Abstract
The Bacillus subtilis antiterminator LicT regulates the expression of bglPH and bglS, which encode the enzymes for the metabolism of aryl-β-glucosides and the β-glucanase BglS. The N-terminal domain of LicT (first 55 amino acids) prevents the formation of ρ-independent terminators on the respective transcripts by binding to target sites overlapping these terminators. Proteins of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) regulate the antitermination activity of LicT by phosphorylating histidines in its two PTS regulation domains (PRDs). Phosphorylation at His-100 in PRD-1 requires the PTS proteins enzyme I and HPr and the phosphorylated permease BglP and inactivates LicT. During transport and phosphorylation of aryl-β-glucosides, BglP is dephosphorylated, which renders LicT active and thus leads to bglPH and bglS induction. In contrast, phosphorylation at His-207 and/or His-269 in PRD-2, which requires only enzyme I and HPr, is absolutely necessary for LicT activity and bglPH and bglS expression. We isolated spontaneous licT mutants expressing bglPH even when enzyme I and HPr were absent (as indicated by the designation “Pia” [PTS-independent antitermination]). Introduced in a ptsHI+ strain, two classes of licT(Pia) mutations could be distinguished. Mutants synthesizing LicT(Pia) antiterminators altered in PRD-2 still required induction by aryl-β-glucosides, whereas mutations affecting PRD-1 caused constitutive bglPH expression. One of the two carbon catabolite repression (CCR) mechanisms operative for bglPH requires the ρ-independent terminator and is probably prevented when LicT is activated by P∼His-HPr-dependent phosphorylation in PRD-2 (where the prefix “P∼” stands for “phospho”). During CCR, the small amount of P∼His-HPr present in cells growing on repressing PTS sugars probably leads to insufficient phosphorylation at PRD-2 of LicT and therefore to reduced bglPH expression. In agreement with this concept, mutants synthesizing a P∼His-HPr-independent LicT(Pia) had lost LicT-modulated CCR.
Bacillus subtilis LicT is an antiterminator of the BglG/SacY family regulating the expression of bglS, which forms an operon together with licT, and of the bglPH operon (16, 25). bglP codes for an aryl-β-glucoside-specific enzyme, IIBCA; bglH codes for a 6-P-β-glucosidase (16); and bglS codes for an extracellular β-glucanase (also called LicS) (21). LicT is composed of an N-terminal RNA binding domain (4) and two regulatory domains controlled by proteins of the phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS). These two domains were therefore called PTS regulation domains (PRDs). The PTS proteins form a phosphorylation cascade not only catalyzing the uptake and concomitant PEP-dependent phosphorylation of carbohydrates but also regulating numerous metabolic functions. The PTS is composed of the general proteins enzyme I and HPr and the sugar-specific enzyme II complexes (for a review, see reference 23).
The crystal structure of the PRDs of LicT, which exhibit significant similarity to each other, has recently been resolved (34). Each PRD contains two conserved histidyl residues (100 and 159; 207 and 269), which are all phosphorylated by PEP, enzyme I, and HPr (18, 33). BglP, the aryl-β-glucoside-specific enzyme II, participates in bglPH (16) and probably also bglS induction. P∼BglP (where the prefix “P∼” stands for “phospho”) is thought either to directly phosphorylate LicT at His-100 or to stimulate the P∼His-HPr-mediated phosphorylation at this site (33). Replacing His-100 with a nonphosphorylatable alanine causes constitutive bglPH expression, suggesting that phosphorylation at this position leads to inactivation of LicT. One model proposes that P∼BglP might also interact with LicT phosphorylated at His-100 (33) and thus prevent the binding of LicT to its targets, the ribonucleic antitermination (RAT) sites (1) on the nascent bglPH and licT-bglS transcripts. If functional LicT is not available, terminator structures are formed on the mRNAs, leading to the synthesis of short transcripts, which in the case of bglPH are composed of only about 110 nucleotides (14). In the presence of an aryl-β-glucoside, BglP is probably primarily present in its unphosphorylated form, since the phosphoryl group accepted from P∼His-HPr is rapidly transferred to its sugar substrate. In the absence of P∼BglP, LicT will not be phosphorylated at His-100. As a consequence, the presence of aryl-β-glucosides allows the synthesis of full-length bglPH and bglS transcripts, as under these conditions LicT can bind to the RAT sites and thus prevent the formation of the terminator partially overlapping the RAT sequences. However, to be able to bind to its RAT sites, LicT needs first to be phosphorylated by P∼His-HPr at His-207 and/or His-269 in PRD-2. When this phosphorylation was prevented by either inactivating enzyme I (18) or HPr (14) or by replacing His-207 and/or His-269 by alanines (33), the corresponding mutants were devoid of LicT antitermination activity.
Diminished phosphorylation in PRD-2 during the uptake of a rapidly metabolizable carbohydrate such as glucose has been proposed to play a role in one of the two carbon catabolite repression (CCR) mechanisms controlling the bglPH operon (14). bglPH expression is regulated by catabolite control protein A (CcpA)-mediated CCR, which is operative in most gram-positive bacteria (6, 30). CcpA is a member of the LacI/GalR repressor family (11). It needs to form a complex with its corepressor seryl-46-phosphorylated HPr (P-Ser-HPr) (7, 10) to be able to bind to most of its DNA targets, the catabolite response elements (cre) (22) located in front or at the beginning of catabolite-repressed transcription units. The bifunctional HPr kinase/phosphatase (HprK/P)—the sensor enzyme of CCR responding to changes in the concentration of ATP, glycolytic intermediates, and inorganic phosphate—catalyzes the ATP-dependent phosphorylation of HPr at Ser-46 (8) as well as the dephosphorylation of P-Ser-HPr (13) and thus indirectly regulates CcpA binding to its DNA targets. However, B. subtilis ccpA mutants (15) or strains in which the bglPH cre was deleted (14) still exhibited four- to sevenfold repression of the bglPH operon by glucose. This residual CCR disappeared in mutants in which the terminator sequence preceding bglPH had been deleted (14, 15), suggesting that this second CCR might be mediated by inactivation of LicT. The CcpA-independent CCR also disappeared in ptsH1 mutants, which synthesize S46A mutant HPr and which are therefore unable to form P-Ser-HPr (9). P-Ser-HPr is a poor substrate for the PEP-dependent phosphorylation by enzyme I (5). The uptake of rapidly metabolizable PTS sugars by B. subtilis wild-type cells leads to high concentrations of P-Ser-HPr and therefore to low amounts of P∼His-HPr (20), the phosphoryl donor for LicT and the enzyme IIAs. As LicT is less efficiently phosphorylated than the enzyme IIAs (18), it will be present primarily in its unphosphorylated, inactive form when a PTS sugar is transported and phosphorylated, which leads to reduced expression of the bglPH operon. The inability of ptsH1 mutants to produce P-Ser-HPr was assumed to allow these strains to form sufficient P∼His-HPr to effectively phosphorylate LicT even when a rapidly metabolizable PTS sugar is taken up, thus preventing CcpA-independent CCR.
