Abstract
FtsH, a membrane-bound metalloprotease, with cytoplasmic metalloprotease and AAA ATPase domains, degrades both soluble and integral membrane proteins in Escherichia coli. In this paper we investigated how membrane-embedded substrates are recognized by this enzyme. We showed previously that FtsH can initiate processive proteolysis at an N-terminal cytosolic tail of a membrane protein, by recognizing its length (more than 20 amino acid residues) but not exact sequence. Subsequent proteolysis should involve dislocation of the substrates into the cytosol. We now show that this enzyme can also initiate proteolysis at a C-terminal cytosolic tail and that the initiation efficiency depends on the length of the tail. This mode of degradation also appeared to be processive, which can be aborted by a tightly folded periplasmic domain. These results indicate that FtsH can exhibit processivity against membrane-embedded substrates in either the N-to-C or C-to-N direction. Our results also suggest that some membrane proteins receive bidirectional degradation simultaneously. These results raise intriguing questions about the molecular directionality of the dislocation and proteolysis catalyzed by FtsH.
Energy-dependent proteolysis plays important roles in controlling and maintaining cellular functions. Whereas the 26S proteasome is a key player of these events in eukaryotic cells, HslUV, ClpAP, ClpXP, Lon, and FtsH and their homologs are known as ATP-dependent proteases in bacteria (13, 23, 38) as well as in the mitochondria and the chloroplasts (1, 23). The ATPase domains or subunits of these proteases belong to the AAA+ ATPase family (29, 34, 38). Many of them are known to form ring-like assemblies of subunits (31, 43). The proteolytic active sites of these enzymes are thought to be located in the interior cavity of the assemblies; experimentally this has been shown for the proteasome, HslUV, ClpAP, and ClpXP (8, 15, 26, 44). For the substrate peptide bond to be hydrolyzed, it must be presented to the catalytic center within the cavity. This implies that the substrate protein itself must be unfolded and translocated into the catalytic chamber (18, 33, 35). Presumably, these proteases utilize their ATPase functions to facilitate unfolding and translocation of substrate proteins.
A characteristic feature of the energy-dependent proteolytic reactions is that they occur in processive manners (24). Once the enzyme initiates the reaction, it continues to work along the polypeptide chain to hydrolyze the substrate successively. Crucial questions here include (i) how the enzyme recognizes the initiation site on the substrate, (ii) how it initiates unfolding or translocation of the substrate, and (iii) how it continues the reaction along the polypeptide chain, expelling the product small peptides from the catalytic chamber. Studies on the Clp proteases demonstrated that these enzymes can recognize either the N- or C-terminal end of substrate polypeptides for the initiation of unfolding or proteolysis (12, 14, 17).
Among the ATP-dependent proteases in Escherichia coli, FtsH is unique in that it is an integral membrane protein and it acts against both membrane-embedded and soluble substrates (3, 5, 19, 23). It has two N-terminally located transmembrane segments and a cytoplasmic region, which includes a zinc-metalloproteinase active site sequence and an AAA ATPase domain (42). Membrane-embedded substrates of FtsH include the SecY subunit of protein translocase (19), subunit a of the F0 component of the H+ ATPase (4), and the product of the yccA open reading frame (21). While its enzymatic active sites are located in the cytoplasmic domain, FtsH exerts processive proteolysis over the entire regions of a substrate, including transmembrane and periplasmic segments. Presumably, this is accomplished by the ability of FtsH to dislocate the extracytoplasmic regions of a substrate to the cytoplasm (22). We reason that such a mechanism evolved to enable hydrolysis of peptide bonds that are otherwise embedded within the hydrophobic environment of the membrane.
Although electron microscopic analysis of the soluble Clp protease complex revealed a central conduit for the entry and passage of a substrate (18, 33), nothing is known about the substrate entry site of FtsH. Particularly intriguing and important is a question of how a membrane-integrated substrate gets access to the active sites of this enzyme, which must be shared by soluble substrates. The following two observations led us to suggest that these two classes of substrates are presented to FtsH via different pathways. First, some mutant forms of HflKC (an FtsH-interacting membrane protein complex) and of YccA (an FtsH substrate) interfere specifically with the degradation of membrane-bound substrates (20, 21). Secondly, membrane integration of FtsH itself is essential for its action against membrane-integrated substrates (3). To explore the membrane protein recognition mechanisms, we characterized the substrate regions required for the initiation of degradation. Our results showed that FtsH can initiate degradation of membrane proteins from N-terminal cytosolic tails (9). For this initiation to occur, an N-terminal tail should have 20 or more amino acid residues. Presumably, FtsH captures such a region to initiate the dislocation reaction.
On the other hand, it was reported that at least one soluble protein is degraded by FtsH when the SsrA tag had been attached to the C terminus (16). These different results together with our contention that soluble and membrane-integrated substrates are presented to FtsH via different pathways prompted us to examine whether the soluble pathway and the membrane pathway share the same directionality in the processive proteolytic reactions.
The primary objective of this study was to clarify whether the N-terminal length recognition is the sole mechanism by which FtsH initiates proteolysis against a membrane-embedded substrate. Experiments using a series of engineered variants of membrane proteins revealed that FtsH-mediated membrane protein degradation can be initiated at either end or both, with a common requirement for the length of the cytosolically exposed residues.
MATERIALS AND METHODS
Plasmids.
Plasmid pCH244 (carrying SecY′-PhoA-′SecY) was constructed as follows. A secY fragment was amplified from pCM4 (27) such that it was flanked by the KpnI and HindIII recognition sequences and was cloned into pTWV228, a pBR322-based lac promoter vector (Takara Shuzo) after digestions with these enzymes. This plasmid was named pCH221. The SpeI recognition sequence was inserted between the codons for Gly313 and Gln314 in the P4 region of SecY, using a mutagenic primer (36), 5′-TGTATTTGCAGCCTGGGACTAGTCAACCGCTTTATGTGTT-3′ (the SpeI sequence is underlined). The resulting plasmid was named pCH238. A DNA fragment, in which the sequence for the PhoA mature sequence was flanked by the SpeI recognition sequences (22), was inserted into the SpeI site of pCH238. The resulting plasmid, carrying an in-frame SecY′-PhoA-′SecY fusion protein, was named pCH244.
