as Escherichia coli phage λ) as a result of DNA-DNA heteroduplex experiments starting in the late 1960s (64, 117). Based on these experiments it was evident both that these phages are genetic mosaics with respect to each other and that the mosaic boundaries are found preferentially at certain sites along a given phage genome. Susskind and Botstein (126) proposed a “modular theory” of phage evolution in which it was suggested that special “linker” sites located between genes facilitated frequent recombination attributable to either short stretches of conserved sequence or a site-specific recombination mechanism, thereby generating genomes with new combinations of gene alleles. Further refinements of this model (17-19) proposed that recombination may occur at any location along the genome but that phages experiencing recombination events that incur deleterious effects (e.g., lying within important genes) would be counterselected.
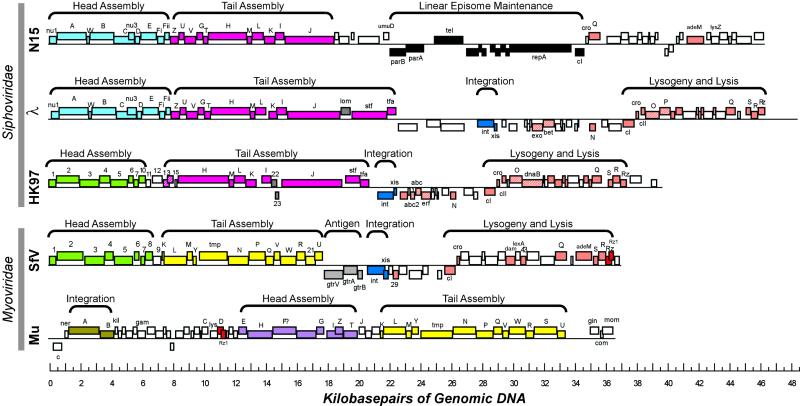
Comparative genomic analysis of temperate coliphages shows the remarkable degree to which these phages can be described as genetic mosaics (Fig. 3). With shared colors indicating robustly similar sequences, we see that phages λ and N15 have closely related sets of head genes, whereas phages HK97 and SfV have head genes that are different from these but homologous to each other; the Mu head genes belong to a sequence family not related to the other two. The tail genes also fall into related groups, but they partition the five phages illustrated here differently, with phages N15, λ, and HK97 forming one group and phages SfV and Mu forming a second.
FIG. 3.
Mosaicism of gene cassettes among dsDNA-tailed bacteriophages. Genes in cassettes bearing homologous genes are coordinately colored; striped genes indicate that protein products perform analogous functions. The sequences for bacteriophages N15 (112), λ (116), HK97 (70), and Mu (94) were obtained from public databases; the SfV sequence (3) was kindly provided by N. Verma prior to publication.
Mosaicism is manifested at different levels of organization in this group of phages; in the early expressed gene regions the meaningful modules of mosaicism are single genes or small groups of genes, rather than the larger groups that characterize the late-expressed head and tail genes. Yet even in these regions we can see that λ, HK97, and SfV share functional order of genes through the region, albeit with many allelic substitutions, and show many examples of frank sequence similarity, while the early regions of N15 and Mu are different from these and from each other. In addition to these extensive mosaic relationships, there are a number of examples in which a novel gene appears in one phage where it is absent in the otherwise homologous region of another phage (e.g., HK97 genes 15, 22, and 23, the SfV gtr genes, and the λ lom gene); there are also a few examples (not evident in Fig. 3) of nonorthologous replacements of a gene or small group of genes within the head gene region or the tail gene region. For example, gene Z1886 of prophage CP933-X of E. coli O157:57 (108) encodes a head protease similar to λ C protein (corresponding to the blue cassette in Fig. 3), yet φCP933-X encodes HK97-like head proteins (corresponding to the green cassette in Fig. 3) in the regions flanking this gene, including, for example, the gp7 adapter protein (encoded by gene Z1902). Such substitutions show that while these genes most often stay together as a coevolving group, the groups are not invariably monolithic.
The difficulties these complex mosaic relationships present for making a hierarchical taxonomy that adequately represents the available biological information should be obvious. If we were to take the relationships among the tail genes as the basis for the highest level taxonomic division (or the virion tail morphology, as is the current practice for defining “families” in the ICTV taxonomy [137]), we would find head gene types inappropriately split among sister taxa, and early genes split differently but equally inappropriately. If we were instead to base the division on an average or integrated representation of the virus as a whole—that is, a phenetic approach—we would achieve a taxonomy that was self-consistent with respect to the averaged value for each virus pair but at the expense of discarding much of the decipherable biological information available for each virus. If one goal of a hierarchical taxonomy is that viruses in one taxon should have more in common with other viruses in that taxon than they do with any virus in a sister taxon, then this goal is impossible to achieve for viruses as pervasively mosaic as these.
Scale and scope: hierarchies in mosaicism.
Genetic mosaicism is not restricted to the coliphages shown in Fig. 3 but is much more widespread. For example, the dsDNA-tailed mycobacteriophages L5, D29, Bxb1, and TM4 show clear mosaicism and do so at different levels of organization, as seen with the lambda-like phages (44, 45, 59, 89). All four of these phages share a related set of late genes involved in viral structure and assembly arranged in a colinear order but interspersed with several obvious instances of mosaic substitutions (e.g., Bxb1 gp24, Bxb1 gp28, TM4 gp20, and TM4 gp22). In the putative early genes of these phages, the mosaicism is extensive, with numerous examples of genes or groups of genes that are present in one genome but absent from others. For example, TM4 gp64 and gp87 are clearly related to Bxb1 gp56 and gp7, respectively, whereas the remaining genes of this segment of the TM4 genome bears little overall similarity to Bxb1 genes. Likewise, there are at least 20 mosaic substitutions within the right arms of the otherwise very similar L5 and D29 genomes (44, 59), and homologues of many of these can be found in other newly characterized mycobacteriophage genomes (unpublished data). In addition, mosaicism is not limited to phages with large genomes. Two relatives of coliphage P4 are found as prophages in different strains of Salmonella enterica; although they are nearly identical over the majority of their genomes, the region containing the gob gene and two other genes of indeterminate function of P4 has been replaced with cassettes encoding a PvuII-like type II restriction endonuclease system in the P4-like prophage in serovar Paratyphi, whereas this region in the prophage in serovar Typhi contains three genes with no identifiable homologues in the database (36).
A number of “dairy” phages that infect the lactic acid bacteria Lactococcus lactis, Streptococcus thermophilus, and Lactobacillus delbrueckii have been studied (12). Although these phages appear to be more homogeneous than the lambda-like phages and exhibit significant similarity at their nucleotide level, there is also evidence of mosaicism (13). For example, the lactococcal phages c2 and sk1 have little nucleotide similarity and different genome organizations, but there are at least eight noncontiguous genes that are shared by both phages (12). Similarly, the lactococcal phage r1t has a cluster of structural genes that are related to those in the Streptococcus pyogenes prophage SF370.3, although the remainder of the genomes are not obviously related (28). The greater degree of similarity among these groups of bacteriophages may reflect their isolation under more highly controlled conditions than pertain to collections of lambdoid phages.
