Abstract
Palindromic units (PUs) are intergenic repeated sequences scattered over the chromosomes of Escherichia coli and several other enterobacteria. In the latter, IS1397, an E. coli insertion sequence specific to PUs, transposes into PUs with sequences close to the E. coli consensus. Reasons for this insertion specificity can relate to either a direct recognition of the target (by its sequence or its structure) by the transposase or an interaction between a specific host protein and the PU target DNA sequence. In this study, we show that for Yersinia pestis, a species deprived of PUs, IS1397 can transpose onto its chromosome, with transpositional hot spots. Our results are in favor of a direct recognition of target DNA by IS1397 transposase.
Despite their small size, microorganisms contain a wide variety of repeated sequences, sometimes accounting for 10% or more of the total genome. Bacterial interspersed mosaic elements (BIMEs) are a family of small extragenic sequences scattered over the Escherichia coli chromosome (2). They could play a role in the functional organization of the bacterial nucleoid. Their mosaic structure associates palindromic units (PUs) (or repetitive extragenic palindromic sequences) (10, 28) with extra-PU motifs. PUs are imperfect palindromic sequences of about 40 bp, transcribed but not translated (4), which can confer a stem-loop secondary structure to mRNA. A complete description of BIMEs can be found on the World Wide Web (http://www.pasteur.fr/recherche/unites/pmtg/repet/index.html).
Insertions sequences (ISs) are small (800 to 2,500 bp) DNA segments capable of inserting into target DNA molecules with a more or less pronounced target specificity. More than 500 ISs have been identified so far for more than 40 bacterial species and classified into 17 families on the basis of open reading frame (ORF) organization, length of the target duplications, similarity of their terminal inverted repeats (IRs), and signature motifs among the transposases (see reference 16 for a review). They play important roles in DNA translocations and other rearrangements in bacteria (9, 13).
Insertion sequence IS1397 has been found in several E. coli natural isolates, always inserted into the central part (loop) of a PU. This 1,432-bp insertion sequence belongs to the IS3 family. It has 25-bp-long terminal inverted repeats (IRL and IRR) and encodes two proteins, OrfA and OrfB, which are in phase 0 and −1, respectively. A −1 translational frameshifting leads to a fusion protein, OrfAB, which is the transposase. Members of our group have shown that IS1397 is able to transpose into PUs with sequences close or identical to the E. coli consensus, even in other enterobacteria (Salmonella Enterica serovar Typhimurium, Klebsiella pneumoniae, and Klebsiella oxytoca) (6, 31).
The nature of PU recognition by IS1397 is still unclear. The transposase could directly bind PU DNA sequence. Alternatively, since PUs were shown to interact specifically with integration host factor (5, 20), DNA gyrase (8, 32), and DNA polymerase (11), they could be the central part of complex nucleoproteic structures which could be recognized by IS1397 transposase. Yersinia pestis is an enterobacterium phylogenetically close to E. coli but in which no sequences homologous to E. coli PU were found. This offered the opportunity to analyze whether a PU-specific host factor is required for IS1397 transposition. Furthermore, if transposition could take place in this host, the analysis of target sites could reveal possible structural or functional PU homologs.
MATERIALS AND METHODS
Media and bacterial strains.
Luria-Bertani (LB) medium was used for bacteria growth. Kanamycin (KAN) and chloramphenicol (CHL) were used at concentrations of 25 and 50 μg/ml, respectively. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a final concentration of 10−3 M, except for the transposition with pBLOCKS, where it was used at a final concentration of 10−6 M. LBS medium is LB medium without NaCl and containing 10% sucrose. Hemin was added at a concentration of 25 μg/ml in Y. pestis culture plates. E. coli strains were grown at 37°C, and Y. pestis was grown at 28°C.
Strains used were the following: Escherichia coli BW19610 [DE3(lac)X74 ΔuidA::pir-116 recA1 ΔphoA532 Δ(phnC?D-P)33-30] (18) and Yersinia pestis 6/69c, a derivative of Yersinia pestis 6/69 (12), which was cured of the pYV virulence plasmid.