A series of fusion proteins, denoted PhoA-TM8-Cn, consisted of exported PhoA domain followed by the TM8-C5 region of SecY with the expected number (indicated by n) of C5 residues protruding into the cytosol. Plasmids carrying them under the control of the lac promoter control were pCH309 (PhoA-TM8-C30), pCH308 (PhoA-TM8-C10), pCH307 (PhoA-TM8-C5) and pCH306 (PhoA-TM8-C−5). DNA fragments were amplified from pCH244 using an upstream primer, 5′-GGCACAGGCGCGTGGTTATCAG-3′, and the respective downstream primers, 5′-CGGGGTACCTTAGATATACTTCGCCGTTTGCT-3′, 5′-CGGGGTACCTTACAGGTTATCTGCTGTTTCAC-3′, 5′-CGGGGTACCTTATTCACGCGGGTTGAAAAC-3′, and 5′-CGGGGTACCTTAGAAGAAACAGAAGAAGAT-3′ (the KpnI sequence is underlined). They were inserted into a 3′ site of phoA on pMS102 (40) after digestions with NcoI (within the phoA region) and KpnI. Each of the XbaI-KpnI fragments of the resulting plasmids was then cloned into pTWV229. pCH312 was a control phoA plasmid and constructed by cloning the XbaI-KpnI phoA fragment from pMS102 into pTWV229. pCH317 (carrying PhoA[SSSS]-TM8-C30) was constructed by replacing a BclI fragment of pCH309 with the corresponding fragment from pTY7 (40).
pCH276 (carrying SecE′-C16), pCH293 (carrying SecE′-C6), and pCH292 (carrying SecE′-C4) were constructed as follows. DNA fragments were amplified from pCM134 (9) using an upstream primer, 5′-ACAATTTCACACAGGAAACAGCTATG-3′, and downstream primers 5′-CGGGGTACCTCAGACTTCGGTACGCGCTTCAC-3′ for pCH276, 5′-CGGGGTACCTCAAACGGTAGCTTTACCTTT-3′ for pCH293, and 5′-CGGGGTACCTCAAGCTTTACCTTTTGTCGT-3′ for pCH292 (the KpnI sequence is underlined) and were cloned into pTWV229 after digestions with HindIII and KpnI.
All the sequences for the plasmid regions that experienced in vitro replication during constructions have been confirmed or have been replaced by the original sequences.
E. coli strains and media.
Strains AR5087 (zad-220::Tn10 sfhC21) and AR5090 (ΔftsH3::kan zad-220::Tn10 sfhC21) were derivatives of JM103 (Δlac-pro endA sbcB15 hsdR thi rpsL supE/F′ lacIq lacZΔM15 [3]). Growth of AR5090 without FtsH was enabled by the sfhC21 suppressor mutation (32). Strains AD1635 (zad-220::Tn10 sfhC21 ΔdegP-1615::cm) and AD1625 (ΔftsH3::kan zad-220::Tn10 sfhC21 ΔdegP-1615::cm) were constructed as follows. A fragment including degP was amplified from the chromosomal DNA of MC4100, using PCR primers 5′-CGGGATCCAGAACACAGCAATTTTGCG-3′ (the BamHI sequence is underlined) and 5′-GGAATTCCTGCATTAACAGGTAGATG-3′ (the EcoRI sequence is underlined), and cloned into pTYE007 (6) after BamHI-EcoRI digestions, yielding pSTD539. A HindIII-HindIII segment within degP on pSTD539 was replaced by a HindIII fragment from pHP45ΩCm (11), which carries a chloramphenicol resistance determinant (Ωcm), to yield pSTD546. Finally, strain DY330 (45) was transformed with the degP::cm BamHI-EcoRI fragment prepared from pSTD546, and the resulting strain (DY330 ΔdegP-1615::cm) was named AD1615. The transformation was performed by electroporation (Bio-Rad gene pulser). Subsequently, the ΔdegP-1615::cm marker was introduced by P1 transduction into the ftsH+ and ΔftsH pair of strains (AR5087 and AR5090), and the resulting products were named AD1635 and AD1625, respectively.
Media used were L (10) and M9 (28)-amino acids (20 μg/ml each, other than cysteine and methionine). They were further supplemented with ampicillin (50 μg/ml) for growing plasmid-bearing cells. For induction of lac promoter-controlled genes, cells were first grown in the presence of glucose (0.4%) to an exponential phase, and then 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) and 5 mM cyclic AMP were added.
Pulse-chase and immunoprecipitation.
In vivo stability of the PhoA, SecE, and SecY derivatives was examined by pulse-chase and immunoprecipitation experiments, essentially as described previously (41). Sodium dodecyl sulfate (SDS)-solubilized whole-cell proteins were diluted 33-fold with 50 mM Tris-HCl (pH 8.1) containing 0.1 mM EDTA, 0.15 M NaCl, and 2% Triton X-100 for immunoprecipitation. Anti-PhoA serum was obtained from 5 Prime 3 Prime, Inc. Antiserum against a sequence of the SecY residues Asn338 to Ala351 (anti-SecYC5 [30]) was provided by H. Mori. Rabbit antiserum against the N-terminal 13 residues of SecE was prepared by custom services provided by Hokkaido System Science; chemically synthesized oligopeptide MSANTEAQGSGRGC was conjugated to keyhole limpet hemocyanin and injected into a rabbit. [35S]methionine-labeled immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by a BAS1800 phosphorimager (Fuji Film).
RESULTS
FtsH can initiate degradation of a model membrane protein at the C-terminal cytosolic region.