Two classes of PRD-containing antiterminators exist in bacteria. Antiterminators of the first class, including LicT, are controlled by the PTS in an antagonistic manner: negatively via the respective substrate-specific enzyme II of the PTS at PRD-1 and positively via the general PTS protein HPr at PRD-2. In contrast, antiterminators of the second class, including SacY and GlcT of B. subtilis, are only negatively controlled by the PTS (for reviews, see references 6 and 29). They are functional without P∼His-HPr-mediated phosphorylation in their PRD-2 and are therefore naturally PTS independent (2, 31). As a consequence, SacY and GlcT do not exhibit antitermination-mediated CCR, and interestingly, the transcription units controlled by SacY and GlcT, sacB and ptsGHI, are also not subject to CCR via CcpA. In order to better understand the molecular details of the stimulating effect caused by phosphorylation at PRD-2 in LicT, we attempted to obtain P∼His-HPr-independent mutant LicT antiterminators resembling the naturally PTS-independent antiterminators SacY and GlcT. By setting up an appropriate screening system, we were indeed able to isolate several licT(Pia) mutants (“Pia” stands for “PTS-independent antitermination”). The effect of these mutations on LicT activity, antitermination-mediated CCR, and inducibility will be discussed in light of the recently determined crystal structure of the PRDs of LicT (34).
MATERIALS AND METHODS
Strains, growth conditions, and transformation procedures.
The B. subtilis strains used in this study are listed in Table 1. They were grown in Luria-Bertani (LB) medium or in minimal medium of the following composition: 50 mM Tris-HCl buffer (pH 7.5), 15 mM (NH4)2SO4, 8 mM MgSO4, 27 mM KCl, 7 mM Na3-citrate, 0.4 mM KH2PO4, 2 mM CaCl2, 1 mM FeSO4, 10 mM MnSO4, 4.5 mM K-glutamate, and 0.1% ribose. To obtain solid media, 1.5% agarose was added to LB or minimal medium. To transform Escherichia coli NM522 and the various B. subtilis strains, we followed previously described procedures (12, 24). To select transformants, the following antibiotics were added to the growth media at the indicated concentrations as appropriate: chloramphenicol, 5 μg/ml; kanamycin, 10 μg/ml; neomycin, 20 μg/ml; spectinomycin, 100 μg/ml; and phleomycin, 0.2 μg/ml (for B. subtilis) and ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; spectinomycin, 150 μg/ml; and kanamycin, 25 μg/ml (for E. coli). When indicated, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was present at a final concentration of 40 μg/ml.
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Genotype or structurec | Source, reference, or construction |
|---|---|---|
| Strains | ||
| BGC5 | trpC2 lys-3 bglS::cat′-erm | pCm::Era→BGW7 (27) |
| BGC9 | GM1133 with ΔlicT-bglS::erm | pSK3→GM1133 |
| BGC12 | GM1210 with amyE::(bglP′-lacZ cat) | pSL3→GM1210 |
| BGC13 | BGC12 with bglPH′-aphA3 phl | pCL18→BGC12 |
| BGC14 | BGC13 with ΔptsGHI::tet | GM1220→BGC13 |
| BGC14-pia1 | BGC14 with licT(Pia) (Phe211Val) | This study |
| BGC14-pia2 | BGC14 with licT(Pia) (Phe211Val) | This study |
| BGC14-pia3 | BGC14 with licT(Pia) (Arg124Trp) | This study |
| BGC14-pia4 | BGC14 with licT(Pia) (Asp 99Asn) | This study |
| BGC14-pia5 | BGC14 with licT(Pia) (Phe211Cys) | This study |
| BGC14-pia6 | BGC14 with licT(Pia) (Arg124Leu) | This study |
| BGC14-pia7 | BGC14 with licT(Pia) (Arg124Trp) | This study |
| BGC14-pia8 | BGC14 with licT(Pia) (Arg124Leu) | This study |
| BGC14-pia9 | BGC14 with licT(Pia) (Arg124Trp) | This study |
| BGC14-piaXb | BGC14-piaX with amyE::(bglP′-lacZ Δcat) | This study |
| BGC50 | BGC12 with amyE::(bglP′-lacZ Δcat) | This study |
| BGC51 | BGC50 with ΔlicT-bglS::erm | BGC9→BGC50 |
| BGC54-piaX | BGC41-piaX with bglS::cat | BGW7→BGC41-piaX |
| BGC72 | BGC50 with bglS::cat | BGW7→BGC51 |
| BGC72-piaX | BGC50 with licT(PiaX) bglS::cat | BGC54-piaX→BGC51 |
| BGC88 | BGC72 with ccpA::Tn917spc | GM1225→BGC72 |
| BGC88-piaX | BGC72-piaX with ccpA::Tn917spc | GM1225→BGC72-piaX |
| BGW7 | trpC2 lys-3 bglS::cat | 25 |
| GM1133 | sacXYΔ3 sacBΔ23 sacTΔ4 amyE::(sacB′-′lacZ phl) bglPΔ3 | 16 |
| GM1210 | ΔlacA sacXYΔ3 sacTΔ23 sacTΔ4 | Laboratory stock |
| GM1220 | GM1210 with ΔptsGHI::tet | Laboratory stock |
| GM1225 | trpC2 pheA1 Δ(bgaX) amyE::(gntRK′-lacZ phl) ccpA::Tn917 spc | 9 |
| Plasmids | ||
| pBGW3 | Ampr Tetr pBR322 derivative containing licT-bglS | 25 |
| pCL18 | Spcr containing the bglPH′-aphA3 phl construction | This study |
| pCL46 | E. coli: Ampr; B. subtilis: Ermr with the temperature-sensitive origin of pE194, containing the amyE::(bglP′-lacZ) fusion of pSL3, but cat is deleted | This study |
| pIC22 | pUC18 derivative with Phlr of pUB110 | 27 |
| pIC239 | Ampr Camr promoterless aphA3 | M. Steinmetz |
| pIC254 | E. coli: Ampr; B. subtilis: Ermr with the temperature-sensitive origin of pE194 containing an 11-kb B. subtilis sequence including bglPH | 26 |
| pIC408 | Spcr ori pUC multiple cloning site of pBluescript II SK(+) | M. Steinmetz |
| pSK3 | Ampr Tetr, pBGW3 with licT-bglS::erm | 15 |
| pSL3 | AmpramyE::(bglP′-lacZ) cat | 15 |
Arrows indicate construction by transformation.
“piaX” stands for pia1, pia4, pia5, pia6, and pia9.
Ampr, Tetr, Spcr, and Ermr, resistance against ampicillin, tetracycline, spectinomycin, and erythromycin, respectively.
β-Galactosidase assays.
To carry out β-galactosidase tests, B. subtilis cells were grown in 0.1% ribose-containing minimal medium. When indicated, 0.05% salicin was added to induce bglPH expression, whereas 0.2% glucose was added to elicit CCR. When the cultures had reached an optical density at 550 nm of 0.8 to 1.0, cells were harvested and β-galactosidase activities were measured as previously described (19). Three to five independent experiments were carried out, and their mean values are presented.
Oligonucleotides and PCR.
To amplify the various licT(Pia) alleles, PCRs were carried out using oligonucleotides LicSma, 5′-d(CAAAAACAGCCCGGGCAGC)-3′, and LicTrev, 5′-d(CCTACCATTTTCATATGTGAATG)-3′, as primers. Amplification of DNA fragments from chromosomal DNA of the various licT(Pia) mutants was carried out in a total volume of 100 μl containing 1 μg of template DNA, a 200 μM concentration of each deoxynucleoside triphosphate, 50 pmol of each primer, and 2.5 U of Taq polymerase (Stratagene). The samples were covered with 60 μl of light mineral oil, and usually 20 cycles of amplification were run.
