Abstract
Pathogenic bacteria are specifically adapted to bind to their customary host. Disease is then caused by subsequent colonization and/or invasion of the local environmental niche. Initial binding of Haemophilus influenzae type b to the human nasopharynx is facilitated by Hib pili, filaments expressed on the bacterial surface. With three-dimensional reconstruction of electron micrograph images, we show that Hib pili comprise a helix 70 Å in diameter with threefold symmetry. The Hib pilus filament has 3.0 subunits per turn, with each set of three subunits translated 26.9 Å along and rotated 53 degrees about the helical axis. Amino acid sequence analysis of pilins from Hib pili and from P-pili expressed on uropathogenic Escherichia coli were used to predict the physical location of the highly variable and immunogenic region of the HifA pilin in the Hib pilus structure. Structural differences between Hib pili and P-pili suggest a difference in the strategies by which bacteria remain bound to their host cells: P-pili were shown to be capable of unwinding to five times their original length (E. Bullitt and L. Makowski, Nature 373:164-167, 1995), while damage to Hib pili occurs by slight shearing of subunits with respect to those further along the helical axis. This capacity to resist unwinding may be important for continued adherence of H. influenzae type b to the nasopharynx, where the three-stranded Hib pilus filaments provide a robust tether to withstand coughs and sneezes.
Binding of Haemophilus influenzae type b to the human nasopharynx via Hib pili precedes the onset of otitis media, meningitis, and epiglottitis in infants and young children and the onset of pneumonia in the elderly. The incidence of H. influenzae type b disease in infants and children has been reduced dramatically by strong vaccination programs in the United States and Europe. However, while H. influenzae type b disease has been eradicated in Iceland and Finland, significant health risks remain in developing countries where the vaccine is not readily available and in undervaccinated regions of Western society. For example, H. influenzae type b disease in the United States has not been eradicated due to undervaccination of at-risk populations (16, 24). Furthermore, vaccines have not yet been developed for any of the “nontypeable” strains of H. influenzae, which remain a health risk worldwide. An understanding of the structure of Hib pili may provide opportunities to develop novel antibacterial agents against disease caused by H. influenzae.
H. influenzae type b pili have structural and gene sequence similarities (4, 11, 13, 17) to pili described as class I (18) but not to those in the type IV pilus superfamily. In particular, strong sequence similarity is found between Hib pilins and pilins that comprise P-pili (13, 17). P-pili are expressed on the surface of uropathogenic Escherichia coli that cause pyelonephritis, urinary tract infections that involve the kidneys. Conversely, there is no significant sequence similarity between H. influenzae pilins and the type IV pilins involved in both twitching motility and bacterial adhesion, nor is there morphological similarity between the atomic structures of known pilins from E. coli (3, 23) and the type IV pilins from Neisseria gonorrhea (21) or Pseudomonas aeruginosa (14).
The structures of the adhesin subunits of type 1 pili and P-pili and a minor pilin of P-pili have been determined in complex with their periplasmic chaperone proteins: X-ray crystallography has been used to investigate the structures of FimH/FimC (3), PapG/PapD (6), and PapK/PapD (23). These pilins all contain subunits with an immunoglobulin-like fold and a donor strand complementation mechanism in which a beta strand donated from the chaperone fills the groove in the pilin subunit. In the modeled pilus filaments, it is a beta strand from the subsequent pilin monomer that inserts into the subunit groove (3, 23). Each of these pilins has sequence similarity with their respective major structural pilins (FimH with FimA, PapK with PapA) as well as similarity to HifA, the major structural pilin of the Hib pilus. The sequence similarities between HifA and PapK, in combination with morphological similarities of the helical filaments that comprise the pili, provide evidence for donor strand complementation of HifA, as seen in type 1 and P-pili (17). Structural information on the similarities and differences between assembled pili can be used to investigate the range of structural polymorphism that can be accommodated by this mechanism.
Immunologic cross-reactivity and sequence analysis are aiding our understanding of assembled pilus structures. Studies on the hydrophilicity of HifA were used to define three regions that were expected to be surface exposed and that might therefore constitute antigenic epitopes (8). Subsequently, it was shown that antibodies to 14- to 15-residue peptides of these regions do not react with assembled pili (10). Even strains of H. influenzae with 99% identity can be immunologically distinct, and researchers have constructed chimeras to enhance reactivity to conformational epitopes, thereby defining noncontiguous residues that are structurally associated (20). Studies such as these, in combination with sequence analyses that define variable regions of pilin subunits (4, 17), provide data for fitting known structures from homologous pilins into new structural data.