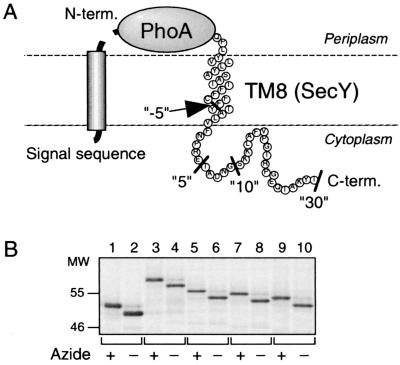
To study whether FtsH can initiate degradation of membrane proteins from the C-terminal cytoplasmic region, we constructed a model substrate protein having a cytosolically disposed C-terminal tail and a periplasmic N-terminal end. SecY has 10 transmembrane (TM1 to TM10), six cytoplasmic (C1 to C6) and five periplasmic (P1 to P5) regions, with both termini facing the cytoplasm (2). Its segment encompassing TM8 and the following 30 amino acid residues of C5 was fused to the C-terminal end of alkaline phosphatase (PhoA). The resulting model protein was named PhoA-TM8-C30 (Fig. 1A). We expected that the PhoA part of this fusion protein is translocated to the periplasm, whereas TM8 acts as a stop-transfer sequence, generating the topological disposition as shown in Fig. 1A. In this case, it is expected that the PhoA signal sequence is cleaved off by the leader peptidase. That this was indeed the case was shown by pulse-labeling PhoA-TM8-C30 in the presence and absence of NaN3, a potent inhibitor of the SecA translocation ATPase. As shown in Fig. 1B (lanes 3 and 4), the protein was produced as a higher-molecular-weight species in the presence of NaN3 than in its absence. Thus, PhoA-TM8-C30 responded to the SecA inhibitor quite similarly to the authentic PhoA protein, which was detected as a precursor form in the presence of NaN3 and as the mature form in the normal cells (Fig. 1B, lanes 1 and 2). These results confirmed the periplasmic localization of the PhoA part.
FIG. 1.
Signal sequence cleavage indicates the periplasmic localization of the N termini of the PhoA-TM8 model proteins. (A) Schematic representation of PhoA-TM8-Cn fusion proteins. The C-terminal ends for the constructs used (n = −5, 5, 10, and 30) are indicated by the black bars. (B) Demonstration of signal sequence-processed and unprocessed species. Strain AR5087 was transformed with either pCH312 (carrying PhoA; lanes 1 and 2), pCH309 (carrying PhoA-TM8-C30; lanes 3 and 4), pCH308 (carrying PhoA-TM8-C10; lanes 5 and 6), pCH307 (carrying PhoA-TM8-C5; lanes 7 and 8), or pCH306 (carrying PhoA-TM8-C−5; lanes 9 and 10). Cells were grown in M9 medium at 37°C, induced for the lac transcription for 10 min, and treated with (lanes with odd numbers) or without (lanes with even numbers) 0.02% NaN3 for 1 min. They were then pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1 min. Labeled proteins were immunoprecipitated with anti-PhoA serum, separated by SDS-PAGE, and visualized.
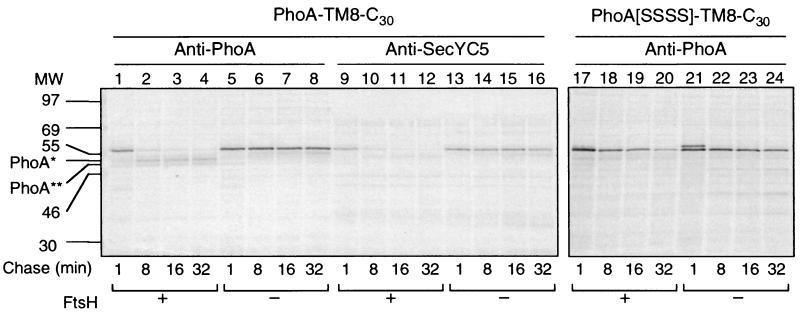
In vivo stability of PhoA-TM8-C30 was studied by pulse-chase experiments, using a pair of ftsH+ and ΔftsH strains, AR5087 and AR5090, respectively. In the ftsH+ cell, the full-length product of PhoA-TM8-C30 (∼58 kDa), which was precipitated with both antibodies against PhoA (Fig. 2, lanes 1 to 8) and SecYC5 (lanes 9 to 16), was found to be degraded during the chase. The half-life of this degradation was about 5 min (Fig. 2, lanes 1 to 4 and 9 to 12). Concomitantly with this degradation, fragments of about 50 kDa appeared (PhoA* and PhoA** in Fig. 2, lanes 1 to 4), which reacted with anti-PhoA but not with anti-SecYC5 (Fig. 2, lanes 9 to 12). These results indicated that in the wild-type cells, PhoA-TM8-C30 was converted to a form that lacked the SecY-derived C-terminal tail. This partial degradation proved to depend on FtsH, since in the ΔftsH cells the intact band remained stable without any detectable generation of the PhoA* or PhoA** products (Fig. 2, lanes 5 to 8 and 13 to 16).
FIG. 2.
Degradation profiles of PhoA-TM8-C30 (with a folded PhoA domain) and PhoA[SSSS]-TM8-C30 (with an unfolded PhoA domain). Plasmid pCH309 (PhoA-TM8-C30) was introduced into AR5087 (ftsH+; lanes 1 to 4 and 9 to 12) and AR5090 (ΔftsH; lanes 5 to 8 and 13 to 16), whereas plasmid pCH317 (PhoA[SSSS]-TM8-C30; lanes 17 to 24) was introduced into AD1635 (ftsH+ ΔdegP; lanes 17 to 20) and AD1625 (ΔftsH ΔdegP; lanes 21 to 24). Cells were grown in M9 medium at 37°C and induced for lac transcription for 10 min. They were then pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1 (lanes 1, 5, 9, 13, 17, and 21), 8 (lanes 2, 6, 10, 14, 18, and 22), 16 (lanes 3, 7, 11, 15, 19, and 23), and 32 (lanes 4, 8, 12, 16, 20, and 24) min. Labeled proteins were immunoprecipitated with anti-PhoA (lanes 1 to 8 and lanes 17 to 24) or anti-SecYC5 (lanes 9 to 16) antibodies, separated by SDS-PAGE, and visualized. PhoA∗ and PhoA∗∗ indicate degradation products produced from PhoA-TM8-C30. PhoA-TM8-C30 gave essentially the same degradation patterns as shown in lanes 1 to 4, when it was expressed in AD1635 lacking DegP.