It has been suggested (1, 92, 114, 131) that T4, a large lytic phage of E. coli, shows less mosaicism than some of the other phage groups, such as the coliphages illustrated in Fig. 3 or the mycobacteriophages discussed above. This is an interesting point, because there are theoretical reasons to suspect that strictly lytic phages such as T4 will have less opportunity for illegitimate exchange than do temperate phages (see below). Although the data to provide a critical test of these issues are just now beginning to appear (e.g., among lactococcal phages [21]), it is clear that T4 does exhibit mosaicism at least on a small scale. For example, a portion of the T4 gp37 tail fiber protein makes a very close match to the corresponding part of the phage λ gpStf tail fiber protein (47, 130); similar mosaicism is found among such diverse phages as P1, P2, Mu, K3, and T2 (56), and genes encoding T4 virion components are shared with a marine cyanophage (57). Another strictly lytic E. coli phage, N4, has a very different genome organization and lifestyle from those of any other known virus, but it has homologues of the T4 rIIA and rIIB genes, inserted in a different flanking context than in T4, as well as a homologue of a gene in the Salmonella temperate phage P22 (L. B. Rothman-Denes, R. W. Hendrix, G. F. Hatfull, et al., unpublished data). Mosaicism may be evident among phages of remarkably different hosts; for example, phage SIO1, infecting the marine bacterium Roseobacter, shares several genes with coliphages T3 and T7 (115).
In total, these examples show that genetic mosaicism is a common feature among bacteriophage genomes. The apparent degree of mosaicism, however, is quite variable and may reflect differences in the lifestyle of the phage (e.g., T4) or the methods of sampling (e.g., the dairy phages), in addition to undescribed variables and constraints that may limit exchange within or between particular groups of phages.
Scale and scope: recombination and mosaicism in ssDNA phages.
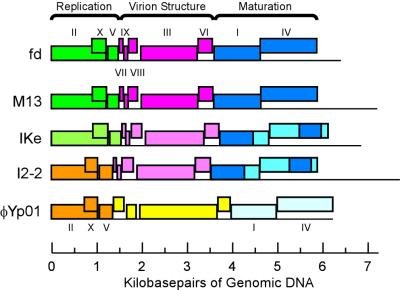
The reservations we raise here about the use of hierarchical taxonomy for viruses apply only to the extent that the viruses indulge in exchange of genetic material and are therefore not strictly clonal. While we have tested the assumption of nonclonality in detail only for the dsDNA-tailed phages, we have also analyzed a small group of ssDNA phage genomes regarding this question (Fig. 4). Examination of even this small subset of phages reveals evidence for both homologous recombination (involving genes I and IV of the phage maturation cassette [106]) and nonorthologous gene replacement (both for the replication gene cassette, noted earlier [123], and for the structural protein cassette). Thus, these phages, as with the tailed phages, cannot appropriately be described by a hierarchical taxonomy. We anticipate that, given sufficient numbers of phage genomes, similar mosaicism will be found among other groups of bacteriophages.
FIG. 4.
Evidence for homologous recombination and mosaicism of gene cassettes among ssDNA filamentous bacteriophages. Genes in cassettes bearing homologous genes are coordinately colored; orthologous gene cassettes with various degrees of similarity are represented by various shades of the same color, whereas nonorthologous replacements are denoted by differently colored cassettes. The darker regions in genes I and IV of phages IKe and I2-2 indicate homologous recombination events with M13/fd-like sequences first noted by Peeters et al. (106). The sequences of bacteriophages fd (8), M13 (140), Ike (106), and I2-2 (123) were obtained from the public database; the φYP01 sequence represents an inferred prophage in the Yersinia pestis genome (104) beginning at gene YP02274 and continuing through downstream genes.
How they do it: mechanisms of gene exchange.
Mosaicism may be a pervasive feature of bacteriophage genomes, but what mechanism gives rise to this characteristic feature? The initial suggestion that it occurs by homologous recombination at linker sequences (74) is unattractive since these are not seen in the numerous complete genome sequences that have been determined. It is more likely that this mosaicism results from nonhomologous (illegitimate) recombination between these phages, not at specific sites but profusely and essentially at random with respect to position along the genome. It is also likely that this recombination occurs often—perhaps usually—out of register with respect to the gene organization of the recombining partners. For example, the homologue of HK97 gene 7 (encoding the head-tail linker) was clearly interrupted in HK022, where gene 7 has been truncated; a fragment bearing gene 8 and gene 9 (the new, but more distantly related head-tail linker protein) was introduced—a gene and a half out of register—at that site (60). Similarly, the HK97 N gene is followed by the 3′ end of a distantly related N gene (70), illustrating an additional “sloppy” recombination event within this bacteriophage. Although it is tempting to think that these events might be extremely rare, it is clear that phage-encoded recombination systems can mediate exchange within very short segments of sequence relatedness (26, 95, 96).
The expected outcome of these processes is a heterogeneous melange of recombinant types, almost all of which are defective and presumably immediately eliminated by natural selection. The few recombinants that survive—perhaps after a number of additional illegitimate or homologous exchanges—are the ones in which the functions of essential genes are not disrupted and whose genomes are suitably sized for packaging into capsids. As a consequence of this harsh filter for function, the discontinuities that result from these recombination events are observed between coding regions, at positions corresponding to domain boundaries in the encoded proteins, or at the boundaries of entire group of genes, especially those like the head genes whose products must interact intimately. The overall result is a group of surviving recombinants with mosaic boundaries (recombination loci) located at a restricted number of points corresponding for the most part to the boundaries of functional genetic modules.
Although the novel junctions formed by illegitimate events that give rise to functional genomes may be relatively rare, they are expected to have considerable evolutionary longevity and be propagated through the population via homologous recombination, as described above. In this way, homologous recombination occurring between genes of identical or nearly identical sequence can produce the same relationships among genomes that were observed in the early heteroduplex experiments and ascribed to recombination between linker sequences located at module boundaries. Interestingly, Clark et al. (22) have recently reported short conserved sequences located between genes at corresponding positions in a group of lambdoid phages, reminiscent of the initial proposal of linker sequences. These “boundary sequences” likely do sponsor homologous exchange at these positions, although it is probably less important quantitatively than homologous recombination in the longer regions of opportunity provided by genes with highly similar sequences.
Where they do it: opportunities for gene exchange.