Plasmid constructions.
Plasmids used in this study are listed in Table 1. To integrate the promoterless kanamycin-neomycin resistance gene aphA3 fused to bglH into the chromosomal bglPH locus of B. subtilis strains, plasmid pCL18 was constructed by first cloning the 2.1-kb NotI-HindIII fragment of pIC254 (26), which contains the 3′ part of bglP and nearly the entire bglH, into pIC408 cut with the same enzymes. pIC408 is an integrative plasmid for B. subtilis that carries a ColEI origin, the multiple cloning site of pBluescript II SK(+), and a spectinomycin resistance gene, allowing selection in E. coli and B. subtilis. A 1.5-kb HindII fragment containing the phleomycin resistance gene of pIC22 (27) was used to replace the 495-bp SmaI-StuI fragment of bglH in the pIC254/pIC408-derived plasmid before a 1.4-kb HindII-RsaI fragment of pIC239 containing the promoterless aphA3 gene was inserted into its EcoRV site (17). The resulting plasmid was called pCL18 and carried a bglPH′-aphA3 transcriptional fusion. In pCL18, the neomycin and the following phleomycin resistance gene are transcribed in the same direction. They are preceded by a 1-kb bglPH and followed by a 0.5-kb bglH fragment from B. subtilis. Since the two antibiotic resistance cassettes are separated by only 100 bp, they could be simultaneously integrated into bglPH of the B. subtilis chromosome, leaving the bglP gene intact and disrupting only bglH and therefore leading to aryl-β-glucoside-inducible neomycin resistance.
To construct plasmid pCL46, plasmid pSL3 (15) was cut with SmaI to remove a 1.5-kb fragment containing the cat gene. The shortened pSL3 was subsequently religated and cut with NruI/EcoRV, and the resulting 2.8-kb fragment was ligated with a 5-kb EcoRI fragment from pIC254, providing plasmid pCL46. Before carrying out this ligation, the 5-kb EcoRI fragment had been treated with the Klenow fragment of DNA polymerase I.
Selection of licT(Pia) mutants.
Strain BGC12 was constructed by integrating the bglP′-lacZ fusion present in ScaI-linearized plasmid pSL3 into the amyE locus of GM1210, a B. subtilis strain devoid of the antiterminators SacY and SacT and of the β-galactosidase LacA (formerly BgaX). After transformation of BGC12 with plasmid pCL18, phleomycin-resistant integrants were isolated and screened for spectinomycin sensitivity. Phleomycin-resistant and spectinomycin-sensitive clones were expected to contain a transcriptional fusion between the promoterless aphA3 gene and bglH. One integrant, called BGC13, was isolated. The presence of salicin in the growth medium conferred neomycin resistance to this strain as well as the ability to form blue colonies on X-Gal plates. The isogenic tetracycline-resistant ΔptsGHI strain BGC14 was constructed by transforming BGC13 with chromosomal DNA of strain GM1220 carrying a tetracycline resistance cassette integrated in ptsGHI. Even when grown in the presence of salicin, strain BGC14 was neomycin sensitive and formed white colonies on X-Gal plates. BGC14-derived mutants, in which the LicT function was independent of enzyme I and HPr, were expected to be resistant to neomycin and to form blue colonies on X-Gal plates. To isolate such mutants, BGC14 was grown for 9 h at 37°C with strong agitation in LB medium containing chloramphenicol, phleomycin, and tetracycline. Aliquots were placed on plates of solid minimal medium supplemented with 0.1% glucitol or on solid LB medium. Both media contained neomycin and X-Gal. Plates were incubated at 30 or 37°C, and at both temperatures several spontaneous blue-colony-forming neomycin-resistant mutants could be isolated.
Construction of ptsHI+ strains containing either wild-type licT or one of the licT(Pia) alleles.
The licT+ strain BGC12 and the BGC14-pia1, -pia4, -pia5, -pia6, and -pia9 mutants were first subjected to deletion of the cat gene located upstream of the bglP′-lacZ fusion integrated at the amyE locus by using a slightly modified version of previously described procedures (2, 16). BGC12 and the five BGC14-pia strains were transformed with pCL46. This plasmid confers erythromycin resistance to the transformants and contains a bglP′-lacZ fusion identical to the one present in BGC12 and the various BGC14-pia strains, except that the cat gene located in BGC12 and the licT(Pia) strains upstream of bglP′-lacZ was absent in pCL46. This plasmid contains also the replication origin of pE194, which functions in B. subtilis only at temperatures below 42°C. Erythromycin-resistant transformants carrying the plasmid integrated into the chromosome by single crossing over were selected at 45°C. Subsequent growth at 30°C for two passages allowed excision of the plasmid, which in some cases led to the replacement of the chromosomally located bglP′-lacZ fusion and the adjacent cat gene with the cat-less bglP′-lacZ fusion present in the integrative plasmid pCL46. Chloramphenicol-sensitive convertants were isolated and incubated on antibiotic-free LB medium at 45°C, followed by the isolation of erythromycin-sensitive segregants which had lost plasmid pCL46. The resulting BGC41-pia1, -pia4, -pia5, -pia6, and -pia9 strains are devoid of the cat gene but are otherwise identical to the corresponding BGC14-pia strain. The BGC41-pia strains were transformed with chromosomal DNA of strain BGW7, carrying an insertion of the cat gene in bglS. A double crossing over with one recombination taking place between the corresponding licT(Pia) point mutation and the cat gene inserted in bglS and the other downstream of the cat gene provided the corresponding chloramphenicol-resistant BGC54-pia transformants. They were devoid of functional BglS and therefore of β-glucanase activity, as was demonstrated with the Congo red plate test (28), and exhibited the licT(Pia) phenotype (neomycin resistance and formation of blue colonies on X-Gal plates).
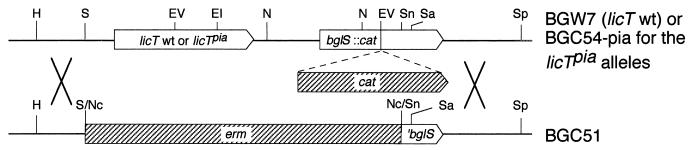
Strain BGC51, which carries the erm gene integrated into licT-bglS (Fig. 1) and which had been obtained by transforming BGC50 with chromosomal DNA from BGC9, was transformed with chromosomal DNA isolated from the BGC54-pia strains. Chloramphenicol-resistant transformants were screened for erythromycin sensitivity resulting from a double crossing over, with one recombination taking place upstream of licT and the other downstream of the cat gene. This procedure allowed cotransformation of the licT(Pia) alleles with the cat gene as outlined in Fig. 1 and provided strains BGC72-pia1, -pia4, -pia5, -pia6, and -pia9. To obtain an isogenic chloramphenicol-resistant licT wild-type strain, BGC51 was transformed with chromosomal DNA from BGW7, providing the chloramphenicol-resistant strain BGC72. The correct nucleotide sequence of the licT alleles in BGC72 and the BGC72-pia strains was verified by DNA sequencing.