In our previous study of degradation of the SecY-(P5)-PhoA fusion protein (without the C-terminal region of SecY), a PhoA-containing fragment with a size similar to that of PhoA** was observed (22). In this case, the degradation proceeded presumably from an N-terminal cytoplasmic region(s) and stopped before the folded PhoA domain. The degradation continued into the PhoA region when it was unfolded due to the lack of the intramolecular disulfide bonds in it. In the present experimental system, in which degradation of PhoA-TM8-C30 appeared to be initiated from the C-terminal tail, the PhoA domain may also have acted as an obstacle in the degradation process. We examined degradation of a cysteine-less variant of the fusion protein, PhoA[SSSS]-TM8-C30, in which no disulfide bond could be made. It was still degraded in an FtsH-dependent manner; its half-life was ∼6 min in ftsH+ cells and ∼30 min in ΔftsH cells. However, degradation of PhoA[SSSS]-TM8-C30 was not accompanied by the PhoA* or the PhoA** products (Fig. 2, lanes 17 to 24). Although the bacterial strains used had deletions of degP, which encodes a major periplasmic protease, some other periplasmic protease may have contributed to the slow degradation of PhoA[SSSS]-TM8-C30 in the absence of FtsH. Essentially the same results were obtained when disulfide bond formation in PhoA-TM8-C30 was suppressed by addition of 20 mM dithiothreitol (DTT) (data not shown). Thus, the unfolded PhoA part was degraded FtsH dependently. In contrast, when the periplasmic PhoA domain was folded tightly, it resisted the FtsH-mediated proteolysis that was initiated at the C-terminal cytosolic region.
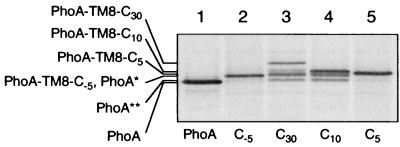
To further characterize the PhoA* and the PhoA** fragments, their apparent molecular masses were compared with those of some additional model proteins. PhoA-TM8-C−5 was one of them, and it was constructed by removing the C-terminal 35 amino acid residues from PhoA-TM8-C30. Thus, it has a deletion of not only the entire cytoplasmic tail but also the C-terminal five residues of the TM8 region. This protein also was produced as the signal sequence-processed form (Fig. 1B, lane 10), which was stable in the ftsH+ cells (Fig. 3A, lanes 1 to 4); the lack of the cytoplasmic domain renders it nonsusceptible to FtsH (see below). Its mobility in SDS-PAGE was almost identical to that of the PhoA* fragment generated from PhoA-TM8-C30 (Fig. 4, lanes 2 and 3). Both PhoA* and PhoA** showed appreciably lower electrophoretic mobilities than the authentic PhoA mature protein (Fig. 4, lane 1). These data suggest that the FtsH-mediated proteolysis of PhoA-TM8-C30 removed not only the entire cytoplasmic region but also a few residues of the TM8 segment. FtsH may have tried to dislocate the transmembrane and periplasmic region of the substrate into the cytoplasm but did so only with a limited success due to the tight folding of the PhoA domain.
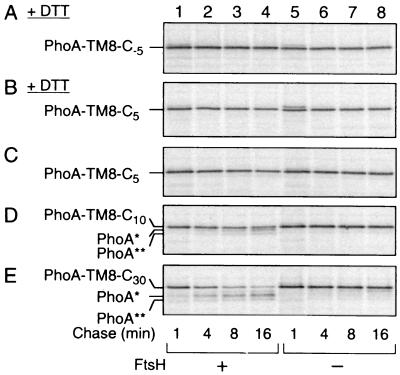
FIG. 3.
FtsH susceptibility of PhoA-TM8-Cn fusion proteins with different lengths of the cytoplasmic tail. (A and B) Resistance of PhoA-TM8-C−5 and PhoA-TM8-C5 in the absence of PhoA folding. AD1635 (ftsH+ ΔdegP; lanes 1 to 4) and AD1625 (ΔftsH ΔdegP; lanes 5 to 8) were transformed with pCH306 (PhoA-TM8-C−5) (A) or pCH306 (PhoA-TM8-C5) (B). Cells were induced for the lac transcription for 10 min and treated with 20 mM DTT for 2 min. They were then pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1, 4, 8, and 16 min, as indicated. (C to E) Degradation of PhoA-TM8 derivatives in the presence of PhoA folding. AR5087 (ftsH+; lanes 1 to 4) and AR5090 (ΔftsH; lanes 5 to 8) were transformed with pCH307 (carrying PhoA-TM8-C5) (C), pCH308 (carrying PhoA-TM8-C10) (D), or pCH309 (carrying PhoA-TM8-C30) (E). Cells were induced for the lac transcription for 10 min and pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1, 4, 8, and 16 min, as indicated. Labeled proteins (all panels) were immunoprecipitated with anti-PhoA, separated by SDS-PAGE, and visualized.
FIG. 4.
Degradation products produced from PhoA-TM8-Cn proteins. Strain AR5087 (ftsH+) was transformed with either pCH312 (carrying PhoA; lane 1), pCH306 (carrying PhoA-TM8-C−5; lane 2), pCH309 (carrying PhoA-TM8-C30; lane 3), pCH308 (carrying PhoA-TM8-C10; lane 4), or pCH307 (carrying PhoA-TM8-C5; lane 5). Cells were induced for the lac transcription and pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 8 min. Labeled proteins were immunoprecipitated with anti-PhoA, separated by SDS-PAGE, and visualized. PhoA∗ indicates a degradation product consisting of the PhoA domain and some TM8 residues, whereas PhoA∗∗ indicates a degradation product containing fewer TM8 residues.