Plasmids.
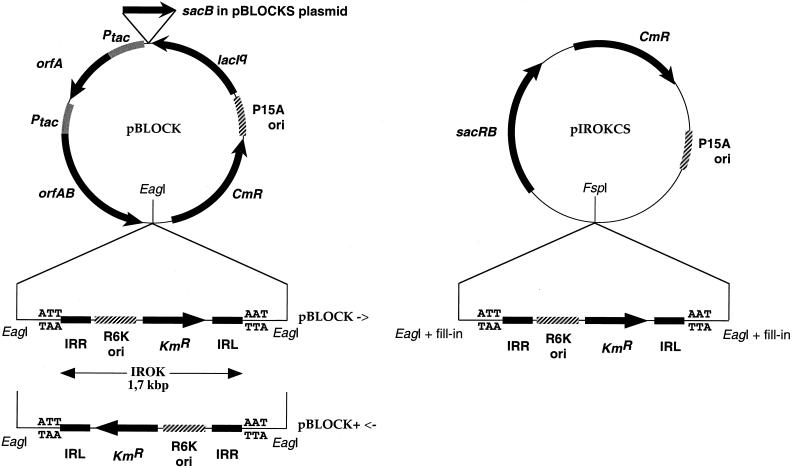
pBLOCK (Fig. 1) has already been described (31). This derivative of pACYC184 carries the P15A origin of replication placed between lacIq and the CHL resistance gene. The Ptac promoter is found twice, upstream of orfA and upstream of orfAB, allowing overproduction of the two proteins upon IPTG induction. OrfA and OrfAB are both toxic to the cell (6). The borders of the IROK transposable module (delineated with arrows on Fig. 1) are IS1397 inverted repeats flanking the KAN resistance gene and the R6K origin of replication (26, 27), which is functional in strains expressing Pir protein, such as BW19610. In pBLOCK+ (Fig. 1), the EagI DNA fragment containing IROK is in the opposite orientation from that in pBLOCK.
FIG. 1.
Plasmids pBLOCK, pBLOCK+, pBLOCKS, and pIROKCS. pBLOCK (31) contains the transposase coding gene orfAB and a transposable module, IROK, which is delineated by the two IS1397 inverted repeats (IRL and IRR). This module is flanked by two EagI sites, and it is in the opposite orientation in pBLOCK+. pBLOCKS contains the sacB gene cloned between orfA and lacIq. pIROKCS contains IROK, but no transposase gene.
pBLOCKS (Fig. 1) is a derivative of pBLOCK in which the sacB gene was cloned at the NheI site. The sacB gene was amplified by PCR with oligonucleotides SacB1 (5′CCCCCCGCTAGCGGCCCGTAGTCTGCAAATCC3′) and SacB2 (5′CCCCCCGCTAGCGCTGTTCACCATGAACAGATCG3′), using pKO3 (15) as a template.
pIROKCS (Fig. 1) is a derivative of the pSU2718 cloning vector (17), in which sacRB genes have been cloned at the HindIII site and the IROK transposable module has been cloned at the SmaI site.
DNA technologies.
Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs or Boehringer Mannheim and used as recommended. Plasmid DNA manipulations were carried out by using standard procedures (24). Extraction of total cellular DNA was performed with the DNA Easy Tissue kit (Qiagen).
DNA sequencing.
DNA sequencing was performed by MWG-Biotech AG and ESGS (Cybergene). The two oligonucleotides used to sequence the junctions of IS1397 chromosomal insertions were purchased from Genset. Their sequences are the following: seqIRL, 5′CGGTTGTGGACAACAAGCCAGGG3′ (complementary to a region of R6K origin of replication); and Kmseqout, 5′CACGAGGCAGACCTCAGCGC3′ (corresponding to a region located between the end of the KAN resistance gene and IRR of the module as found on pBLOCK).