We present results from our studies of the structure of Hib pili with sequence analysis of the major structural pilin, HifA, and electron microscopy and image processing of the structure of Hib pili assembled into a helical filament. A comparison of the Hib pilus structure to that of P-pili elucidates significant structural differences within class I pili, a superfamily defined (18) by sequence similarity of the major structural pilins. Subunits in Hib pili appear to be oriented almost vertically along the filament axis, while subunits are oriented approximately horizontally in models of P-pili (23) and type 1 pili (3). Our data provide a basis for understanding bacterial strategies that facilitate survival of H. influenzae type b bacteria in the nasopharynx, preceding invasion of the human host and systemic disease.
MATERIALS AND METHODS
Amino acid sequence alignments.
Initial alignments of pilin sequences were performed with the program ALIGN (22). These results were compared with alignments reported by Girardeau et al. (13) and Krasan et al. (17), alignments that were made between HifA of different strains (13), and between HifA and a minor adapter P-pilin, PapK, not the major structural pilin PapA (17). The great strength of these reference alignments is that they have taken into account additional data beyond the primary sequence, including results from biochemical, mutational, and evolutionary studies on H. influenzae pili and E. coli pili. Therefore, we have manually adjusted the sequence alignments of HifA/Eagan, HifA/M43p+, and PapA/J96 predicted by ALIGN, utilizing amino acid similarity tables (22) and the results reported by these two groups.
Sample preparation.
Hib pili from H. influenzae strain Eagan were a generous gift from Joseph St. Geme III (Washington University School of Medicine, St. Louis, Mo.), purified as described previously (25). Briefly, H. influenzae was grown overnight in supplemented brain heart infusion broth at 37°C and then centrifuged to pellet the bacteria. Pili were sheared from the cell surface with a blender, and the bacterial cells were removed by centrifugation. Pili were purified by cycles of precipitation in ammonium sulfate and resuspension in 0.5 M Tris, pH 7. Final resuspension was in 50 mM Tris, pH 7.
Grids for transmission electron microscopy were made by adsorption of the sample onto carbon-coated grids for 3 min, washed with 8 drops of 50 mM Tris (pH 7), and stained with 1% uranyl acetate for 15 s. Samples were imaged with a Philips CM12 transmission electron microscope operated at 120 kV with a LaB6 filament. Images were recorded under minimal electron dose conditions at a magnification of ×60,000 and digitized at 3.5 Å/pixel.
Image processing.
Three-dimensional reconstructions of Hib pili were computed with single-particle data analysis (9) and the iterative helical real space reconstruction (IHRSR) method (7). After each cycle, the helical symmetry of the average structure was calculated and imposed, providing an improved reference particle for each subsequent iteration (7).
Specifically, overlapping lengths of Hib pili were masked from images of negatively stained sample for a total of approximately 165,000 subunits. The single-particle analysis program package SPIDER (9) was used to align each filament fragment to a reference particle, both rotationally and translationally. An initial reference particle comprising the P-pilus helical reconstruction (2) with 3.28 subunits per turn was not an appropriate starting model, as the iterative procedure did not converge to a stable solution for the Hib pilus structure.
Computed Fourier transforms of the best H. influenzae type b filament images were indexed, and found to have threefold symmetry (3.0 subunits per turn), with an approximately 300 Å repeat (data not shown). Therefore, a three-stranded rope model was constructed, and projections of this model perpendicular to the helical axis, computed every 5 degrees, were used as the initial references. Each particle was fit to the references with cross-correlation, and back projections were computed to produce an average structure. The helical symmetry of the result was computed and imposed, providing improved reference particles for each subsequent cycle of refinement. After 50 cycles, the structure had converged to a stable solution. Three back projection algorithms (SPIDER command bp3f or bp32, bprp, and the radon back projection algorithm written and generously provided by Michael Radermacher) were tested to ensure that the resultant reconstruction was reliable.