From all of these results, we conclude that FtsH can initiate proteolysis of PhoA-TM8-C30 from the C-terminal tail. In wild-type cells, in which PhoA is folded normally, the proteolysis stops after it goes into a few residues of the TM segment. The degradation of the unfolded PhoA domain could have represented processive proteolysis by FtsH, with active dislocation of the periplasmic region to the cytosol (22). However, a formal possibility remains that FtsH only degrades the cytoplasmic tail and that some other protease is activated upon elimination of the tail. In this context, PhoA-TM8-C−5, which lacks the C-terminal tail, could be regarded as a substrate of the other protease discussed above. We examined stability of PhoA-TM8-C−5 in the presence of 20 mM DTT. It was found that PhoA-TM8-C−5 was completely stable (Fig. 3A). Similar results were observed with PhoA-TM8-C5, which has five residues in the putative cytosolic tail (Fig. 3B). These results disfavor the possibility that FtsH and some unknown enzymes act successively for the degradation of PhoA-TM8-C30.
Features of the C-terminal cytoplasmic tail required for the degradation initiation.
Given the fact that the enzymatic active sites of FtsH reside cytosolically, it is likely that FtsH recognizes the cytosolically exposed C-terminal tail of PhoA-TM8-C30 for initiating its degradation. In the N-terminal initiation mode studied previously, the N-terminal tail of 20 residues or more was required (9). In addition to PhoA-TM8-C−5 and PhoA-TM8-C5, we constructed PhoA-TM8-C10 by deleting the C-terminal 20 resides from PhoA-TM8-C30. These proteins were all produced as signal sequence-cleaved mature products (Fig. 1B, lanes 5 to 10). In the pulse-chase experiments reported in Fig. 3C, D, and E, the host cells used were degP+. PhoA-TM8-C30 exhibited degradation profiles that were essentially identical to those reported in Fig. 2. It was FtsH-dependently converted to the PhoA* and PhoA** states with a half-life of ∼5 min. PhoA-TM8-C10 was also degraded in the ftsH+ cells (Fig. 3D, lanes 1 to 4), but not in the ΔftsH cells (Fig. 3D, lanes 5 to 8). The degradation was slower (half-life, about 12 min) than that of PhoA-TM8-C30 and was accompanied by the appearance of smaller bands with mobility identical to those of PhoA* and PhoA** (Fig. 4, compare lanes 3 and 4).
In contrast to the above proteins having a putative cytosolic tail of 10 or more residues, both PhoA-TM8-C5 and PhoA-TM8-C−5 were stable in the ftsH+ cells. This was true even in the presence of DTT, provided that the cells were deficient in DegP (Fig. 3A, B, and C). The electrophoretic mobility of PhoA-TM8-C5 was slightly lower than that of PhoA* or PhoA-TM8-C−5, suggesting that no proteolytic reaction had been initiated for this protein. This is consistent with the results that this protein resisted FtsH even when its PhoA part was kept unfolded by DTT (Fig. 3B). These results suggest that the C-terminal tail has an essential role in the initiation of degradation of PhoA-TM8-C10 and PhoA-TM8-C30 proteins. The fact that the stability of the extracellular domain can be affected by manipulating the cytoplasmic region may be best explained by the processive degradation that is initiated at the C-terminal tail and involving a dislocation process, as has been proposed for the N-terminally initiated degradation (9).
C-terminal-region-dependent degradation of SecE derivatives.
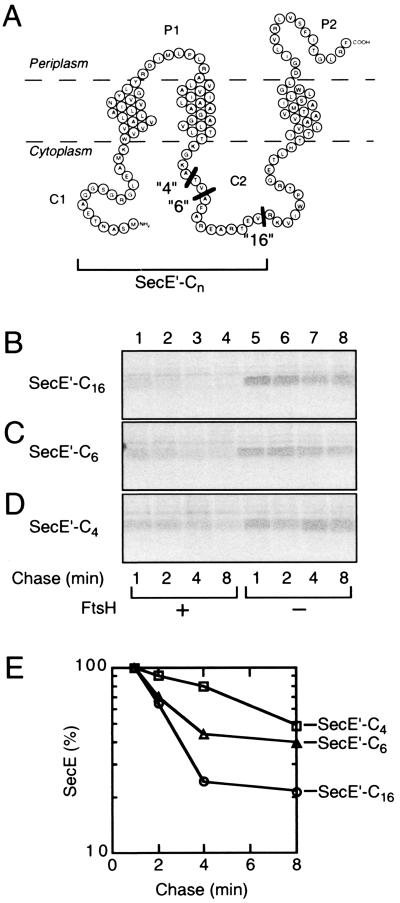
SecE contains three transmembrane segments, N-terminal and central cytoplasmic regions, and a C-terminal periplasmic tail (37). This membrane protein, which is normally stable, was sensitized to FtsH when the N-terminal tail was extended (9). Here, we constructed C-terminally truncated SecE derivatives, having the truncation points within the central cytoplasmic region. The resulting constructs were named SecE′-C16, SecE′-C6, and SecE′-C4, having expected C-terminal cytoplasmic tails of 16, 6, and 4 residues, respectively (Fig. 5A). Their intracellular stability, in the pair of ftsH+ and ΔftsH cells, was examined by pulse-chase experiments. While all of them were stable in the ΔftsH cells (Fig. 5, lanes 5 to 8), SecE′-C16 was degraded in the ftsH+ cells quite rapidly (Fig. 5B, lanes 1 to 4; Fig. 5E). SecE′-C6 was also degraded FtsH-dependently but less rapidly than SecE′-C16 (Fig. 5C, lanes 1 to 4; Fig. 5E). SecE′-C4 was much more stable than the other SecE derivatives (Fig. 5D, lanes 1 to 4; Fig. 5E). These results show that the C-terminal region can determine the FtsH susceptibility of the N-terminally adjacent SecE region. A correlation was observed between the length of the C-terminal tail and the efficiency of the degradation.