FIG. 1.
Insertion of wild-type licT and the licT(Pia) alleles in the ptsHI+ strain BGC51 containing the bglPH′-aphA3 and the bglP′-lacZ fusions. DNA from strain BGW7 or the five BGC54-pia strains, which all carry a cat gene inserted in bglS, was used to transform strain BGC51. Double crossing over upstream of licT and downstream of the cat insertion allowed cotransformation of the licT alleles with the chloramphenicol resistance cassette, providing strains BGC72 and BGC72-pia, which are isogenic except for the licT(Pia) mutations. The following abbreviations were used for restriction sites: EI, EcoRI; EV, EcoRV; H, HindIII; N, NdeI; Nc, NciI; S, SmaI; Sa, SalI; Sn, SnaBI; Sp, SphI.
Construction of ccpA mutants containing wild-type licT or one of the licT(Pia) alleles.
Chromosomal DNA isolated from strain GM1225, in which the ccpA gene was disrupted by a Tn917 insertion marked with a spectinomycin cassette, was used to transform BGC72 and the five BGC72-pia strains, and spectinomycin-resistant clones were isolated. The resulting licT+ and licT(Pia) strains were called BGC88 and BGC88-pia1, -pia4, -pia5, -pia6, and -pia9.
RESULTS
licT mutants exhibiting enzyme I- and HPr-independent bglPH expression.
To isolate mutants in which the antiterminator LicT was active even when the general PTS proteins enzyme I and HPr were not functional, a ΔptsGHI strain carrying a bglPH′-aphA3 fusion at the chromosomal bglPH site and a bglP′-lacZ fusion integrated at the amyE locus was constructed. This strain, BGC14, was also devoid of LacA to prevent interference with intrinsic β-galactosidase activity and of the antiterminators SacY and SacT to avoid cross talk between members of the BglG/SacY family. Expression of both transcriptional fusions was under control of the bglPH promoter, the bglPH RAT sequence recognized by LicT, and the crebglPH recognized by P-Ser-HPr/CcpA. Owing to the absence of enzyme I and HPr, BGC14 was neomycin sensitive and formed white colonies on X-Gal plates even in the presence of salicin. Mutations leading to enzyme I- and HPr-independent bglPH expression were expected to allow the expression of the bglPH′-aphA3 fusion of BGC14 and therefore to confer neomycin resistance to the ΔptsGHI strain. In fact, we isolated two classes of BGC14-derived spontaneous neomycin-resistant mutants: (i) white-colony-forming mutants carrying cis-acting mutations in the regulatory region preceding the bglPH′-aphA3 fusion, which led to PTS-independent expression of only this fusion, and (ii) blue-colony-forming mutants affected in a bglPH-controlling trans-acting factor, most likely LicT. The second class of mutants allowed PTS-independent expression of both bglPH fusions. Mutants of the first and second class were obtained at a ratio of about 50:1. A total of nine mutants of the second class were isolated and further characterized. As these strains exhibited PTS-independent bglPH expression, they were called BGC14-pia1 to BGC14-pia9. To quantify the effect of the mutations in the BGC14-pia strains on the expression of the bglP′-lacZ fusion, β-galactosidase assays were carried out with crude extracts of BGC13, BGC14, and the various BGC14-pia mutants. Strain BGC13, which is isogenic to BGC14 except that it is ptsGHI+, exhibited salicin-inducible β-galactosidase activity (Table 2). In contrast, due to the absence of functional enzyme I and HPr, very low β-galactosidase activity was measured in strain BGC14 irrespective of whether the cells were grown in the absence or presence of salicin. However, although functional enzyme I and HPr are also absent in the BGC14-pia mutants, they exhibited β-galactosidase activities elevated between 250- and nearly 1,500-fold compared to the activity detected in strain BGC14 (Table 2).
TABLE 2.
Effect of point mutations in licT on the expression of a bglP′-lacZ fusion in strains carrying a deletion of the ptsGHI genes
| Strain | Relevant genotypea | β-Galactosidase activityb (Miller units/mg of protein) |
|---|---|---|
| BGC13 | Wild type | 6 (1,707)c |
| BGC14 | ΔptsGHI::tet | 3 (3) |
| BGC14-pia1 | ΔptsGHI::tet licT(Pia1) | 2,500 |
| BGC14-pia2 | ΔptsGHI::tet licT(Pia2) | 2,660 |
| BGC14-pia3 | ΔptsGHI::tet licT(Pia3) | 4,190 |
| BGC14-pia4 | ΔptsGHI::tet licT(Pia4) | 790 |
| BGC14-pia5 | ΔptsGHI::tet licT(Pia5) | 2,170 |
| BGC14-pia6 | ΔptsGHI::tet licT(Pia6) | 3,060 |
| BGC14-pia7 | ΔptsGHI::tet licT(Pia7) | 3,750 |
| BGC14-pia8 | ΔptsGHI::tet licT(Pia8) | 2,930 |
| BGC14-pia9 | ΔptsGHI::tet licT(Pia9) | 3,840 |
All strains carried the bglP′-lacZ fusion of strain BGW46 (15) integrated at the amyE locus. Cells were grown at 37°C in minimal medium with 0.1% ribose as the carbon source.
The mean values of at least three independent experiments are presented. Deviations were less than ±15%.
Values in parentheses represent β-galactosidase activity measured with cells grown in the presence of 0.05% salicin.
Mapping of the mutations in the BGC14-pia strains.
The BGC14-pia strains were transformed with chromosomal DNA from strain BGC9 carrying the licT gene disrupted by an erythromycin resistance cassette. Erythromycin-resistant and therefore licT-disrupted transformants were obtained with each of the nine BGC14-pia strains. They exhibited neomycin sensitivity and formed white colonies on X-Gal plates, indicating that the pia phenotype is LicT-related. To test whether the mutations map in licT itself, the nine BGC14-pia mutants were transformed with chromosomal DNA of BGC5, in which bglS, the gene downstream of licT, was disrupted with an erythromycin resistance cassette. If licT was mutated in the BGC14-pia strains, a high percentage of the erythromycin-resistant transformants was expected to have lost the pia phenotype owing to cotransformation of the wild-type licT allele of BGC5 with the erm gene integrated into bglS. Indeed, between 64 and 76% of the erythromycin-resistant transformants of each mutant had lost their pia phenotype, suggesting that all BGC14-pia strains carried mutations in licT. The licT alleles and the mutant LicT antiterminators were therefore termed licT(Pia1) to licT(Pia9) and LicT(Pia1) to LicT(Pia9), respectively.
Nucleotide sequence of the licT(Pia) alleles.