In the final reconstruction, approximately 145,000 subunits were retained. The resolution of the reconstruction is at least 19 Å, as calculated by splitting the data set in half and using a Fourier shell coefficient of 0.5 to determine self-similarity. If the less stringent criterion of the crossing point of the noise curve (noise factor 3) and the Fourier shell coefficient curve was used, the resolution would be estimated to be 12 Å. As a check for the validity of imposing threefold symmetry, data were analyzed with the P1 space group (no imposed symmetry), resulting in a reconstruction that again showed strong threefold symmetry (data not shown).
Images shown are of the resultant reconstructions computed with the SPIDER weighted back projection algorithm bp32f (9). Surface views in Fig. 4 were calculated with WEB (9). Electron density maps of the model H. influenzae type b filament were computed with Gaussian-weighted mass distributions from atomic coordinates of the minor P-pilin PapK (PDB 1PDK, chain B) (23). Isocontour surfaces of the reconstruction and of the model filament were computed with SITUS (28). Surface views in Fig. 6 are displayed with the visual molecular dynamics program (VMD) (15).
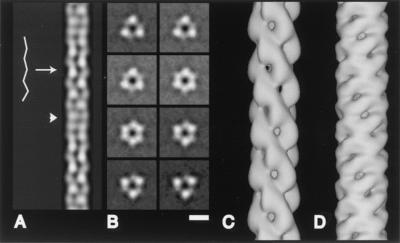
FIG. 4.
Results of three-dimensional reconstruction of Hib pili. (A) In projection, the central channel around the helical axis has a zigzag appearance along part of its length (arrow) and regions in which the central channel is indistinct (arrowhead), as is observed in electron micrographs of negatively stained Hib pili (see Fig. 2). (B) Cross section views show the threefold symmetry about the helical axis and a central channel straight up the filament axis. Cross-sections shown are spaced 2.7 Å apart. (C) Surface of the three-dimensional reconstruction of Hib pili. (D) For comparison, the surface of the three-dimensional reconstruction of P-pili; see also reference 2. Bar: 50 Å (A and B), 25 Å (C and D).
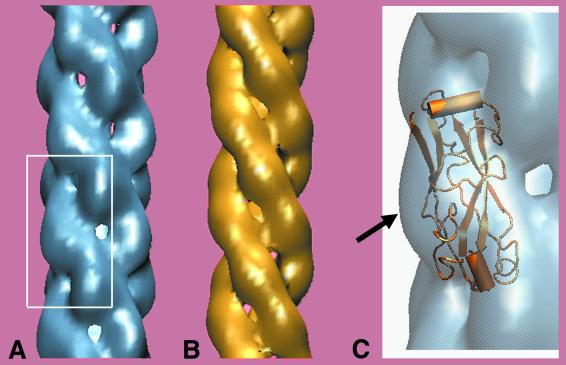
FIG. 6.
Three-dimensional reconstruction and model of Hib pilus. (A) Three-dimensional reconstruction computed from electron microscope data of H. influenzae type b pilus. Note the strong connections running along the left-handed three-start helix. The filament is ≈7 nm in diameter, with scalloped edges. (B) One model of the Hib pilus constructed from subunits of the homologous minor P-pilin PapK. The PapK structure was taken from PDB file 1PDK B (23) and placed on the Hib pilus helix. (C) An enlarged view of the three-dimensional reconstruction from the boxed region in A, with a cartoon of the PapK monomer included to illustrate the proposed position of the extra density of HifA compared to PapK. In our model, the extra 20% mass of HifA is seen as the surface-exposed region that extends furthest from the helical axis (arrow in C), corresponding to regions of highly variable amino acid sequence of the HifA subunit.
RESULTS
Sequence similarities and differences between major structural proteins of Hib pili and P-pili.
To examine the similarities and differences between the major structural pilins of Hib pili and P-pili, we first aligned sequences from two strains of HifA, strain Eagan and strain M43p+, with the software program ALIGN (22), producing an alignment identical to that in reference 17. Excluding the three variable regions of HifA (17, 20), HifA/Eagan and HifA/M43p+ have 82% identity and 95% similarity.
The Hib pilus major subunit, HifA, is 20% larger than PapA, the major structural pilin in P-pili expressed on the surface of pyelonephritis-causing E. coli. Thus, alignment of the 196-amino-acid HifA sequence to the 163-amino-acid PapA sequence necessarily includes at least 33 amino acids in HifA with no corresponding amino acid in PapA. These amino acid “insertions” may be distributed randomly or may appear as a loop(s).