The pathways by which viruses and bacteriophages exchange genetic material vary dramatically in concert with their lifestyles or host ranges. Exchange of genes among bacteriophages is often viewed as a three-body problem, where two viral particles infect the same cell, and homologous recombination allows for gene exchange during the infection. Although this scenario stems from the way bacteriophage genetics is performed in laboratory settings, its low probability of success in natural environments likely translates into little impact for many groups of bacteriophages. An alternate route to gene exchange would involve an incoming bacteriophage and a prophage resident in the host genome (17). Surveys of completely sequenced bacterial genomes show that most harbor prophages, averaging 2.6 per genome for free-living bacteria (76). In these cases, two events are possible. First, resident prophages may serve as gene donors for incoming phages, acting as recipients. Although this is possible, a severe constraint is imposed in that the resulting recombinant phage must not only be functional but also have a genome size appropriate for packaging into its capsid. This constraint limits such exchanges to events in which regions of similar length are transferred.
Alternatively, the incoming bacteriophage may act as a gene donor and the prophage may act as the recipient. Here, there is no constraint on the recombinant to be either functional or to have a genome size appropriate for its capsid. Numerous events, including multiple gene acquisitions and deletions, may occur before a viable recombinant phage is produced. Moreover, both incoming bacteriophages (which do not mount a successful lytic infection), as well as other resident prophages, may serve as gene donors. These mechanisms would allow for high levels of gene exchange among temperate bacteriophages but would not be available for lytic bacteriophages. As a result, we may expect that genome mosaicism will be more profound among temperate bacteriophages; results consistent with this hypothesis have been reported among temperate and virulent lactococcal phages (21).
Eukaryotic viruses are constrained in an additional way. Although proviruses may be formed, the recombinant virus must be formed and released during the lifetime of the individual, unless the provirus is present in the germ line. This limited opportunity for gene exchange reduces the likelihood of successful recombinants being formed by illegitimate means, in which case the progeny viruses may be of inappropriate size or genome composition. Yet homologous recombination events, whereby allelic information is exchanged, would occur among proviruses. Therefore, one may expect that viruses of multicellular eukaryotes will show less genome mosaicism than do temperate bacteriophages.
DISCUSSION
Sampling the continuum.
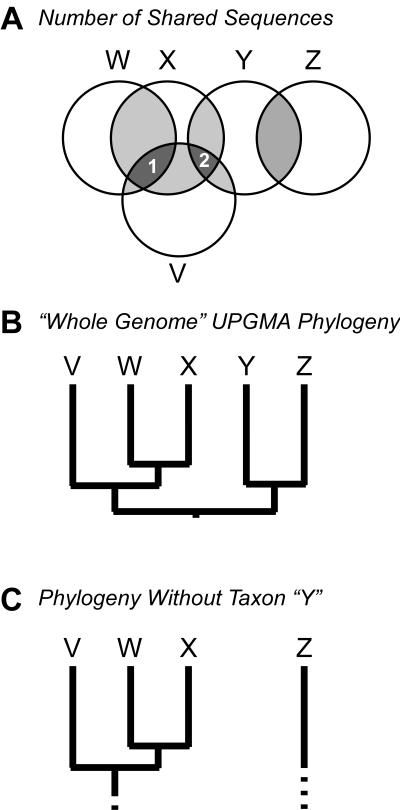
In constructing a taxonomy for any group of real biological organisms, we are working with incomplete information about the natural population. If sampling of information about the organisms is sparse relative to the population's diversity, significant relationships can be missed. This is illustrated in Fig. 5, in which the circles represent the genomes of individual viruses and the area of overlap between two circles represents shared genes. Note that the inclusion of the genome of phage Y in the analysis (Fig. 5B) reveals relationships between phages V, W, and X and phage Z; without phage Y, phage Z would be considered unrelated. This situation is illustrated by the inclusion of phage SfV in Fig. 3, which shows that Mu-like bacteriophages exchange gene cassettes with λ-like bacteriophages.
FIG. 5.
Problems arising in using phenetic methods for bacteriophage taxonomy. (A) Relationships inferred from overall similarity (e.g., number of shared genes, DNA sequence similarity, DNA-DNA hybridization, proteomic overlap) are here depicted in a Venn (141) diagram. These data can be misleading in two ways (see the text). (B) A phylogeny is drawn for all taxa in part A by using the robust phenetic approach UPGMA (122, 127). (C) Relationships are contingent upon the taxa included in the analysis; here, the elimination of taxon “Y” eliminates the connection between taxon “Z” and the remaining taxa.
The magnitude of the problem of sparse sampling of the population is different for different groups of viruses; it appears to be especially severe for the tailed phages. These are the most abundant and diverse group of viruses, but all have been isolated on host bacteria that can be grown in the laboratory, and these bacteria represent a tiny slice of bacterial diversity (32, 66). Furthermore, it is becoming clear that there are groups of tailed phages that fail to plaque under the conditions commonly used for isolating new phages, and these groups are presumably poorly represented in extant collections (65). Even for the relatively well sampled phages of E. coli and its allies, the sparseness of sampling can be seen by the fact that a phage of the N4 type has been isolated only once or that genome sequencing so frequently turns up entirely new types of phages (e.g., N15) or novel relationships among known groups of phages (e.g., SfV connecting λ and Mu). A consequence of sparse sampling is that a group of phages that in reality forms a continuous network of genetic interactions can appear to be multiple noninteracting groups if the sampling is poor (Fig. 5). Currently, it is not yet clear whether the tailed phages can be divided into multiple, genetically coherent groups, or whether they will be seen to be an indivisible continuum of types when sufficient data are available.
Failures of phenetics.
The mosaicism of phage genomes depicted in Fig. 3, 4, and 5 exemplifies the failings of phenetic methods—those based on measures of overall similarity—in reconstructing viral phylogeny and in serving as a framework for viral taxonomy. The sampling problem discussed above illustrated this weakness, when the relationship between phages V, W, and X and phage Z was overlooked until phage Y was included (Fig. 5B and C). Furthermore, the relationships one may infer from overall levels of similarity can be tremendously misleading, in that one may conclude that phages Y and Z represent members of one group and phages V, W, and X represent members of a related group. However, phenetic phylogenetic methods will always supply the user with a particular set of relationships, without regard to the biological relevance of these answers. If one uses phages HK97, SfV, and Mu in Fig. 3 to represent phages X, Y, and Z in Fig. 5A, these relationships become apparently absurd.
Sets of mosaic genomes that share different gene cassettes cannot be represented by a single, simple phylogenetic tree which reduces this genetic complexity to a single measure of overall relatedness. Rather, the segments of these complex genomes have distinct evolutionary histories that are blended when phenetic approaches are employed, thereby producing a set of relationships that does not reflect any aspect of the genome's true history. This is illustrated by gene sets 1 and 2 in Fig. 5A, which predict different sets of relationships among these phages. One cannot expect that a single, hierarchical taxonomy can accurately represent complex, reticulate relationships; only with a reticulate, multidimensional phylogenetic approach (see, for example, reference 7) can the complex relationships among bacteriophage genomes be explicated and appreciated.
Limitations of vertical inheritance.