The nucleotide sequence of the different licT(Pia) alleles was determined by sequencing PCR products containing the complete licT(Pia) alleles. These PCR products were obtained by using chromosomal DNA of the BGC14-pia strains as template and the oligonucleotides LicSma and LicTrev as primers. DNA sequencing of the licT(Pia) alleles present in the PCR fragments revealed that in the three strains BGC14-pia3, -pia7, and -pia9, arginine 124 of LicT was replaced with a tryptophan (Table 3). Arginine 124 was also affected in strains BGC14-pia6 and -pia8, where it was replaced with a leucine. Aspartate 99 was mutated to asparagine in strain BGC14-pia4. Phenylalanine 211 was replaced with valine in the two strains BGC14-pia1 and BGC14-pia2 but was changed to a cysteine in strain BGC14-pia5 (Table 3). The following five strains, each containing one of the five distinct licT(Pia) alleles, were chosen for further experiments: BGC14-pia1 (Phe-211-Val), -pia4 (Asp-99-Asn), -pia5 (Phe-211-Cys), -pia6 (Arg-124-Leu), and -pia9 (Arg-124-Trp). Please, note that the DNA sequence published in reference 25 differs from the licT sequence present on the SubtiList server (http://genolist.pasteur.fr/SubtiList/) in three positions: positions 235 and 236 are AG and position 672 is A in reference 25, whereas they are CA and G in the SubtiList sequence. In all PCR products we found AG for positions 235 and 236 as in reference 25, but G for position 672 as in the SubtiList sequence, suggesting that the sequence differences between reference 25 and SubtiList are due to sequence errors.
TABLE 3.
Point mutations in the licT gene responsible for PTS-independent antitermination activity of LicT in various BGC14-pia strains
| Straina | Nucleotide exchange in the affected codonsb | Amino acid exchange |
|---|---|---|
| BGC14-pia1 | TTC→GTC | Phe-211→Val |
| BGC14-pia2 | ||
| BGC14-pia3 | CGG→TGG | Arg-124→Trp |
| BGC14-pia7 | CGG→TGG | Arg-124→Trp |
| BGC14-pia9 | CGG→TGG | Arg-124→Trp |
| BGC14-pia4 | GAC→AAC | Asp-99→Asn |
| BGC14-pia5 | TTC→TGC | Phe-211→Cys |
| BGC14-pia6 | CGG→CTG | Arg-124→Leu |
| BGC14-pia8 | CGG→CTG | Arg-124→Leu |
Strains in italics were used for further genetic characterization.
Boldface letters indicate the exchanged nucleotide.
Inducible and constitutive licT(Pia) mutants.
Genetic experiments had suggested that the phosphorylation state of the aryl-β-glucoside-specific PTS transporter BglP is important for bglPH expression (16) and that dephosphorylation of P∼BglP by one of its substrates provides the induction signal. Since the isolated licT(Pia) mutants are devoid of enzyme I and HPr, P∼BglP cannot be formed in these strains. In agreement with the predicted concept, these strains exhibited inducer-independent expression of the bglP′-lacZ fusion integrated at the amyE site (Table 2). In order to test whether in a background allowing P∼BglP formation the licT(Pia) mutations would still lead to inducer-independent expression of the bglP′-lacZ fusion, the licT(Pia) alleles were transferred into a ptsGHI+ strain by using an approach combining gene conversion, double crossing over, and cotransformation (see Materials and Methods). The resulting strains BGC72-pia1, -pia4, -pia5, -pia6, and -pia9 as well as the isogenic licT deletion strain BGC51 and the licT+ strain BGC72 were grown in minimal medium containing 0.1% ribose in the presence or absence of 0.05% salicin, and their β-galactosidase activities were measured (Table 4). As expected, virtually no bglP′-lacZ expression could be detected in the ΔlicT strain BGC51. In contrast, bglP′-lacZ expression in the isogenic licT+ strain BGC72 was about 100-fold inducible by salicin. For the five BGC72-pia strains, two different phenotypes could be observed. Similar to strain BGC72, expression of the bglP′-lacZ fusion in strains BGC72-pia1 and BGC72-pia5 was found to require induction by salicin. The low β-galactosidase activity measured in these two strains in the absence of salicin was increased about 100-fold when salicin was added to the growth medium (Table 4). In contrast, already when salicin was absent expression of the bglP′-lacZ fusion in strains BGC72-pia4, -pia6, and -pia9 was 1.5- to 6.4-fold higher than bglP′-lacZ expression measured in induced BGC72. The licT(Pia4) mutation does not seem to cause complete PTS independence, as it led to the lowest β-galactosidase activity in the ptsGHI deletion strain (790 units), which was 5.6-fold higher in the ptsGHI+ background. When salicin was present in the growth medium, it significantly lowered β-galactosidase activity in strain BGC72-pia4 but increased β-galactosidase activity in strain BGC72-pia6 (Table 4).
TABLE 4.
Effects of point mutations in licT on induction and glucose repression of a bglP′-lacZ fusion in a ptsHI+ background
| Strain | Relevant licT allele or amino acid exchange | β-Galactosidase activitya (Miller units/mg of protein) in strain grown with:
|
||
|---|---|---|---|---|
| No salicin | Salicinb | Salicin + glucoseb | ||
| BGC51 | ΔlicT | NDc | 3 | ND |
| BGC72 | Wild-type licT | 7 | 690 | 19 |
| BGC72-pia1d | Phe211Val | 11 | 960 | 46 |
| BGC72-pia5d | Phe211Cys | 6 | 800 | 32 |
| BGC72-pia4 | Asp99Asn | 4,430 | 2,670 | 150 |
| BGC72-pia6 | Arg124Leu | 930 | 1,930 | 170 |
| BGC72-pia9 | Arg124Trp | 2,740 | 2,180 | 140 |
The mean values of at least three independent experiments are presented. Deviations were less than ±15%.
Salicin and glucose were added at concentrations of 0.05 and 0.2%, respectively.
ND, not determined.
The two salicin-inducible licT(Pia) strains are listed first, followed by the three inducer-independent strains.
Effect of the licT(Pia) mutations on CCR of the bglPH operon.
The BGC72-pia strains are ptsGHI+ and therefore able to efficiently transport glucose and other PTS sugars, which allowed us to measure CCR. When glucose was present in the growth medium together with salicin, it caused strong repression of bglP′-lacZ expression in all BGC72-pia strains, including those in which the bglP′-lacZ fusion was constitutively expressed. The repression was between 12- and 25-fold and was therefore significantly weaker than the >35-fold glucose repression observed for the licT wild-type strain BGC72 (Table 4).
Two CCR mechanisms are operative for the bglPH operon (14) (Fig. 2C ). One is mediated via CcpA, P-Ser-HPr, and the crebglPH. The other requires the terminator preceding bglPH and was proposed to be the consequence of incomplete P∼His-HPr-mediated phosphorylation of LicT at PRD-2 during the uptake of a rapidly metabolizable PTS sugar, which is assumed to lead to low LicT activity (14). If this concept were correct, the terminator-dependent and LicT-controlled CCR was expected to be absent in the licT(Pia) strains, as the various LicT(Pia) proteins were highly active as antiterminators even when they could not be phosphorylated by P∼His-HPr in PRD-2 (Table 2). To test whether LicT-controlled CCR had indeed disappeared in the licT(Pia) strains, CcpA-dependent CCR was prevented in strain BGC72 and its licT(Pia) derivatives by disrupting the ccpA gene. β-Galactosidase activity was measured in the resulting licT+ strain BGC88 and the BGC88-pia derivatives after growth in minimal medium containing either ribose, ribose plus salicin, or ribose plus salicin and glucose. β-Galactosidase activity in the licT+ ccpA disruption strain BGC88 grown in a medium containing ribose, salicin, and glucose was sixfold reduced compared to the β-galactosidase activity measured in cells grown in the presence of only ribose and salicin. In contrast, no or only very slight (<1.3-fold) glucose repression could be detected in the ccpA strains harboring one of the licT(Pia) alleles (Table 5). The three strains exhibiting strong bglP′-lacZ expression already in the absence of salicin were also grown in minimal medium containing ribose and glucose. Again, glucose exerted no or only weak (1.3-fold) repression compared to cells grown only on ribose (Table 5).