Automated sequence alignment of HifA/Eagan and HifA/M43p+ to PapA/J96 with ALIGN (22) predicted significantly different locations for the insertions of HifA amino acids with respect to PapA for each strain of H. influenzae despite the 82% identity and 95% homology between the two HifA sequences. As structural variations of this magnitude are extremely unlikely to occur in vivo from such highly homologous monomers, a manual alignment of the PapA sequence was completed to reconcile the preliminary sequence alignments. In addition, the new alignment was required to account for data available from published alignments of HifA/M43p+ with PapA/F7-2 (13) and of HifA/Eagan with PapK, a minor P pilin with 28% sequence identity and 55% similarity to PapA (17).
First, a shift of the PapA sequence HYTAVVKKSSAVG resulted in the correspondence of a HifA insert with the longest highly variable region in the HifA sequence. This shift reduced the sequence alignments from 23% identical and 61% similar to 22% identical and 57% similar, excluding the HifA inserts. Data from previous alignments of the major structural pilins of class I pili were used to indicate regions of conservation and of variability for predicted secondary structures in HifA (4, 13) and PapA (26). These data were also used as guidelines in our realignment of the HifA/Eagan and PapA/J96 pilin sequences (Fig. 1). Further minor modifications were made to the automated alignment to ensure that the sequence alignment was in best agreement with both the alignment of HifA/Eagan to PapK (17) and the alignment of HifA/M43p+ to PapA/P02972 (13).
FIG. 1.
Alignment of HifA and PapA after accounting for data from previous studies (10, 12, 13, 17, 20, 22, 26). HifA from two strains of H. influenzae, Eagan and M43p+, have 82% identity and 95% similarity or 76% identity and 91% similarity, excluding and including, respectively, the highly variable regions of HifA. HifA/Eagan (from H. influenzae) and PapA/J96 (from E. coli) have 21% identity and 58% similarity or 17% identity and 48% similarity, excluding and including, respectively, the amino acid insertions of HifA with respect to PapA. The alignment of HifA/Eagan and PapK/J96 of Krasan et al. (17) is included.
In the final alignment (Fig. 1), HifA/Eagan and PapA/J96 had 21% identity and 58% similarity, excluding the amino acid insertions of HifA with respect to PapA. The N-terminal 20 amino acids had 40% identity and 55% similarity, and the C-terminal 14 amino acids had 21% identity and 79% similarity.
Morphology of Hib pili.
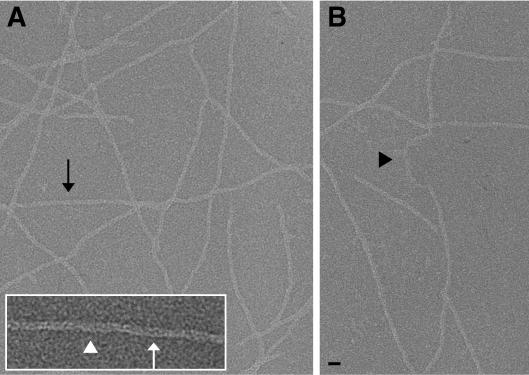
To determine the structure of Hib pili, purified pili were negatively stained and imaged by minimal-electron-dose transmission electron microscopy (Fig. 2). Hib pili did not demonstrate the extended stretches of extremely straight pilus seen in P-pili, but short stretches of reasonably straight segments were found (black arrow, Fig. 2A). The IHSRS method (7) only requires filaments to be approximately straight for the chosen box length, in this case 250 Å, rather than the greater than 1,500-Å-long filament segments required for a sufficient signal-to-noise ratio to use spline fitting and traditional helical reconstruction methods (5) on such a thin filament (70 Å diameter). Damaged pili were seen with apparent shear along their length (black arrowhead in Fig. 2B), and Hib pili did not exhibit the extensive unwinding of the helical filament seen in P-pili (1, 2).
FIG. 2.
Images of isolated, negatively stained H. influenzae type b pili. These pili are straight for short distances along the filament (black arrow in A). When damaged, Hib pili do not unwind into thin fibrillar structures but appear to be sheared perpendicular to the filament axis (black arrowhead in B). Along the length of the filament, there are regions in which a low-density channel is barely visible (white arrowhead) and regions in which the channel appears as a zigzag along the filament axis (white arrow). Bars: 100 Å (A and B), 50 Å (inset).