Despite these caveats, the effects of gene transfer between disparate genomes did not necessitate a complete deconstruction of the hierarchical paradigm for microbial evolution. An alternative was to recognize mosaicism and utilize these data in the context of some overarching phylogenetic framework over which all of the complexities of gene exchange can be draped. In bacterial taxonomy, a “core genome” has been built around the rRNA genes (103, 149, 150) and genes which contribute to the replication-transcription-translation machinery, although even these genes are subject to lateral transfer (67, 97, 144, 148, 152-154). Here, the vast majority of genes inherited in a bacterial lineage are transmitted vertically, from mother to daughter, during replication. As a result, the impact of any one horizontal exchange is minimized since it involves such a small fraction of the total genome. One may view these genes as defining an organismal lineage, whereas all other genes merely provide the means to persist in a particular habitat.
This approach has been manifested in the use of gene content phylogenies, which infer large-scale relationships based on a genome’s overall gene inventory (43, 120, 128). However, this model has its drawbacks. First, it ignores the relationships among many of the genes in the chromosome that are not congruent with the “overall” organismal phylogeny; indeed, it may be that none of the genes follow an ancestry inferred by merging the histories of all genes into a single representation. Second, in this approach one is divorcing the concept of taxonomic relationships from the history of the cell's genetic material, equating organismal evolution to the history of the cytoplasm and not the DNA. Indeed, refinements of this approach recognize gene transfer and reticulation among bacterial genomes (121).
The same problems arise if one attempts an “overall” phylogenetic approach in considering the taxonomy of viruses and bacteriophages. A clear candidate gene around which to build such a consortium would be the bacteriophage capsid gene. While other genes dictate phage lifestyle (tail genes affect host range, integrases and replicases affect modes of reproduction, etc.), the major capsid gene could be viewed as defining a bacteriophage lineage; it has even been proposed that phages could have evolved as consortia of genes which increase their collective fitnesses by allowing more efficient packaging of themselves, including the capsid gene (62). However, the small size of most bacteriophage genomes amplifies the impact of any horizontal transfer event, which now can represent a substantial portion of its genome. In the case of bacteriophage N15, 50% of its genome is clearly related to lambdoid bacteriophages (Fig. 3), whereas the remaining 50% was likely derived from a linear plasmid (112). Such a chimeric organism defies simple categorization into either group (lambdoid phage or plasmid), and assigning a single measure of “overall similarity” with other genomes needlessly ignores the information revealed by its chimeric genome.
If a core genome approach is taken, one must decide which genes formulate such a core—especially considering that most phages lack most components of the replication-transcription-translation apparatus—by cataloging which genes participate in comparable gene consortia. Lacking a consortium, one could construct a phylogeny by using overall gene inventories and use it subsequently to derive a classification system, a counterintuitive but feasible approach. Such an approach would suffer the pitfalls of phenetic classification (Fig. 5) and would likely produce bewildering results, such as grouping ssRNA phages and dsDNA phages on the same phylogenetic tree by virtue of some chain of shared proteins, even though these groups undoubtedly had independent origins. More importantly, such methods always yield a single, hierarchical dendrogram which blends together, thereby obfuscating, the rich set of data showing nonvertical gene inheritance.
Mechanisms for cohesion.
Rather than viewing recombination among bacteriophages as a process defeating the otherwise orderly organization of viruses into neat taxonomic categories, we view recombination as a cohesion mechanism that may identify biologically relevant groups of organisms. Instead of following a “core” genome, one may define biologically significant viral or bacteriophage groups by virtue of their shared pools of gene cassettes. As illustrated above, diverse phages can be drawn together into loose assemblages defined by common sets of gene cassettes that are exchanged among them (Fig. 2, 3, and 4). The constraints on cassette exchange likely reflect many factors, including the limitations of DNA packaging into the virion capsid, utility of the genes for a phage of a given lifestyle, or the ability to express a gene within the host range exploited by a collection of bacteriophages. This view merely extends the Mayrian biological species concept to embrace groups of organisms at higher taxonomic levels.
Additional mechanisms may provide meaningful cohesion among other groups of viruses. For example, the likely independent origins, disparate genome sizes and distinct lifestyles of ssDNA filamentous bacteriophages and dsDNA-tailed bacteriophages translates into few gene cassettes being exchanged among these very different organisms. Similarly, the ecological differences between animal viruses and bacteriophages—which must express their genes and replicate their genomes by using fundamentally different molecular biological systems—also serve to reduce the likelihood of successful gene exchange among these groups. Here, the ecological species concept may be similarly applied at a higher taxonomic level.
Certainly genetic cohesion may be achieved in the absence of gene exchange and recombination; periodic selection—or disruptive frequency-dependent selection (79)—may lead to large groups of organisms that maintain similarity strictly through vertical inheritance, as is the case for influenza viruses (14, 15, 42), allowing for clear delineation of a biologically relevant group. In all cases, we may use the biology of the organisms to define the boundaries (which will often remain quite unclear) between meaningful groups rather than searching for an arbitrary threshold to decide when organisms must be sufficiently different to fall into different groups.
However, host range constraints may not have the broad reach one may suspect. As noted above, marine bacteriophage SIO1 shares several genes with coliphages T3 and T7 (115). More dramatically, phages of the Archaea can strongly resemble either tailed bacteriophages, as is the case with Methanobacterium phage ψM2 (109), or eukaryotic viruses, as with the Sulfolobus rudiviruses SIRV1 and SIRV2 (11, 107). Therefore, strong divisions among eukaryotic viruses (e.g., between most plant and animal viruses) that are made in the absence of sequence information should be viewed with caution, especially since affinity between disparate viral groups has been noted (51).
Taxonomically disruptive homology.
An advantage to considering taxonomy based on shared gene cassettes and other mechanisms of cohesion is that biologically relevant groupings are likely to be revealed; after all, if the genomes can exchange genes, they must share some aspects of their lifestyles. If a simple “core genome” approach were taken, based solely on homology, highly unorthodox groupings could result due to ancient similarities among diverse groups of viruses and bacteriophages that clearly no longer exchange genes. This is especially evident if one considers the major capsid genes of bacteriophages, a likely candidate gene around which to build a “core genome.” That is, there are convincing cases in which homology can be inferred between bacteriophages and viruses of eukaryotic hosts, based on detailed similarities in virion structure (including capsid protein folds), mechanism of virion assembly, and other features of the life cycles (61).
For example, the ssDNA bacteriophage φX174 shares its β-barrel capsid protein fold with many ssRNA plant and animal viruses (see http://mmtsb.scripps.edu/viper/viper.html) but not with the capsid protein of the ssDNA filamentous phages. Also, adenoviruses show a clear similarity to the bacteriophages exemplified by E. coli phage PRD1 (9), Pseudomonas phage φ6 of the Cystoviridae (16) appears unequivocally related to the reoviruses (52) of plant and animal hosts, and the herpesviruses and the tailed bacteriophages are too similar in features of their virion structure and virion assembly mechanisms to be explained by anything short of common ancestry (61). Yet these eukaryotic/prokaryotic virus pairs are now clearly isolated from each other both genetically and ecologically. Furthermore, they have evolved apart from each other to such an extent that it would be unreasonable to group them together on any but the highest taxonomic level, despite the undeniable homology of some of their most salient phenotypic traits.