FIG. 5.
FtsH susceptibility of SecE derivatives having a C-terminal cytosolic tail due to a truncation at the central cytosolic domain. (A) Schematic representation of the SecE derivatives. The truncation points are indicated by black bars. (B to D) Pulse-chase experiments. Strains AR5087 (ftsH+; lanes 1 to 4) and AR5090 (ΔftsH; lanes 5 to 8) were transformed with either pCH276 (SecE′-C16) (B), pCH293 (SecE′-C6) (C), or pCH292 (SecE′-C4) (D). Cells were induced for lac transcription and pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1, 2, 4, and 8 min, as indicated. Labeled proteins were immunoprecipitated with anti-SecE N1 serum, separated by SDS-PAGE, and visualized. (E) Graphical representations of the degradation kinetics in the ftsH+ cells. Radioactivities associated with SecE′-C16 (circles) (from panel B, lanes 1 to 4), SecE′-C6 (triangles; from panel C) (lanes 1 to 4) and SecE′-C4 (squares) (from panel D, lanes 1 to 4) are plotted as values relative to that at the 1-min chase point for each protein.
Taken together with our previous results with the SecE derivatives with extended N termini (9), it can be stated that the degradation of the same SecE segment can be initiated at either side of it, depending on the length of the cytoplasmic tails. Our results also indicate that different amino acid sequences, the SecY C5 sequence in the case of PhoA-TM8 constructs and the SecE C2 sequence in the case of SecE′ constructs, of the C-terminal tail can confer FtsH susceptibility.
Multiple initiation of bidirectional degradation.
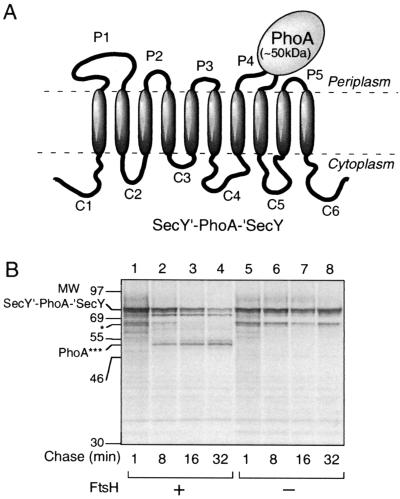
The results so far presented suggest that FtsH can initiate degradation of a membrane protein either from the N-terminal tail or from the C-terminal tail. However, it has not been shown whether a particular substrate protein can receive both modes of degradation simultaneously. Both the N- and C-terminal tails of SecY are cytoplasmic and longer than 20 amino acids. Thus, degradation of SecY could be initiated at both termini. In addition, degradation initiation at some of the internal loops is conceivable. To test whether multiple initiations indeed occur for SecY degradation, we constructed a SecY derivative (SecY′-PhoA-′SecY) in which PhoA had been sandwich fused within the P4 region. Pulse-chase experiments showed that SecY′-PhoA-′SecY was degraded FtsH dependently (Fig. 6, lanes 1 to 4). This degradation was accompanied by the production of an ∼50-kDa fragment, which reacted with anti-PhoA antibodies (Fig. 6, lanes 1 to 4; PhoA***). Its mobility was similar to that of the PhoA* fragment observed with the PhoA-TM8-C30 and the PhoA-TM8-C10 proteins (Fig. 4).
FIG. 6.
FtsH-dependent degradation of SecY′-PhoA-′SecY yields a PhoA∗-like fragment. (A) Schematic representation of SecY′-PhoA-′SecY. PhoA was sandwich fused into the P4 region of SecY. (B) Degradation of SecY′-PhoA-′SecY. Plasmid pCH244 (carrying SecY′-PhoA-′SecY) was introduced into strains AR5087 (ftsH+; lanes 1 to 4) and AR5090 (ΔftsH; lanes 5 to 8). Cells were induced for lac transcription and pulse-labeled with [35S]methionine for 1 min, which was followed by a chase with unlabeled methionine for 1, 8, 16, and 32 min, as indicated. Labeled proteins were immunoprecipitated with anti-PhoA, separated by SDS-PAGE, and visualized. PhoA∗∗∗ indicates a product, composed mostly of PhoA, generated by the bidirectional degradation by FtsH. The band indicated by a single asterisk might have been a product of internal translation initiation within secY (22).
Thus, both the N-terminal and the C-terminal sides of PhoA in the SecY′-PhoA-′SecY fusion protein were removed by the FtsH-dependent degradation. This should be contrasted with another PhoA sandwich fusion protein, YccA-(P3)-PhoA-His6-Myc, whose C-terminal tail was oriented periplasmically. In this case, only the N-terminal side of PhoA was degraded (provided that the PhoA moiety was folded normally [22]). These results are consistent with a notion that processive degradation of membrane proteins can proceed from either end depending on the locations of the initiation region. Presumably, this degradation involves a dislocation process, which is incompatible with a tightly folded periplasmic domain.
DISCUSSION
The FtsH-mediated membrane protein degradation is a processive process. To enable the degradation over the topological barrier of the membrane, FtsH may actively dislocate the substrate protein (22). Similar modes of degradation were also proposed for the proteolysis mediated by the Saccharomyces cerevisiae mitochondrial FtsH homologs, i-AAA and m-AAA proteases (25). To understand the molecular mechanisms of these enzymes, it is important to elucidate how they initiate the reaction and what directions they move along or how they translocate the substrate polypeptide chain. The issue of directionality is a crucial point in understanding energy-dependent proteolytic reactions.