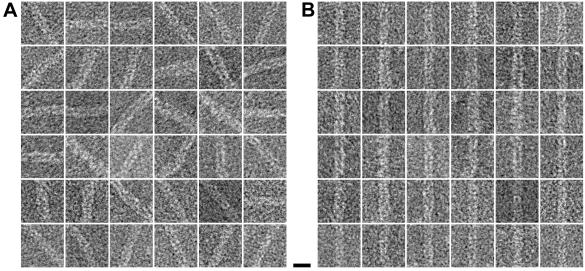
For three-dimensional reconstruction, Hib pili were treated as discrete single particles, 250 Å in length. These short filament segments were aligned to a reference and averaged; 36 such “particles” are shown as raw images (Fig. 3A) and after alignment (Fig. 3B). After three-dimensional reconstruction, the helical symmetry was determined. This symmetry was imposed on the average structure, thereby creating a new reference model for the next cycle of alignment and averaging (see Materials and Methods) (7).
FIG. 3.
Images of Hib pili treated as single particles. (A) Filaments were selected and boxed as overlapping segments; shown are 36 of the ≈5,000 selected areas that were used in the data analysis. (B) The same 36 images after in-plane rotation and translational alignment to a reference particle. Bar, 100 Å.
The resultant structure of Hib pili, after 53 cycles of alignment, had threefold symmetry, with a rise of 26.9 Å and a rotation of 53 degrees for each set of three subunits (Fig. 4, 5, and 6). This solution was stable, as 50 additional cycles did not alter the resultant structure (data not shown). The sense of the 53-degree rotation was a left-handed helix (25), determined previously by St. Geme et al. (25), with a 50-Å pitch. The pili had an approximate helical repeat containing nine sets of three subunits, or 242 Å (9 × 26.9 Å). Thus, a unit cell contains three HifA subunits in a ring (Fig. 4B), and this ring is translated and rotated to form the next unit cell of the helix. In this circumstance, three subunits in identical vertical positions along the helix axis are positioned symmetrically, separated by 120° with respect to each other. The next ring of three subunits is positioned 26.9 Å above, rotated 53° from the preceding ring so as to generate a left-handed three-start helix.
A central channel of approximately 20 Å in diameter is visible in the Hib reconstruction, straight up the filament axis. The straight channel was unexpected, as images of Hib pili often show low density with a zigzag appearance in the center of the filament (white arrow, Fig. 2A inset), as well as regions where the central low density is much less pronounced (white arrowhead, Fig. 2A inset). This variation in appearance along the filament length is due to the nature of transmission electron microscopy; images are a projection through the sample, with all planes of the object in focus; they are not a surface view. When such a projection image is calculated from the H. influenzae type b reconstruction, the zigzag and diffuse appearances of the central channel are both clearly apparent (white arrow and white arrowhead, respectively, in Fig. 4A). Similarly, our current results show that while only two strands are discernible at any time in a projection image (Fig. 4A), the filament comprises a three-start helix with threefold symmetry (Fig. 4B and 6). Connectivity of density in Hib pili is strongest along this three-start (Fig. 4C and 6). The outer edge of the filament has a scalloped appearance (Fig. 4C and 6). Thus, the measured diameter varies from 60 to 70 Å as the projected density narrows and widens along the length of the filament.
DISCUSSION
Our current data show that Hib pili are threefold symmetric about the helical axis, with a repeat distance of ≈242 Å. It was reported previously (25), based upon fast-freeze/deep-etch electron microscopy, that the Hib pilus contained a left-handed one-start and a right-handed two-start helix, with a crossover distance of 260 Å. How can these observations be reconciled with our current description of the Hib pilus structure? Shadowed samples provide a view of a surface-coated structure, whereas negatively stained samples provide a projection image through the protein density. The apparent one-start helix seen in shadowed images may be a visualization of the ring of density formed by filling the groove between each set of three subunits that comprise a unit cell. Three-dimensional reconstruction provides information about the protein density through the interior of a macromolecular assembly. Thus, at our current resolution of better than 20 Å, additional details are now available for a more complete understanding of the Hib pilus structure.