A modest proposal for bacteriophage taxonomy.
We have discussed some problems that have emerged in the classification of viruses and how genomic sequence analyses show that the systems currently employed are discordant with viral evolutionary histories. Although we recognize that a major reevaluation of viral taxonomy requires broad involvement of the virology community, we will propose here a modest framework—along with some specific guidelines—within which a new taxonomy might be molded.
We suggest that the formulation of any viral taxonomy must satisfy three basic tenets. First, members of a group should exhibit similarity that has resulted from one or more clearly defined cohesion mechanisms, examples of which are described above. Second, for any given virus, a significant amount of sequence data should be available—preferably the genome—for meaningful taxonomic assignment; this is not unreasonable given the ease, speed, and cost of DNA sequencing methodologies. At least among bacteriophages, features obtained in other ways (host of isolation, general morphology, and genome size) offer little value in deducing the evolutionary relationships that must form the foundation of any meaningful taxonomic system. Third, groups may be reticulate; that is, whereas all viruses within a particular group will bear the salient features of that group, any one virus could—and in most cases will—belong to more than one group at the same taxonomic level. The reticulate nature of these groups is clearly a radical departure from the strictly hierarchical Linnaean classification system but is attractive in that it embraces the most obvious feature of many viral genomes: their pervasive mosaicism. Moreover, there is no restriction in choosing how to delineate groups or how to classify hybrid organisms; these viruses are merely placed into more than one group.
Even so, at the highest taxonomic levels a hierarchical system appears to be appropriate; we suggest that the highest tier—here designated a “domain”—segregates organisms with different forms of genetic material: dsDNA, ssDNA, ssRNA, and dsRNA, as proposed earlier (6). This organization seems reasonable since there is little evidence of any significant genetic exchange between these groups, and it seems quite likely that they arose independently. Domains may be subdivided into hierarchically distinct “divisions,” which encompass viruses that also exhibit little or no evidence for genetic exchange across division boundaries (Table 1). For many eukaryotic viruses, these groups may be defined by strikingly different host ranges, modes of infection, or other ecological features that confer cohesion on their constituent members. Whereas a division reflects cohesion mechanisms uniting its members, other features—such as morphological distinctiveness—may result from these cohesion mechanisms and be useful touchstones for navigating this hierarchical portion of the classification system.
TABLE 1.
Taxonomic scheme incorporating reticulate groups
| Virus | Possible taxonomy of virus
|
|||
|---|---|---|---|---|
| Hierarchical portion
|
Reticulate portion (modi)a | |||
| Domain | Division | Family | ||
| N15 | dsDNA | Tailed bacteriophages | NAb | (a) λ-like head genes |
| (b) Genes for λ-like flexible tail | ||||
| (c) Linear-episome-mediated temperate phage | ||||
| λ | dsDNA | Tailed bacteriophages | NA | (a) λ-like head genes |
| (b) Genes for λ-like flexible tail | ||||
| (d) Integrase-mediated temperate phage | ||||
| HK97 | dsDNA | Tailed bacteriophages | NA | (e) HK97-like head genes |
| (b) Genes for λ-like flexible tail | ||||
| (d) Integrase-mediated temperate phage | ||||
| SfV | dsDNA | Tailed bacteriophages | NA | (e) HK97-like head genes |
| (f) Genes for Mu-like contractile tail | ||||
| (d) Integrase-mediated temperate phage | ||||
| Mu | dsDNA | Tailed bacteriophages | NA | (g) Mu-like head genes |
| (f) Genes for Mu-like contractile tail | ||||
| (h) Transposase-mediated temperate phage | ||||
| Newcastle disease virus | ssRNA | Negative-sense genomic RNA | Paramyxoviruses | (i) Respirovirus-like RNA editing |
| (j) Rubulavirus-like glycoproteins | ||||
| (k) Encodes P and V but not C proteins | ||||
| Ebola virus | ssRNA | Negative-sense genomic RNA | Filoviruses | (l) Glycoproteins require transcriptional editing |
| (m) Filamentous virions | ||||
| M13 | ssDNA | Filamentous phages | NA | (n) M13-like structural genes |
| (o) M13-like replication genes | ||||
| (p) M13-like maturation genes | ||||
| I2-2 | ssDNA | Filamentous phages | NA | (n) M13-like structural genes |
| (q) I2-2-like replication genes | ||||
| (p) M13-like maturation genes | ||||
| φYP01 | ssDNA | Filamentous phages | NA | (r) φYP01-like structural genes |
| (q) I2-2-like replication genes | ||||
| (p) M13-like maturation genes | ||||
| φX174 | ssDNA | Icosahedral phages | NA | (s) External scaffolding protein |
| (t) Lysis via inhibition of MraY protein | ||||
Modi, designated here by lowercase letters in parentheses, are groups that may contain different subsets of bacteriophages (see the text).
NA, not applicable.
For bacteriophages, it is not clear whether hierarchical groups are justified below the level of the division. Within divisions, organisms may be sorted into groups that share common sets of features, but these groups are not exclusive from one another. We term such a reticulate group a “modus,” reflecting the exchange of genetic modules among bacteriophage genomes (Fig. 3 and 4); viruses within a modus would share a particular module or phenotypic character which typifies that group. For example (Table 1), phage SfV might belong to the domain of dsDNA viruses, the division of tailed bacteriophages, but to at least three modi, including (i) phages with HK97-like head proteins and maturation processes, (ii) phages with Mu-like contractile tails, and (iii) integrase-mediated temperate phages. In principle, SfV would belong to many other modi as well, including some defined after the isolation of phages that reveal hitherto undiscovered relationships. Clearly, the boundaries of modi may sometimes be difficult to define whether sequence similarity is a criterion, necessitating revisiting the sequence profiles defining modi as additional data become available. The accumulation of large numbers of modi that could be used to classify viruses, as well as the need for facile navigation of a reticulate classification system, clearly necessitates implementation of new technology—that is, computer database systems and sophisticated data retrieval tools—to allow effective and efficient management and use of such a classification system. The utilization of sophisticated data retrieval tools relieves a potential downside of this system, that is, the accumulation of large numbers of modi for any one phage, which result from the elimination of the seemingly information-rich, but biologically misleading, multitiered hierarchical classification system
As illustrated in Table 1, different phages will in general be assigned to different—but overlapping—sets of modi. In this way, the taxonomy of the phage carries a wealth of biological information, does not unnecessarily discard any information by forcing a phage into a strictly hierarchical classification system, and clearly shows the degree and nature of relatedness between any two phages by the sets of modi that they share. For example, tailed bacteriophages λ and Mu share fewer modi than the closely knit dairy phages, reflecting different degrees of genetic relatedness. Although it may be tempting to assign a name to a group of modi to allow more facile descriptions in conversation and in text, formal adoption of such names would defeat the purpose of avoiding such arbitrary naming (that is, it would obscure the underlying reticulate nature of modus groups) and would impose an unnecessary additional level of taxonomic complexity.