FtsH is unique in that it degrades both membrane-integrated and soluble proteins. This poses interesting questions as to the mechanisms by which it recognizes membrane proteins for degradation initiation. On the membrane, the movement of both the enzyme and the substrate will be restricted to the lateral dimension, at least until they encounter each other. In the subsequent reactions, they will interact properly and this interaction will somehow lead to more dynamic events, in which vertical dislocation movement of the substrate as well as processive proteolytic reactions take place with concomitant ATP hydrolysis cycles. Thus, it is imaginable that a specialized mechanism exists for the recognition of membrane-integrated substrate molecules.
We showed that degradation initiation of certain membrane proteins requires the presence of an N-terminal tail of 20 amino acid residues or more, whose length but not exact sequences is important (9). In contrast, Herman et al. showed that FtsH can recognize the SsrA tag attached to the C-terminal end of a soluble protein to initiate its degradation (16). Since soluble protein substrates and membrane-embedded substrates of FtsH seem to be presented to this enzyme via different pathways (20, 21), it is important to examine the similarity and the difference between the soluble pathway and the membrane pathway in the substrate recognition and the directionality of proteolysis.
Our present results indicate that FtsH-dependent processive degradation of membrane proteins can be initiated not only at an N-terminal tail but also at a C-terminal cytosolic region. In the N-terminal initiation of proteolysis studied previously, a crucial feature that determines the FtsH susceptibility was the length of the cytosolic tail. Our systematic studies showed that the threshold length is about 20 residues and that several different sequences can confer the FtsH sensitivity (9). Thus, it is likely that a certain extent of protrusion into the cytosol is required for FtsH to initiate degradation as well as dislocation of the substrate. In the present examinations, at least two different C-terminal sequences, those in the SecY C5 and SecE C2 regions, functioned as degradation initiation signals. Experiments using a different series of PhoA-SecY fusion protein indicated that the C4 sequence of SecY is also recognized by FtsH for degradation initiation (S. Chiba, Y. Akiyama, and K. Ito, unpublished data). Thus, the C-terminal initiation can occur with a large variety of amino acid sequences. However, this does not mean that any sequence can confer the sensitivity. We speculate that the cytosolic protrusion should not be folded tightly or does not interact strongly with other components.
Like the N-terminal initiation studied previously (9), the C-terminal initiation requires a certain length of cytosolic tail. However, the apparent length required was about 10 residues, whereas a region of 5 residues was too short. These numbers also hold for the SecY C4 sequence-dependent degradation (Chiba et al., unpublished data). The apparently different length recognition between the N-terminal and the C-terminal modes of initiation raises several questions about substrate recognition by FtsH. Is the same substrate recognition site utilized for the two modes of initiation? If this is the case, does the local substrate or enzyme orientation differ between the two modes? On the other hand, in view of the intrinsic polarity of the polypeptide molecule, there may be a fundamental mechanistic difference between the N-terminal tail recognition and the C-terminal tail recognition. It should also be pointed out, however, that without structural information it is difficult to say definitively whether the difference we noted is real or only apparent. For instance, it is possible that there is some deviation in the assignment of the transmembrane regions, from the real structure, and such deviation could differ systematically between an export signal and a stop transfer element.
The bidirectionality in the degradation process does not imply that the catalytic mechanism of peptide bond hydrolysis is also bidirectional. FtsH may be a processive endopeptidase, since its degradation products are not amino acid monomers but oligopeptides of several residues (7, 39). Thus, within the putative catalytic chamber of FtsH, the substrate polypeptide could have some degree of flexibility such that the peptide bond to be hydrolyzed is always presented to the active site with a fixed directionality. To accomplish this, FtsH might actively redirect the polypeptide using its unfolding activity. The oligomeric structure might enable this kind of mechanism.
Although membrane-embedded and soluble proteins may be recognized differently by the FtsH-HflKC complex (20), there may be no fundamental difference in the two modes of proteolytic reactions with respect to the overall directionality. Thus, we predict that some soluble protein substrates may be recognized via their N-terminal regions by FtsH. It is already known that the Clp proteases recognize either the N- or C-terminal tail of the substrate for the proteolytic reaction to be initiated (12, 14, 17). Molecular understanding of the bidirectional processivity exhibited by the energy-dependent proteases is an important problem that remains for future studies.
Given that FtsH is an endopeptidase, it could initiate proteolysis not only at a tail but also at an internal region of a polypeptide. However, the C2 region of SecE is only recognized by FtsH as a degradation substrate after genetic truncation to produce the cytoplasmic end. Thus, tails rather than loops may be the preferred initiation sites. However, we experienced some cases where an extended cytosolic loop sensitizes the protein to FtsH (S. Chiba et al., unpublished data), suggesting that degradation can indeed be initiated at a loop.
We have shown that FtsH-dependent degradation of SecY′-PhoA-′SecY occurs at least from two regions, one N-terminal to and the other C-terminal to the PhoA domain. The PhoA-like end product indicates that the former proceeds in the N-to-C direction and the latter proceeds in the C-to-N direction. Multiple initiation of processive proteolysis may be beneficial for FtsH, since this mechanism will enable it to accomplish the very rapid degradation of unwanted membrane proteins.
Acknowledgments
We thank K. Mochizuki, M. Yamada, and M. Sano for technical support and H. Mori, N. Saikawa, K. Kanehara, N. Shimohata, and H. Nakatogawa for discussion.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from CREST, Japan Science and Technology Corporation.