H. influenzae type b pili and P-pili are fundamentally similar; they are helical adhesive filaments extending from the outer cell membrane of pathogenic gram-negative bacteria. In both H. influenzae and E. coli, structural pilins are transported across the periplasm by chaperone proteins and exit via a homo-oligomeric usher protein that sits at the outer membrane. The pili each have lengths on the order of 1 μm and diameters of ≈70 Å (Fig. 4C and D). In addition, the major structural pilin for each pilus, HifA for Hib pili and PapA for P-pili, is expected to be structurally similar, and there is evidence for donor strand complementation based on morphological similarities of the pili and on sequence alignment between HifA and PapK, a minor structural P-pilin (17).
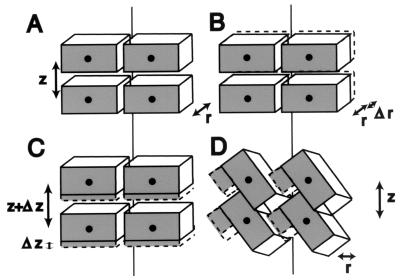
As seen in structures of filamentous phages, similar monomers assembled into filaments with different symmetry can produce dramatic alterations in functional properties. For example, a difference in the packing of the capsid proteins from Pf1 (19) versus Pf3 (27) produces phage that differ by a factor of 2.4 in their DNA-to-protein ratios. Despite their overall similarity in length and width, Hib pili and P-pili also have significant morphological differences. For example, Hib pili are not straight for extended stretches as are P-pili, necessitating the use of new image analysis methods (7). In addition, the amino acid sequence insertions in the HifA subunit compared with the PapA subunit must alter the resultant helical filament. That is, one cannot add mass to an object without changing it. By analogy, if a building supplier delivers bricks that are 20% larger than those sent previously, a house built with the new bricks cannot be structurally identical to one constructed with the older (smaller) bricks (Fig. 5).
FIG. 5.
Possible ways for a helix to accommodate a 20% increase in mass. (A) Initial rectangular “subunit,” oriented approximately horizontally, with radius r and rise per subunit z. The helix is viewed after slicing it down the back and laying it flat on the page. (B) The subunit could increase in thickness, thereby increasing the filament radius, r. (C) The subunit could increase in height, increasing the rise per subunit, z. (D) As appears to occur in Hib pili compared with P-pili, the subunits could tip up, making space for the additional mass without increasing z or r; the number of subunits per turn is reduced.
The extra mass of HifA compared with PapA appears to be accommodated in the Hib pilus structure by a rotation of each subunit and concomitant reduction in the number of subunits per turn (3.0 compared with 3.28). While there are a variety of ways to build a filament with larger subunits, there are experimental data restricting the possibilities for Hib pili. First, the diameter of the filament could increase, making the walls thicker (r + Δr, Fig. 5B). Data do not support this possibility, as both P-pili and Hib pili have diameters of ≈70 Å. Second, the cavity within the fiber could be reduced in size. Again, data do not support this possibility; the central channel in Hib pili is 20 Å in diameter, while that in P-pili has an elliptical cross-section, with major and minor axes of 25 and 15 Å, respectively, yielding an insufficient difference in volume (<7%). Third, the rise per subunit could increase (z + Δz, Fig. 5C). This does occur; there is a 7.6-Å rise per subunit for P-pili, compared with a 9.0-Å rise per subunit for H. influenzae type b (calculated from a 26.9-Å rise per three subunits). This increase could be due to an increase in the height, along the helical axis, of each subunit. To calculate the expected change, the new height for a cylinder with constant diameter (70 Å) and central channel (20 Å) and an increased volume of 20% was determined. The increase in height would be from 24.9 Å in P-pili (2) to 29.1 Å in Hib pili. This does not fit the existing data, as the pitch in Hib is increased compared to P-pili, but only to 26.9 Å. Fourth, the subunits may tip up, creating a space for the extra mass to be inserted at the surface (shown in Fig. 5D). This possibility need not change the filament radius (r), the channel size, or the rise per subunit (z) and is consistent with our structural data. It is also consistent with biochemical and immunochemical data from other laboratories, which predict that the insertions of HifA occur at surface-exposed regions of high sequence variability (4, 13, 17).