Table 1 shows no “family” assignment for dsDNA-tailed phages, reflecting our belief that all of these phages exchange modules. However, if groups of phages were not to share any modules (e.g., λ-like and T4-like phages), they could be placed unambiguously into distinct families. For such a hierarchy to be robust, the collections of modi describing phages must be separable into nonoverlapping groups: that is, no described phage links the groups of phages encompassed by these seemingly distinct groups of modi. Yet this situation is analogous to that shown in Fig. 5C, where a single new isolate may reveal relationships between groups previously thought unconnected (Fig. 5B); thus, newly formed family-level hierarchies may collapse in the face of new data. In contrast, if a group of viruses were to be purely clonal, all members would share the same sets of modi, recapitulating a purely hierarchical taxonomic system. In practice, then, such a system can accommodate the addition or subtraction of hierarchical levels of classification, commensurate with available data.
Conclusions.
The pervasive genomic mosaicism exhibited by bacteriophages argues strongly that the Linnaean, hierarchical paradigm for biological classification—while in place for 250 years—is insufficient and potentially misleading when used to describe the relationships among these organisms. Although it has been the sole taxonomic model used for the 80 years since their discovery (30), the categorization of bacteriophages by this scheme often focuses on features encoded by a minority of their genes (e.g., Fig. 1) and serves to underrepresent their complexity (Fig. 3 and 4). Rather than delineating viral lineages by how different they must be to comprise a distinct category (134), we propose that mechanisms leading to cohesion among groups—including independent origins, frequency of genetic exchange, ecological isolation, and periodic selection—be identified for each group of viruses and be used to form groupings (either hierarchical or potentially reticulate) that more accurately represent meaningful biological relationships among these diverse organisms.
Acknowledgments
We thank N. Verma for kindly providing the SfV genome sequence prior to publication and R. Edwards and E. A. Presley for insightful discussions.
This work was supported by a grant from the David and Lucile Packard Foundation to J.G.L. and NIH grants GM51975 to G.F.H., R.W.H., and J.G.L.; AI28927 to G.F.H.; and GM47795 to R.W.H.
REFERENCES
- 1.Ackermann, H. W., and H. M. Krisch. 1997. A catalogue of T4-type bacteriophages. Arch. Virol. 142:2329-2345. [DOI] [PubMed] [Google Scholar]
- 2.Alejska, M., A. Kurzyniska-Kokorniak, M. Broda, R. Kierzek, and M. Figlerowicz. 2001. How RNA viruses exchange their genetic material. Acta Biochim. 48:391-407. [PubMed] [Google Scholar]
- 3.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltimore, D. 1971. Expression of animal virus genomes. Bacteriol. Rev. 35:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandelt, H.-J., P. Forster, and A. Rohl. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37-48. [DOI] [PubMed] [Google Scholar]
- 8.Beck, E., R. Sommer, E. A. Auerswald, C. Kurz, B. Zink, G. Osterburg, H. Schaller, K. Sugimoto, H. Sugisaki, T. Okamoto, and M. Takanami. 1978. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 5:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 10.Bishop, D. H. L. 1985. The genetic basis for describing viruses as species. Intervirology 24:79-93. [DOI] [PubMed] [Google Scholar]
- 11.Blum, H., W. Zillig, S. Mallok, H. Domdey, and D. Prangishvili. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281:6-9. [DOI] [PubMed] [Google Scholar]
- 12.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 13.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 14.Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. [DOI] [PubMed] [Google Scholar]
- 15.Bush, R. M., W. M. Fitch, C. A. Bender, and N. J. Cox. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457-1465. [DOI] [PubMed] [Google Scholar]
- 16.Butcher, S. J., T. Dokland, P. M. Ojala, D. H. Bamford, and S. D. Fuller. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J. 16:4477-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell, A., and D. Botstein. 1983. Evolution of the lambdoid phages, p. 365-380. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Campbell, A. M. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 19.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Virology 3:383-397. [Google Scholar]
- 20.Chatton, E. 1937. Titres et travaux scientifique (1906-1937). Sottano, Sète, France.
- 21.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark, A. J., W. Inwood, T. Cloutier, and T. S. Dhillon. 2001. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Mol. Biol. 311:657-679. [DOI] [PubMed] [Google Scholar]
- 23.Cohan, F. M. 2001. Bacterial species and speciation. Syst. Biol. 50:513-524. [DOI] [PubMed] [Google Scholar]
- 24.Copeland, H. F. 1956. The classification of lower organisms. Pacific Books, Palo Alto, Calif.
- 25.Darwin, C. 1859. On the origin of species by means of natural selection or the preservation of favored races in the struggle for life. John Murray, London, England.
- 26.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado, E., M. M. Thomson, M. L. Villahermosa, M. Sierra, A. Ocampo, C. Miralles, R. Rodriguez-Perez, J. Diz-Aren, R. Ojea-de Castro, E. Losada, M. T. Cuevas, E. Vazquez-de Parga, R. Carmona, L. Perez-Alvarez, L. Medrano, L. Cuevas, J. A. Taboada, and R. Najera. 2002. Identification of a newly characterized HIV-1. BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29:536-543. [DOI] [PubMed] [Google Scholar]
- 28.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 29.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.d'Herelle, F. 1924. Immunity in natural infectious disease. The Williams & Wilkins Co., New York, N.Y.