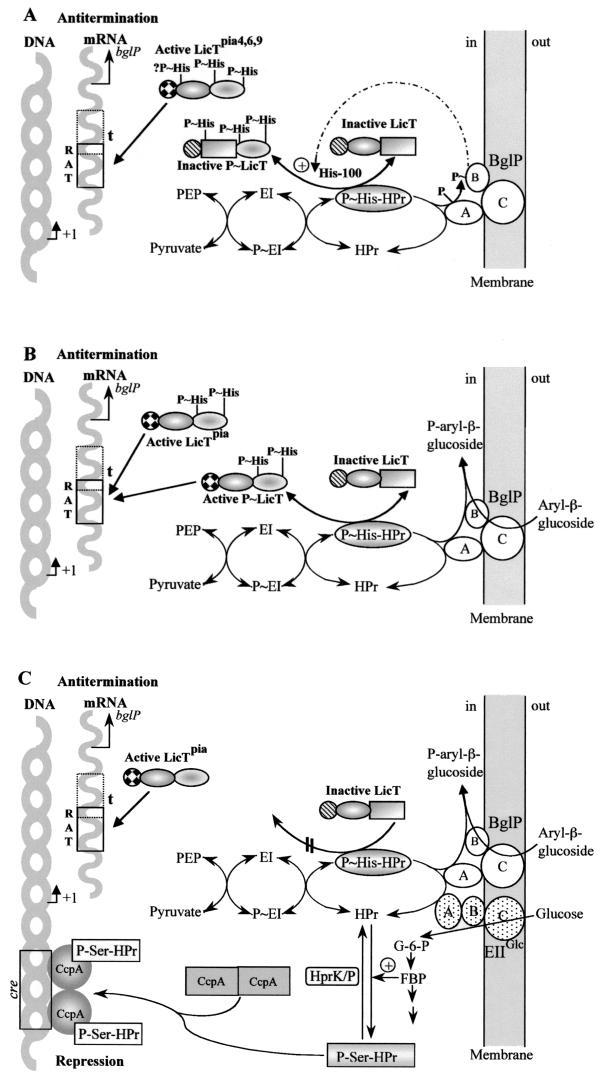
FIG.2.
Proposed mechanisms regulating the expression of the B. subtilis bglPH operon at the DNA (P-Ser-HPr/CcpA-dependent CCR) and RNA (LicT-controlled induction and CCR) level. The PRDs of LicT are represented either by boxes (nonactivated PRD) or ellipses (activated PRD), whereas striped circles represent inactive and checkered circles represent active RNA binding domains. (A) Absence of aryl-β-glucosides and other PTS sugars. In the absence of a PTS sugar, part of HPr is present as P∼His-HPr (20), which will phosphorylate LicT at His-207 and/or His-269. This phosphorylation in PRD-2 is necessary for LicT antitermination activity. However, P∼His-HPr will also phosphorylate the domains EIIA and EIIB of BglP. The presence of P∼BglP will allow phosphorylation of LicT also in PRD-1 at His-100, which renders LicT inactive, even when it is phosphorylated in PRD-2. Nevertheless, bglP mutants (16) and strains synthesizing LicTH100A (33) or LicT(Pia4), LicT(Pia6), and LicT(Pia9) express the bglPH operon under these conditions. They exhibit inducer-independent LicT antitermination activity, as their mutations either hinder phosphorylation at His-100 or prevent the negative effect of phosphorylation at His-100 on LicT antitermination activity. (B) Presence of aryl-β-glucosides. In the presence of an aryl-β-glucoside, the phosphoryl group of P∼BglP will be transferred to its substrate and no or only very little P∼BglP will be present in the cell. LicT will therefore not be phosphorylated at His-100. As the uptake of aryl-β-glucosides is slow compared to the transport of, for example, glucose or mannitol, sufficient P∼His-HPr will be present to phosphorylate LicT in PRD-2 at His-207 and/or His-269. Under these conditions, LicT and of course also the various LicT(Pia) antiterminators will be active. This model requires that during PTS-catalyzed aryl-β-glucoside phosphorylation, the transfer of the phosphoryl group from P∼His-HPr to the EIIB domain represent the rate-limiting step. (C) Additional presence of glucose. The presence of glucose in an aryl-β-glucoside-containing medium exerts a repressive effect on bglPH expression, which is mediated by two different mechanisms. (i) The metabolism of glucose causes an increase of the fructose-1,6-bisphosphate concentration, which activates the kinase activity of HprK/P. The resulting P-Ser-HPr forms a complex with CcpA, which allows the catabolite repressor to prevent expression of the bglPH operon by binding to the cre overlapping the −35 region. (ii) As a large amount of P-Ser-HPr, which is a poor substrate for phosphorylated enzyme I, is formed during the metabolism of glucose and as its rapid transport causes dephosphorylation of HPr at His-15, very little P∼His-HPr is present in glucose-metabolizing B. subtilis cells (20). Under these conditions, P∼His-HPr will primarily phosphorylate glucose via PtsG, and LicT will be barely phosphorylated in PRD-2 and therefore remain inactive. The approximately sixfold repression of bglPH expression caused by glucose in ccpA mutants (15) is due to diminished LicT phosphorylation and activation. Strains synthesizing one of the LicT(Pia) antiterminators do not exhibit this second CCR mechanism, as the LicT(Pia) antiterminators are active without being phosphorylated in PRD-2.
TABLE 5.
Effect of licT(Pia) mutations on LicT-modulated CCR of a bglP′-lacZ fusion in a ccpA background
| Strain | licT allele | β-Galactosidase activitya (Miller units/mg of protein) in strain grown with:
|
|||
|---|---|---|---|---|---|
| No salicin | Glucoseb | Salicinb | Salicin + glucoseb | ||
| BGC88 | Wild-type | 10 | NDc | 800 | 130 |
| BGC88-pia1d | Phe211Val | 53 | ND | 1,850 | 1,820 |
| BGC88-pia5d | Phe211Cys | 14 | ND | 1,830 | 1,560 |
| BGC88-pia4 | Asp 99Asn | 4,510 | 3,450 | 4,130 | 3,140 |
| BGC88-pia6 | Arg124Leu | 4,690 | 4,290 | 5,120 | 3,940 |
| BGC88-pia9 | Arg124Trp | 5,300 | 5,470 | 4,590 | 4,640 |
The mean values of at least three independent experiments are presented. Deviations were less than ± 15%.
Salicin and glucose were added at concentrations of 0.05 and 0.2%, respectively.
ND, not determined.
The two salicin-inducible licT(Pia) strains are listed first, followed by the three inducer-independent strains.