This predicted addition of mass at the pilus surface is also consistent with the results of our sequence alignment between the major structural pilins of Hib pili and P-pili, HifA/Eagan and PapA/J96, respectively. With biochemical, mutational, immunologic, and evolutionary data (4, 8, 10, 12, 13, 17, 20, 26), we have produced an alignment in which the absence of cross-reactivity of antibodies to HifA amino acids 62 to 72 and 97 to 102 (20) is correlated with insertion regions of HifA compared with PapA, and the extended hypervariable region defined by Krasan et al. (17) corresponds to the largest insertion region. This region is expected to be surface exposed. While it is not yet possible to define the boundaries between individual subunits in our reconstruction, our data suggest the location of the large HifA insert to be the mass extending farthest from the helical axis, as can be seen modeled in Fig. 6.
What are the structural consequences of Hib pili having threefold symmetry, with subunits oriented approximately vertically along the filament axis? Both Hib and P-pili have about 3 subunits per turn of their helix; Hib pili contain 3 subunits in each turn, and P-pili contain 3.28 subunits per turn. Thus, only Hib pili comprise a structure with threefold symmetry at all positions along the helical axis. An observed consequence of this difference in symmetry and differences between the pilin subunits (HifA compared to PapA) is that in P-pili the predominant interactions are along the one-start helix, forming a coil that can be unwound under stress. In Hib pili the predominant interactions are along each of three interwound helices. It is therefore not possible to unwind the Hib pilus into a comparable single fibrillar structure. The Hib filament can be described by imagining three elastic ropes wound about each other, each connected to the others by weak cross-bridges. If the cross-bridges begin to peel apart, separating one rope from the other two, that rope may bulge out but will not easily become a thin distinct fiber; this would require either that the damage be propagated all the way to the end of the filament or that the strand be severed, conditions that were not observed in images of thousands of Hib pili.
Overextension of the helical filament into thin fibrillae, as shown for P-pili (1, 2), was not observed in Hib pili. The comparative velocity of flow in each host environment provides a plausible argument for this structural difference. A cough or a sneeze can reach a velocity of 150 km/h, whereas urine flows at ≈0.002 km/h and mucociliary clearance occurs at ≈0.0005 km/h. Continued adherence of H. influenzae to the nasopharynx most likely occurs in niches about which there is no detailed information, whereas initial adherence may occur in exposed, relatively unprotected locations. The bacteria must then maintain contact long enough to create or locate a more protected environment. Consequently, while typical environmental velocities in the urinary tract and the nasopharynx differ by a factor of only about 10, intermittent coughs and sneezes may result in an extreme environment for H. influenzae to withstand. The precise shear pressures have not yet been calculated for these two environments, but it seems likely that an extended thin fiber 20 Å in diameter could not survive velocities 75,000 times greater than those experienced in the urinary tract.
We hypothesize that the combination of a three-stranded helix and orientation of the HifA subunits almost vertically along the helical axis provide the structural stability necessary for Hib pili to survive in the nasopharynx prior to invasion of host cells. Thus, while P-pili and Hib pili have an overall morphology of ≈70-Å-diameter helical filaments with a length on the order of 1 μm, the structure of each pilus type is specifically adapted to its local environmental niche.
Acknowledgments
We thank Donna Cabral-Lilly and David G. Morgan for many helpful discussions throughout the course of this work. We thank Rebecca Ray for technical assistance.
This work was funded, in part, by NIH/NIGMS grants to E.B. and E.H.E. and a grant from the Medical Foundation to E.B.