- 31.Doherty, D. H., P. Gauss, and L. Gold. 1982. On the role of the single-stranded DNA binding protein of bacteriophage T4 in DNA metabolism. I. Isolation and genetic characterization of new mutations in gene 32 of bacteriophage T4. Mol. Gen. Genet. 188:77-90. [DOI] [PubMed] [Google Scholar]
- 32.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doolittle, W. F. 1999. Lateral genomics. Trends Cell Biol. 9:M5-M8. [PubMed] [Google Scholar]
- 34.Doolittle, W. F. 2000. Uprooting the tree of life. Sci. Am. 282:90-95. [DOI] [PubMed] [Google Scholar]
- 35.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 37.Eigen, M. 1993. Viral quasispecies. Sci. Am. 269:32-39. [DOI] [PubMed] [Google Scholar]
- 38.Eshleman, S. H., M. J. Gonzales, G. Becker-Pergola, S. C. Cunningham, L. A. Guay, J. B. Jackson, and R. W. Shafer. 2002. Identification of Ugandan HIV type 1 variants with unique patterns of recombination in pol involving subtypes A and D. AIDS Res. Hum. Retrovir. 18:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsenstein, J. 1985. Confidence limits on phylogenies with a molecular clock. Syst. Zool. 34:152-161. [Google Scholar]
- 42.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long-term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. USA 94:7712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitz-Gibbon, S. T., and C. H. House. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 45.Ford, M. E., C. Stenstrom, R. W. Hendrix, and G. F. Hatfull. 1998. Mycobacteriophage TM4: genome structure and gene expression. Tuberc. Lung Dis. 79:63-73. [DOI] [PubMed] [Google Scholar]
- 46.Fu, X., H. Wang, and X. Zhang. 2002. High-frequency intermolecular homologous recombination during herpes simplex virus-mediated plasmid DNA replication. J. Virol. 76:5866-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George, D. G., L. S. Yeh, and W. C. Barker. 1983. Unexpected relationships between bacteriophage lambda hypothetical proteins and bacteriophage T4 tail-fiber proteins. Biochem. Biophys. Res. Commun. 115:1061-1068. [DOI] [PubMed] [Google Scholar]
- 48.Gibbs, A. J. 2000. Virus nomenclature descending into chaos. Arch. Virol. 145:1505-1507. [DOI] [PubMed] [Google Scholar]
- 49.Gogarten, J. P., W. F. Doolittle, and J. G. Lawrence. Bacterial evolution in light of transfer. Mol. Biol. Evol., in press. [DOI] [PubMed]
- 50.Gogarten, J. P., H. Kibak, P. Dittrich, L. Taiz, E. J. Bowman, B. J. Bowman, M. F. Manolson, R. J. Poole, T. Date, T. Oshima, J. Konishi, K. Denda, and M. Yoshida. 1989. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc. Natl. Acad. Sci. USA 86:6661-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldbach, R. 1987. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 4:197-202. [PubMed] [Google Scholar]
- 52.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 53.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 55.Haeckel, E. 1866. Generelle Morphologie der Organismen: Allgemeine Grundzüge der organischen Formen-Wissenschaft mechanisch begründet durch die von Charles Darwin reformierte Descendenz-Theorie. Georg Riemer, Berlin, Germany.
- 56.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hambly, E., F. Tétart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison, B. D. 1985. Usefulness and limitations of the species concept for plant viruses. Intervirology 24:71-78. [DOI] [PubMed] [Google Scholar]
- 59.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395-405. [DOI] [PubMed] [Google Scholar]
- 60.Hendrix, R. W. Bacteriophages: evolution of the majority. Theor. Pop. Biol., in press. [DOI] [PubMed]
- 61.Hendrix, R. W. 1999. The long evolutionary reach of viruses. Curr. Biol. 9:R914-R917. [DOI] [PubMed] [Google Scholar]
- 62.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 63.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Highton, P. J., Y. Chang, and R. J. Myers. 1990. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol. Microbiol. 4:1329-1340. [DOI] [PubMed] [Google Scholar]
- 65.Huang, S., S. J. Hayes, K. Lieman, G. A. Griess, and P. Serwer. 2001. Strategies for analysis of the evolution of bacteriophages. Recent Res. Dev. Virol. 3:1-12. [Google Scholar]
- 66.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ibba, M., J. L. Bono, P. A. Rosa, and D. Soll. 1997. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 94:14383-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwabe, N., K.-I. Kuma, M. Hasegawa, S. Osawa, and T. Miyata. 1989. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA 86:9355-9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 71.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage φPVL carrying Panton-Valentine leukocidin genes. Gene 215:57-67. [DOI] [PubMed] [Google Scholar]
- 72.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van Der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 73.Kingsbury, D. W. 1988. Biological concepts in virus classification. Intervirology 29:242-253. [DOI] [PubMed] [Google Scholar]
- 74.Lawrence, J. G. 2001. Catalyzing bacterial speciation: correlating lateral transfer with genetic headroom. Syst. Biol. 50:479-496. [DOI] [PubMed] [Google Scholar]
- 75.Lawrence, J. G. Gene transfer in bacteria: speciation without species? Theor. Pop. Biol., in press. [DOI] [PubMed]
- 76.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 77.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levin, B. 1981. Periodic selection, infectious gene exchange, and the genetic structure of E. coli populations. Genetics 99:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levin, B. R. 1988. Frequency-dependent selection in bacterial populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319:459-472. [DOI] [PubMed] [Google Scholar]
- 80.Linnaeus, C. 1758. Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Holmiae.
- 81.Liu, H. M., D. P. Zheng, L. B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Majewski, J., and F. M. Cohan. 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Majewski, J., P. Zawadzki, P. Pickerill, F. M. Cohan, and C. G. Dowson. 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marturano, J., and L. Fiore. 2002. Investigation of the presence of recombinant polioviruses in the hit population in Albania during the 1996 outbreak. J. Clin. Microbiol. 40:316-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mayr, E. 1963. Animal species and evolution. Harvard University Press, Cambridge, Mass.
- 87.Mayr, E. 1954. Change of genetic environment and evolution, p. 156-180. In J. S. Huxley, A. C. Hardy, and E. B. Ford (ed.), Evolution as a process. Allen and Unwin, London, England.
- 88.Mayr, E. 1942. Systematics and the origin of species. Columbia University Press, New York, N.Y.
- 89.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 90.Milne, R. G. 1985. Alternatives to the species concept for virus taxonomy. Intervirology 24:94-98. [DOI] [PubMed] [Google Scholar]
- 91.Mishler, B. D., and M. J. Donoghue. 1982. Species concepts: a case for pluralism. Syst. Zool. 31:491-503. [Google Scholar]
- 92.Monod, C., F. Repoila, M. Kutateladze, F. Tétart, and H. M. Krisch. 1997. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J. Mol. Biol. 267:237-249. [DOI] [PubMed] [Google Scholar]
- 93.Montavon, C., C. Toure-Kane, J. N. Nkengasong, L. Vergne, K. Hertogs, S. Mboup, E. Delaporte, and M. Peeters. 2002. CRF06-cpx: a new circulating recombinant form of HIV-1 in West Africa involving subtypes A, G, K, and J. J. Acquir. Immune Defic. Syndr. 29:522-530. [DOI] [PubMed] [Google Scholar]
- 94.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage mu genome sequence: analysis and comparison with mu-like prophages in Haemophilus, Neisseria, and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 95.Mosig, G., J. Gewin, A. Luder, N. Colowick, and D. Vo. 2001. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc. Natl. Acad. Sci. USA 98:8306-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 97.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nagy, M., E. Nagy, and T. Tuboly. 2002. Sequence analysis of porcine adenovirus serotype 5 fibre gene: evidence for recombination. Virus Genes 24:181-185. [DOI] [PubMed] [Google Scholar]
- 99.Nilsson, A. S., and E. Haggård-Ljungquist. 2001. Detection of homologous recombination among bacteriophage P2 relatives. Mol. Phylogenet. Evol. 21:259-269. [DOI] [PubMed] [Google Scholar]
- 100.Nimonkar, A. V., and P. E. Boehmer. 2002. In vitro strand exchange promoted by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem. 277:15182-15189. [DOI] [PubMed] [Google Scholar]
- 101.Ochman, H., J. G. Lawrence, and E. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 102.Ohshima, K., Y. Yamaguchi, R. Hirota, T. Hamamoto, K. Tomimura, Z. Tan, T. Sano, F. Azuhata, J. A. Walsh, J. Fletcher, J. Chen, A. Gera, and A. Gibbs. 2002. Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J. Gen. Virol. 83:1511-1521. [DOI] [PubMed] [Google Scholar]
- 103.Olsen, G. J., and C. R. Woese. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7:113-123. [DOI] [PubMed] [Google Scholar]
- 104.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, A. V. Karlyshev, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 105.Paterson, H. E. H. 1985. The recognition concept of species, p. 21-29. In E. S. Vrba (ed.), Species and speciation. Transvaal Museum, Pretoria, South Africa.