DISCUSSION
The activity of PRD-containing antiterminators and transcriptional activators is regulated by multiple PEP-dependent enzyme I- and HPr-catalyzed phosphorylation of conserved histidyl residues (for a review, see references 6 and 29). In most PRD-containing transcriptional regulators, including LicT of B. subtilis, these phosphorylations can exert positive or negative effects on the activity of the antiterminators and transcriptional activators depending on which PRD becomes phosphorylated. Phosphorylation at His-100 in PRD-1 of LicT—which requires P∼BglP as either the phosphoryl donor or the phosphorylation stimulator—inhibits LicT antitermination activity (Fig. 2A). The uptake of aryl-β-glucosides leads to P∼BglP dephosphorylation and therefore to poor phosphorylation at His-100 in LicT, which probably provides the molecular basis for induction of the bglPH operon (Fig. 2B). In contrast, phosphorylation by PEP, enzyme I, and HPr of at least one of the two conserved histidyl residues in PRD-2 is necessary to render LicT active (33). We were able to isolate five distinct mutants in which LicT was functional, although these mutants were missing enzyme I and HPr. Studies of bglPH expression in the licT(Pia) mutants allowed us to confirm that diminished phosphorylation of LicT by enzyme I and HPr in PRD-2 serves as a CCR mechanism. As outlined in Fig. 2C, expression of the bglPH operon is controlled by two CCR mechanisms. The CcpA-independent CCR mechanism was proposed to imply LicT, as it disappeared when the terminator preceding the bglPH operon was deleted (14, 15). As the residual CCR of ccpA mutants was also absent in ptsH1 mutants, which are unable to form P-Ser-HPr, it has been suggested that the second CCR mechanism operative for bglPH might be based on reduced activation of LicT activity due to diminished phosphorylation in PRD-2 during the uptake of a rapidly metabolizable PTS sugar such as glucose (Fig. 2C). During the metabolism of glucose, about 60% of HPr is converted to P-Ser-HPr, and only little P∼His-HPr, the phosphoryl donor for the phosphorylation of LicT in PRD-2, is present (20). In addition, enzyme IIAs of the PTS were found to be more rapidly phosphorylated by P∼His-HPr than LicT (18). As a consequence, LicT will be barely phosphorylated when glucose is taken up and will therefore be inactive (Fig. 2C), except when HPr cannot be converted to P-Ser-HPr, as is the case in ptsH1 mutants. The absence of CcpA-independent CCR in licT(Pia) strains confirmed this concept. LicT(Pia) antiterminators are active without phosphorylation by PEP, enzyme I, and HPr at PRD-2, and as a consequence, strains synthesizing one of the LicT(Pia) proteins did not exhibit CcpA-independent CCR of the bglPH operon (Table 5) (Fig. 2C). Only LicT(Pia4) was not completely PTS independent (compare Tables 2 and 4), and slight CcpA-independent CCR was therefore detectable in the presence and absence of salicin (Table 5). In contrast, weak CCR of β-galactosidase activity in strains synthesizing the slightly salicin-activatable LicT(Pia6) (Table 4) occurred only when salicin was present.
Although all five licT(Pia) alleles prevented the terminator-dependent CCR, we could distinguish two different classes of licT(Pia) mutations when bglPH induction by aryl-β-glucosides was measured in a ptsHI+ background. When the altered amino acid was located in PRD-1 of LicT (Asp-99 replaced with Asn or Arg-124 replaced with Trp or Leu), the corresponding mutants exhibited inducer-independent bglPH expression. These mutations must therefore cause structural changes, which either prevent the negative effect on LicT activity exerted by phosphorylation at His-100 or hinder phosphorylation at His-100. However, these LicT(Pia) antiterminators clearly differ from the previously described H100A mutant LicT. Owing to the absence of the phosphorylatable His-100, this mutant LicT caused constitutive expression from the bglPH promoter in a ptsHI+ background but was inactive when the ptsHI genes were deleted (33). In contrast, the three licT(Pia) mutations affecting Asp-99 or Arg-124 had a dual effect and not only caused constitutive expression from the bglPH promoter but also rendered the mutant LicT antiterminators enzyme I and HPr independent. Interestingly, when the altered amino acid in the LicT(Pia) antiterminators was located in PRD-2 (Phe-211 replaced with Cys or Val), expression from the bglPH promoter required salicin for induction in a ptsHI+ background. These two mutant LicT antiterminators therefore resemble the antiterminators SacY and GlcT, which are both active without phosphorylation by PEP, enzyme I, and HPr in PRD-2 and therefore are naturally PTS independent. Nevertheless, they are still inactivated by phosphorylation in PRD-1 (2, 31, 32). The two licT(Pia) mutations affecting Phe-211 cause similar effects on LicT activity as replacing one of the two phosphorylatable histidyl residues in PRD-2 (His-207 or His-269) with an aspartate. Interestingly, when both conserved histidyl residues in PRD-2 of LicT were replaced with aspartates, the resulting doubly mutated LicT was not only enzyme I and HPr independent but also allowed constitutive expression from the bglPH promoter (33). The double mutation in PRD-2 therefore leads to a phenotype similar to that exhibited by strains synthesizing LicT(Pia) antiterminators affected in PRD-1. Probably due to the different effects exerted by the two classes of licT(Pia) mutations, CCR was less severe when PRD-1 of LicT was affected (Table 4). In addition, in the presence of salicin the inducer-independent LicT(Pia) antiterminators affected in PRD-1 led to two- to threefold-higher β-galactosidase activity compared to wild-type LicT or LicT(Pia) antiterminators affected in PRD-2. In the inducer-independent mutant LicT antiterminators, the activity-stimulating effect caused by phosphorylation at His-207 and/or His-269 might therefore be transferred with higher efficiency to the RNA binding domain.
The crystal structure of the regulatory domains of LicT, composed of wild-type PRD-1 and the mutant PRD-2 with the two phosphorylatable histidyl residues replaced with aspartates, has recently been determined (34). This allowed us to trace the amino acids mutated in the LicT(Pia) antiterminators in the PRD structure and to reason why these mutations lead to the licT(Pia) phenotype. Although Asp-99 is located next to His-100, it is unlikely that the D99N mutation would prevent phosphorylation at the adjacent histidine, which would explain the constitutive LicT activity. Being located on an alpha-helix, Asp-99, which is conserved in all antiterminators of the BglG/SacY family, but not in PRD-containing transcriptional activators (32), is not in contact with the neighboring His-100 but points towards the region connecting the RNA binding domain to PRD-1. It is therefore more likely that the D99N mutation causes structural changes stimulating the RNA binding activity exhibited by the first 55 amino acids of LicT (4). bglPH expression in mutants synthesizing LicT affected in PRD-2 [at Phe-211 in LicT(Pia1) and LicT(Pia5) or replacing His-207 or His-269 with an aspartate (33)] required induction by salicin. This result suggested that the activating signal transferred from PRD-2 via PRD-1 to the N-terminal RNA binding domain can be intercepted by phosphorylation at His-100 (Fig. 2A). As Asp-99 is obviously located behind His-100 in this signal transduction pathway, phosphorylation at His-100 can probably not prevent the activating effect of the D99N mutation on the RNA binding activity of LicT, which might explain why this mutation leads not only to enzyme I- and HPr-independent but also to inducer-independent bglPH expression. In fact, phosphorylation at the neighboring His-100 even slightly enhanced the stimulating effect of the D99N mutation, as in a ptsHI+ background expression of the bglP′-lacZ fusion was almost twofold higher in the absence of salicin (LicT phosphorylated at His-100) than in the presence of salicin (LicT not phosphorylated at His-100) (Table 4).