Southern blot hybridization.
Total DNA was extracted from overnight cultures. One to five micrograms of DNA were loaded on a 1% agarose Tris-acetate gel. DNA transfer onto Hybond N+ membrane (Amersham) was performed as previously described (6). Hybridization was carried out using the DIG Nucleic Acid Labeling and Detection system (Boehringer Mannheim) at 56°C. The probe used was digoxigenin-dUTP-labeled with the Promega Nick-Translation kit.
Selection of transposition events for Y. pestis using pBLOCK.
We slightly modified the strategy used for the selection and the study of transposition events for K. oxytoca and Salmonella serovar Typhimurium (31) in order to minimize liquid culture steps of Y. pestis. Independent clones of Y. pestis 6/69c electroporated with pBLOCK or pBLOCK+ were grown overnight on LB plates containing hemin, KAN, and IPTG to induce transposition. They were then streaked on LB plates containing hemin, CHL, and IPTG to check the loss of the plasmid. Cms clones were grown overnight in LB liquid medium containing KAN and IPTG prior to DNA extraction. Total cellular DNAs were digested with MluI (which has a single site in pBLOCK, in the lacI gene), and 1 to 5 μg was analyzed by Southern blot with IROK (Fig. 1) as a probe. Fragments of DNA digested with MluI or BstEII were circularized with T4 DNA ligase and used to transform BW19610, a strain which allows autonomous replication of circles containing R6K origin of replication present in IROK. Recombinant clones were selected on LB plates containing KAN. After DNA sequencing, chromosomal regions flanking the module were identified using FASTA software (21) from Infobiogen (http://www.infobiogen.fr/) and BLAST (1) from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) or the Sanger Centre (http://www.sanger.ac.uk/Projects/Y_pestis/). Chromosomal locations given thereafter between brackets refer to strain CO-92 biovar Orientalis, which was sequenced there.
Selection of transposition events in E. coli using pBLOCKS.
Transposition has been tested as previously described for Salmonella serovar Typhimurium and K. oxytoca (31), with a slight modification: 1 μl of overnight cultures of independent pBLOCKS-transformed clones was plated on LBS containing KAN or LBS containing KAN and IPTG (10−6 M) (which is not toxic at this concentration). Sucrose-resistant clones were then tested for CHL and IPTG sensitivity. Clones presenting the correct phenotype (i.e., sucrose, KAN, and IPTG resistant and CHL sensitive) were analyzed as before.
Transposition assays using pIROKCS.
Independent clones of E. coli and Y. pestis transformed with pIROKCS were grown overnight in liquid LB medium containing KAN and CHL. Ten microliters for E. coli and 500 μl for Y. pestis of each culture were plated on LBS containing KAN, incubated overnight, and then tested for CHL sensitivity on LBS containing CHL. All clones tested were still CHL resistant, indicating that they had kept the plasmid and that transposition could not be detected in this case.
RESULTS AND DISCUSSION
IS1397 is an IS3 family member discovered in natural isolates of E. coli. It is always inserted into small extragenic repeated sequences scattered on the E. coli chromosome, the PUs (3). PUs are the basic motif of BIMEs, which associate PUs with extra-PU motifs according to precise rules. They have been described for several enterobacteria, where slight sequence variations allowed us to define species-specific consensuses (6, 31). We previously tested IS1397 transposition with PU-containing species (E. coli [6], Salmonella serovar Typhimurium, K. pneumoniae, and K. oxytoca [31]) and showed that it is able to transpose from a donor plasmid into their chromosomes, almost exclusively into PUs. Interestingly, transposition for K. pneumoniae took place preferentially into PUs which were close to the E. coli consensus rather than into characteristic K. pneumoniae PUs (which are more GC rich). The more the PU consensus differs from the E. coli PU consensus, the more we observed non-PU insertions (such as in K. oxytoca). Y. pestis is phylogenetically close to E. coli, but a systematic computer analysis of intergenic regions in this species did not reveal the presence of any repeated sequence similar to PUs or BIMEs. We investigated the ability of IS1397 to transpose onto the chromosome of this PU-free species and analyzed the nature of transposition targets.