- 106.Peeters, B. P. H., R. M. Peters, J. G. G. Schoenmarkers, and R. N. H. Konings. 1985. Nucleotide sequence and genetic organization of the genome of the N-specific filamentous bacteriophage IKe: comparison with the genome of the F-specific filamentous phages M13, fd and f1. J. Mol. Biol. 181:27-39. [DOI] [PubMed] [Google Scholar]
- 107.Peng, X., H. Blum, Q. She, S. Mallok, K. Brugger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226-234. [DOI] [PubMed] [Google Scholar]
- 108.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 109.Pfister, P., A. Wasserfallen, R. Stettler, and T. Leisinger. 1998. Molecular analysis of Methanobacterium phage ψM2. Mol. Microbiol. 30:233-244. [DOI] [PubMed] [Google Scholar]
- 110.Plyusnin, A., S. K. Kukkonen, A. Plyusnina, O. Vapalahti, and A. Vaheri. 2002. Transfection-mediated generation of functionally competent Tula hantavirus with recombinant S RNA segment. EMBO J. 21:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raaijmakers, H., O. Vix, I. Toro, S. Golz, B. Kemper, and D. Suck. 1999. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18:1447-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 113.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 114.Repoila, F., F. Tétart, J. Y. Bouet, and H. M. Krisch. 1994. Genomic polymorphism in the T-even bacteriophages. EMBO J. 13:4181-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rohwer, F. L., A. M. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 116.Sanger, F., A. R. Coulson, G. F. Hong, D. F. Hill, and G. B. Petersen. 1982. Nucleotide sequence of bacteriophage lambda DNA. J. Mol. Biol. 162:729-773. [DOI] [PubMed] [Google Scholar]
- 117.Simon, M. N., R. W. Davis, and N. Davidson. 1971. Heteroduplexes of DNA molecules of lambdoid phages: physical mapping of their base sequence relationships by electron microscopy, p. 313-328. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 118.Smith, D. B., J. McAllister, C. Casino, and P. Simmonds. 1997. Virus “quasispecies”: making a mountain out of a molehill? J. Gen. Virol. 78:1511-1519. [DOI] [PubMed] [Google Scholar]
- 119.Smith, J. M., C. G. Dowson, and B. G. Spratt. 1991. Localized sex in bacteria. Nature 349:29-31. [DOI] [PubMed] [Google Scholar]
- 120.Snel, B., P. Bork, and M. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 121.Snel, B., P. Bork, and M. A. Huynen. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12:17-25. [DOI] [PubMed] [Google Scholar]
- 122.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 28:1409-1438. [Google Scholar]
- 123.Stassen, A. P. M., E. F. P. M. Schoenmarkers, M. Yu, J. G. G. Schoenmarkers, and R. N. H. Konings. 1992. Nucleotide sequence of the genome of the filamentous bacteriophage I2-2: module evolution of the filamentous phage genome. J. Mol. Evol. 34:141-152. [DOI] [PubMed] [Google Scholar]
- 124.Steinbacher, S., R. Seckler, S. Miller, B. Steipe, R. Huber, and P. Reinemer. 1994. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265:383-386. [DOI] [PubMed] [Google Scholar]
- 125.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 126.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tateno, Y., M. Nei, and F. Tajima. 1982. Accuracy of estimated phylogentic trees from molecular data. I. Distantly related species. J. Mol. Evol. 18:387-404. [DOI] [PubMed] [Google Scholar]
- 128.Tekaia, F., A. Lazcano, and B. Dujon. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9:550-557. [PMC free article] [PubMed] [Google Scholar]
- 129.Templeton, A. R. 1987. Species and speciation. Evolution 41:233-235. [Google Scholar]
- 130.Tétart, F., C. Desplats, and H. M. Krisch. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J. Mol. Biol. 282:543-556. [DOI] [PubMed] [Google Scholar]
- 131.Tétart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Regenmortel, M. H. V. 1989. Applying the species concept to plant viruses. Arch. Virol. 104:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Van Regenmortel, M. H. V. 2000. Introduction to the species concept in virus taxonomy, p. 3-16. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 135.Van Regenmortel, M. H. V. 1990. Virus species, a much overlooked but essential concept in virus taxonomy. Intervirology 31:241-254. [DOI] [PubMed] [Google Scholar]
- 136.Van Regenmortel, M. H. V., D. H. L. Bishop, C. M. Fauquet, M. A. Mayo, J. Maniloff, and C. H. Calisher. 1997. Guidelines to the demarcation of virus species. Arch. Virol. 142:1505-1518. [PubMed] [Google Scholar]
- 137.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 138.Van Regenmortel, M. H. V., M. A. Mayo, C. M. Fauquet, and J. Maniloff. 2000. Virus nomenclature: consensus versus chaos. Arch. Virol. 145:2227-2232. [DOI] [PubMed] [Google Scholar]
- 139.Van Valen, L. 1976. Ecological species, multispecies, and oaks. Taxon 25:223-239. [Google Scholar]
- 140.van Wezenbeek, P. M., T. J. Hulsebos, and J. G. Schoenmakers. 1980. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene 11:129-148. [DOI] [PubMed] [Google Scholar]
- 141.Venn, J. 1881. Symbolic logic. Macmillan, London, England.
- 142.Vulic, M., F. Dionisio, F. Taddei, and M. Radman. 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. USA 94:9763-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vulic, M., R. E. Lenski, and M. Radman. 1999. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc. Natl. Acad. Sci. USA 96:7348-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang, Y., Z. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 146.Whittaker, R. H. 1969. New concepts of kingdoms or organisms: evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science 163:150-160. [DOI] [PubMed] [Google Scholar]
- 147.Wiley, E. O. 1978. The evolutionary species concept reconsidered. Syst. Zool. 27:17-26. [Google Scholar]
- 148.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Woese, C. R. 1991. The use of ribosomal RNA in reconstructing evolutionary relationships among bacteria, p. 1-24. In R. K. Selander, A. G. Clark, and T. S. Whittam (ed.), Evolution at the molecular level. Sinauer Associates, Inc., Sunderland, Mass.
- 150.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Woese, C. R., G. J. Olsen, M. Ibba, and D. Soll. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64:202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689-710. [PubMed] [Google Scholar]
- 154.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yonesaki, T., and T. Minagawa. 1985. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 4:3321-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zawadzki, P., M. S. Roberts, and F. M. Cohan. 1995. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140:917-932. [DOI] [PMC free article] [PubMed] [Google Scholar]