REFERENCES
- 1.Bullitt, E., and L. Makowski. 1998. Bacterial adhesion pili are heterologous assemblies of similar subunits. Biophys. J. 74:623-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullitt, E., and L. Makowski. 1995. Structural polymorphism of bacterial adhesion pili. Nature 373:164-167. [DOI] [PubMed] [Google Scholar]
- 3.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 4.Clemans, D. L., C. F. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeRosier, D. J., and P. B. Moore. 1970. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 52:355-369. [DOI] [PubMed] [Google Scholar]
- 6.Dodson, K. W., J. S. Pinkner, T. Rose, G. Magnusson, S. J. Hultgren, and G. Waksman. 2001. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105:733-743. [DOI] [PubMed] [Google Scholar]
- 7.Egelman, E. H. 2000. A robust algorithm for the reconstruction of helical filaments with single-particle methods. Ultramicroscopy 85:225-234. [DOI] [PubMed] [Google Scholar]
- 8.Forney, L. J., J. R. Gilsdorf, and D. C. Wong. 1992. Effect of pili-specific antibodies on the adherence of Haemophilus influenzae type b to human buccal cells. J. Infect. Dis. 165:464-470. [DOI] [PubMed] [Google Scholar]
- 9.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 10.Gilsdorf, J. R., L. J. Forney, and K. W. McCrea. 1993. Reactivity of antibodies against conserved regions of pilins of Haemophilus influenzae type b. J. Infect. Dis. 167:962-965. [DOI] [PubMed] [Google Scholar]
- 11.Gilsdorf, J. R., C. F. Marrs, K. W. McCrea, and L. J. Forney. 1990. Cloning, expression, and sequence analysis of the Haemophilus influenzae type b strain M43p+ pilin gene. Infect. Immun. 58:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilsdorf, J. R., K. McCrea, and L. Forney. 1990. Conserved and nonconserved epitopes among Haemophilus influenzae type b pili. Infect. Immun. 58:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girardeau, J. P., Y. Bertin, and I. Callebaut. 2000. Conserved structural features in class I major fimbrial subunits (pilin) in gram-negative bacteria. Molecular basis of classification in seven subfamilies and identification of intrasubfamily sequence signature motifs which might be implicated in quaternary structure. J. Mol. Evol. 50:424-442. [DOI] [PubMed] [Google Scholar]
- 14.Hazes, B., P. A. Sastry, K. Hayakawa, R. J. Read, and R. T. Irvin. 2000. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 299:1005-1017. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 16.Jafari, H. S., W. G. Adams, K. A. Robinson, B. D. Plikaytis, and J. D. Wenger. 1999. Efficacy of Haemophilus influenzae type b conjugate vaccines and persistence of disease in disadvantaged populations. The Haemophilus Influenzae Study Group. Am. J. Public Health 89:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasan, G. P., F. G. Sauer, D. Cutter, M. M. Farley, J. R. Gilsdorf, S. J. Hultgren, and J. W. St. Geme 3rd. 2000. Evidence for donor strand complementation in the biogenesis of Haemophilus influenzae haemagglutinating pili. Mol. Microbiol. 35:1335-1347. [DOI] [PubMed] [Google Scholar]
- 18.Low, D., B. Braaten, and M. van der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 19.Marvin, D. A., and E. J. Wachtel. 1976. Structure and assembly of filamentous bacterial viruses. Phil. Trans. R. Soc. Lond. B Biol. Sci. 276:81-98. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, K. L., and R. S. Munson, Jr. 1992. Construction of chimaeric genes for mapping a surface-exposed epitope on the pilus of non-typable Haemophilus influenzae strain M37. Mol. Microbiol. 6:2583-2588. [DOI] [PubMed] [Google Scholar]
- 21.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 23.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 24.Schuchat, A., T. Hilger, E. Zell, M. M. Farley, A. Reingold, L. Harrison, L. Lefkowitz, R. Danila, K. Stefonek, N. Barrett, D. Morse, and R. Pinner. 2001. Active bacterial core surveillance of the emerging infections program network. Emerg. Infect. Dis. 7:92-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St. Geme, J. W., J. S. Pinkner III, G. P. Krasan, J. Heuser, E. Bullitt, A. L. Smith, and S. J. Hultgren. 1996. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc. Natl. Acad. Sci. USA 93:11913-11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Die, I., M. Wauben, I. Van Megen, H. Bergmans, N. Riegman, W. Hoekstra, P. Pouwels, and B. Enger-Valk. 1988. Genetic manipulation of major P-fimbrial subunits and consequences for formation of fimbriae. J. Bacteriol. 170:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welsh, L. C., M. F. Symmons, J. M. Sturtevant, D. A. Marvin, and R. N. Perham. 1998. Structure of the capsid of Pf3 filamentous phage determined from X-ray fibre diffraction data at 3.1 A resolution. J. Mol. Biol. 283:155-177. [DOI] [PubMed] [Google Scholar]
- 28.Wriggers, W., R. A. Milligan, and J. A. McCammon. 1999. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125:185-195. [DOI] [PubMed] [Google Scholar]