REFERENCES
- 1.Adam, Z. 2000. Chloroplast protease: possible regulators of gene expression? Biochimie 82:647-654. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y., and K. Ito. 1987. Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J. 6:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama, Y., and K. Ito. 2000. Roles of multimerization and membrane association in the proteolytic functions of FtsH (HflB). EMBO J. 19:3888-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akiyama, Y., A. Kihara, and K. Ito. 1996. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 399:26-28. [DOI] [PubMed] [Google Scholar]
- 5.Akiyama, Y., A. Kihara, H. Tokuda, and K. Ito. 1996. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271:31196-31201. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama, Y., T. Yoshihisa, and K. Ito. 1995. FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 270:23485-43490. [DOI] [PubMed] [Google Scholar]
- 7.Asahara, Y., K. Atsuta, K. Motohashi, H. Taguchi, M. Yohda, and M. Yoshida. 2000. FtsH recognizes proteins with unfolded structure and hydrolyzes the carboxyl side of hydrophobic residues. J. Biochem. 127:931-937. [DOI] [PubMed] [Google Scholar]
- 8.Bochtler, M., L. Ditzel, M. Groll, and R. Huber. 1997. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA 94:6070-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba, S., Y. Akiyama, H. Mori, E. Matsuo, and K. Ito. 2000. Length recognition at the N-terminal tail of membrane proteins as a mode of initiation of FtsH-mediated proteolysis. EMBO Rep. 1:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 11.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 12.Gonciarz-Swiatek, M., A. Wawrzynow, S. J. Um, B. A. Learn, R. McMacken, W. L. Kelley, C. Georgopoulos, O. Sliekers, and M. Zylicz. 1999. Recognition, targeting, and hydrolysis of the λ O replication protein by the ClpP/ClpX protease. J. Biol. Chem. 274:13999-14005. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S. 1999. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 2:142-147. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, S., E. Roche, Y.-N. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groll, M., L. Ditzel, J. Löwe, D. Stock, M. Bochtler, H. D. Bartunik, and R. Huber. 1997. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386:463-471. [DOI] [PubMed] [Google Scholar]
- 16.Herman, C., D. Thévenet, P. Bouloc, G. C. Walker, and R. D'Ari. 1998. Degradation of carboxy-terminal-tagged cytoplasmic protein by the Escherichia coli protease HflB (FtsH). Genes Dev. 12:1348-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoskins, J. R., S. Y. Kim, and S. Wickner. 2000. Substrate recognition by the ClpA chaperone component of ClpAP protease. J. Biol. Chem. 275:35361-35367. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, T., F. Beuron, M. Kessel, S. Wickner, R. Maurizi, and A. C. Steven. 2001. Translocation pathway of protein substrates in ClpAP protease. Proc. Natl. Acad. Sci. USA 98:4328-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kihara, A., Y. Akiyama, and K. Ito. 1995. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. USA 92:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kihara, A., Y. Akiyama, and K. Ito. 1996. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15:6122-6131. [PMC free article] [PubMed] [Google Scholar]
- 21.Kihara, A., Y. Akiyama, and K. Ito. 1998. Differential pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol. 279:175-188. [DOI] [PubMed] [Google Scholar]
- 22.Kihara, A., Y. Akiyama, and K. Ito. 1999. Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18:2970-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langer, T. 2000. AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25:247-251. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C., M. P. Schwartz, S. Prakash, M. Iwakura, and A. Matouschek. 2001. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7:627-637. [DOI] [PubMed] [Google Scholar]
- 25.Leonhard, K., B. Guiard, G. Pellecchia, A. Tzagoloff, W. Neupert, and T. Langer. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 26.Löwe, J., D. Stock, B. Jap, P. Zwickl, W. Baumeister, and R. Huber. 1995. Crystal structure of the 20S proteasome from the archaeon T. acidophilum. Science 268:533-539. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo, E., H. Mori, T. Shimoike, and K. Ito. 1998. Syd, a SecY-interacting protein, excludes SecA from the SecYE complex with an altered SecY24 subunit. J. Biol. Chem. 273:18835-18840. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 30.Nishiyama, K., Y. Kaburayama, J. Akimaru, S. Matsuyama, H. Tokuda, and S. Mizushima. 1991. SecY is an indispensable component of the protein secretory machinery of Escherichia coli. Biochim. Biophys. Acta 1065:89-97. [DOI] [PubMed] [Google Scholar]
- 31.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPase: common structure-diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 32.Ogura, T., K. Inoue, T. Tatsuta, T. Suzaki, K. Karata, K. Young, L.-H. Su, C. A. Fierke, J. E. Jackman, C. R. H. Raetz, J. Coleman, T. Tomoyasu, and H. Matsuzawa. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31:833-844. [DOI] [PubMed] [Google Scholar]
- 33.Ortega, J., S. K. Singh, T. Ishikawa, M. R. Maurizi, and A. C. Steven. 2000. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell 6:1515-1521. [DOI] [PubMed] [Google Scholar]
- 34.Patel, S., and M. Latterich. 1998. The AAA term: related ATPases with diverse functions. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 35.Reid, B. G., W. A. Fenton, A. L. Horwich, and E. U. Weber-Ban. 2001. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl. Acad. Sci. USA 98:3768-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawano, A., and A. Miyawaki. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz, P. J., P. D. Riggs, A. Jacq, M. J. Fath, and J. Beckwith. 1989. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 3:1035-1044. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, M., A. N. Lupas, and D. Finley. 1999. Structure and mechanism of ATP-dependent protease. Curr. Opin. Chem. Biol. 3:584-591. [DOI] [PubMed] [Google Scholar]
- 39.Shotland, Y., A. Shifrin, T. Ziv, D. Teff, S. Koby, O. Kobiler, and A. B. Oppenheim. 2000. Proteolysis of bacteriophage λ CII by Escherichia coli FtsH (HflB). J. Bacteriol. 182:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sone, M., Y. Akiyama, and K. Ito. 1998. Additions and corrections to differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J. Biol. Chem. 273:27756. [DOI] [PubMed] [Google Scholar]
- 41.Taura, T., T. Baba, Y. Akiyama, and K. Ito. 1993. Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J. Bacteriol. 175:7771-7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomoyasu, T., K. Yamanaka, K. Murata, T. Suzaki, P. Bouloc, A. Kato, H. Niki, S. Hiraga, and T. Ogura. 1993. Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175:1352-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vale, R. D. 2000. AAA proteins: lords of ring. J. Cell Biol. 150:F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 45.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]