In the LicT dimer (3), PRD-1 and PRD-2 of opposite subunits interact via helices α3 of PRD-1 and α2 of PRD-2 (34), the latter alpha-helix containing His-207, the major in vitro phosphorylation site in LicT (18). This interaction, which is mainly established via a salt bridge formed between the well-conserved Glu-121 and Lys-209, has been proposed to be the major route of signal transduction from PRD-2 to PRD-1. Intriguingly, the two other amino acids affected in the licT(Pia) mutants, Arg-124 and Phe-211, are located next to the two amino acids forming the intersubunit salt bridge. Being situated four amino acids behind the phosphorylatable His-207 in alpha-helix 2 of PRD-2, Phe-211 is in close contact with the phosphorylation site (one turn on the helix). It is therefore tempting to assume that replacing Phe-211 with valine or cysteine causes structural changes similar to those induced by phosphorylation at His-207. Interestingly, whereas expression of the bglP′-lacZ fusion in ptsHI+ strains synthesizing LicT affected at Phe-211 required induction, it was constitutive or only twofold inducible in strains containing LicT altered at Arg-124 (Table 4). Replacing Arg-124 with bulky hydrophobic amino acids such as tryptophan or leucine must therefore cause structural changes, which not only lead to the stimulation of the RNA binding activity of LicT but presumably also hinder phosphorylation at His-100. The PRD-1 structure in these LicT(Pia) antiterminators might therefore resemble the PRD-1 structure in truncated doubly mutated (H207D, H269D) LicT, where the phosphorylatable histidines in PRD-1 are all buried at the dimer interface, with 0% surface exposure (34). This closed conformation prevents the access of P∼His-HPr to His-100 and His-159 and therefore phosphorylation in PRD-1. In H207D/H269D mutant LicT, PRD-2 is also present in a closed conformation with the aspartates buried in the center of the dimer. This is probably not the case for the histidines in PRD-2 of R124W mutant LicT. R124W mutant LicT has been purified by following the procedure described for wild-type LicT (18). Phosphorylation of R124W mutant LicT with [32P]PEP, enzyme I, and HPr and subsequent separation of tryptic fragments revealed a phosphorylation pattern similar to that obtained with wild-type LicT (C. Lindner and J. Deutscher, unpublished results), with His-207 in PRD-2 being the major in vitro phosphorylation site (18). It will therefore be interesting to determine the structure of the regulatory domains of R124W mutant LicT and the other LicT(Pia) proteins, work which is currently in progress.
Acknowledgments
We are very grateful to the late Michel Steinmetz, who initiated this research, which was supported by the CNRS, the INRA, the INA-PG, the European Union HCM Program (contract ERBCHBGCT93 0459) (to J.D.), PROCOPE 1994, and grant He 1887/1-3 from the Deutsche Forschungsgemeinschaft (to M.H.).
Jörg Stülke, Stéphane Aymerich, and Pablo Tortosa are acknowledged for valuable discussions.
REFERENCES
- 1.Aymerich, S., and M. Steinmetz. 1992. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl. Acad. Sci. USA 89:10410-10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crutz, A.-M., M. Steinmetz, S. A. Aymerich, R. Richter, and D. Le Coq. 1990. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J. Bacteriol. 172:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Declerck, N., H. Dutartre, V. Receveur, V. Dubois, C. Royer, S. Aymerich, and H. van Tilbeurgh. 2001. Dimer stabilization upon activation of the transcriptional antiterminator LicT. J. Mol. Biol. 314:671-681. [DOI] [PubMed] [Google Scholar]
- 4.Declerck, N., F. Vincent, F. Hoh, S. Aymerich, and H. van Tilbeurgh. 1999. RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT. J. Mol. Biol. 294:389-402. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher, J., and R. Engelmann. 1984. Purification and characterization of an ATP-dependent protein kinase from Streptococcus faecalis. FEMS Microbiol. Lett. 23:157-162. [Google Scholar]
- 6.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2001. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 7.Deutscher, J., E. Küster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 8.Deutscher, J., B. Pevec, K. Beyreuther, H.-H. Kiltz, and W. Hengstenberg. 1986. Streptococcal phosphoenolpyruvate-sugar phosphotransferase system: amino acid sequence and site of ATP-dependent phosphorylation of HPr. Biochemistry 25:6543-6551. [DOI] [PubMed] [Google Scholar]
- 9.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 11.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 12.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305-320. [DOI] [PubMed] [Google Scholar]
- 13.Kravanja, M., R. Engelmann, V. Dossonnet, M. Blüggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and W. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol. 31:59-66. [DOI] [PubMed] [Google Scholar]
- 14.Krüger, S., S. Gertz, and M. Hecker. 1996. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J. Bacteriol. 178:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krüger, S., and M. Hecker. 1995. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J. Bacteriol. 177:5590-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Coq, D., C. Lindner, S. Krüger, M. Steinmetz, and J. Stülke. 1995. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J. Bacteriol. 177:1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindner, C. 1998. The bglPH operon of Bacillus subtilis and its regulation by the antiterminator protein LicT. Ph.D. thesis. University of Greifswald, Greifswald, Germany.
- 18.Lindner, C., A. Galinier, M. Hecker, and J. Deutscher. 1999. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol. Microbiol. 31:995-1006. [DOI] [PubMed] [Google Scholar]
- 19.Martin-Verstraete, I., V. Charrier, J. Stülke, A. Galinier, B. Erni, G. Rapoport, and J. Deutscher. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28:293-303. [DOI] [PubMed] [Google Scholar]
- 20.Monedero, V., S. Poncet, I. Mijakovic, S. Fieulaine, V. Dossonnet, I. Martin-Verstraete, S. Nessler, and J. Deutscher. 2001. Mutations lowering the phosphatase activity of HPr kinase/phosphatase switch off carbon metabolism. EMBO J. 20:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, N., D. J. McConnell, and B. A. Cantwell. 1984. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme β-glucanase. Nucleic Acids Res. 12:5355-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson, W. L., and G. H. Chambliss. 1985. Isolation and characterization of a cis-acting mutation conferring catabolite repression resistance to α-amylase synthesis in Bacillus subtilis. J. Bacteriol. 161:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schnetz, K., J. Stülke, S. Gertz, S. Krüger, M. Krieg, M. Hecker, and B. Rak. 1996. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol. 178:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz, M., and R. Richter. 1994. Easy cloning of Mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 28.Stülke, J. 1993. Regulation of β-glucanase synthesis in Bacillus subtilis. Ph.D. thesis. University of Greifswald, Greifswald, Germany.
- 29.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD: a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 30.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 31.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 32.Tortosa, P., S. Aymerich, C. Lindner, M. H. Saier, Jr., J. Reizer, and D. Le Coq. 1997. Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J. Biol. Chem. 272:17230-17237. [DOI] [PubMed] [Google Scholar]
- 33.Tortosa, P., N. Declerck, H. Dutartre, C. Lindner, J. Deutscher, and D. Le Coq. 2001. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol. Microbiol. 41:1381-1393. [DOI] [PubMed] [Google Scholar]
- 34.van Tilbeurgh, H., D. Le Coq, and N. Declerck. 2001. Crystal structure of an activated form of the PTS regulation domain from the LicT transcriptional antiterminator. EMBO J. 20:3789-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]