We studied the transposition of IS1397 in Y. pestis using pBLOCK or pBLOCK+, two plasmids previously used in Salmonella serovar Typhimurium and K. oxytoca (31). Selection of transposition events relied on the Kmr IPTGR Cms phenotype, which should result from transposition of the module into the chromosome followed by the loss of the plasmid. The presence of both orfA and orfAB (both lethal when overexpressed) minimized the recovery of IPTG-resistant clones due to mutations in these genes. A deletion encompassing both these genes and the CHL resistance gene would not allow the maintenance of an autonomously replicating plasmid, since the p15A origin of replication is located between the CHL resistance gene and orfA (Fig. 1).
Forty-eight and forty-four independent Y. pestis clones containing pBLOCK and pBLOCK+, respectively, were streaked on IPTG and KAN plates and subsequently replica plated on CHL and IPTG plates to check for the loss of the plasmid. A total of 57 independent Kmr IPTGr Cms clones were obtained and examined for the presence of IROK on the chromosome by Southern blot hybridization. They displayed one or several bands ranging from 2.5 to 15 kb. Chromosomal DNA fragments encompassing IROK contain a KAN resistance gene and an R6K origin of replication, which is active in E. coli BW19610. After recircularization, 25 plasmids were recovered and used to sequence the new flanking regions of the transposable module. Several classes of transposition events were observed.
Class I: circles.
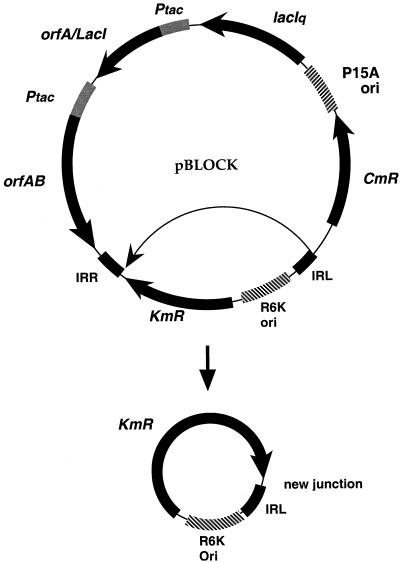
We obtained five clones (I.a to I.e) consisting of circles derived from pBLOCK (Fig. 2). Four of them were a truncated version of the transposable module (IROK). IRL was fused to the DNA region located after the KAN resistance gene. Thus, IRR was deleted and no extra bases were found at the junction. These circles probably correspond to an intramolecular transposition event within pBLOCK. Such a phenomenon has already been observed in the case of IS911 (22, 30) and was interpreted as an alternative pathway to the formation of intermediate transposition circles. These circles could not be detected on Southern blots when the total DNA content (i.e., chromosomal and plasmidic) of the candidates was analyzed (data not shown), indicating that they are in very low copy numbers and/or very transient. This is consistent with the fact that they do not carry a fully functional origin of replication and must be lost at a high frequency. Covalently closed circles have a high transformation potential in an E. coli strain such as BW19610 which can support their replication. If the ligation efficiency of linear MluI or BstEII chromosomal fragments was low, the recovery of circular molecules at the expense of ligated chromosomal fragments was expected. It is interesting that we never observed such circular forms in other enterobacteria (6, 31), and it may be due to the tendency of Y. pestis to keep replicating such suicidal plasmids (E. Carniel, unpublished observation). In clone I.a, which will be discussed below, IRR was present and IRL was modified from 5′TGCAGTTCA3′ to 5′TGCAGTGCA3′ (base change is in italic; the same type of change as observed in clone III.d). IRL and IRR were joined by an AAG-triplet which is originally flanking neither IRR nor IRL in pBLOCK (Fig. 1).
FIG. 2.
Structure of the circles observed in clones I.a to I.e. See the legend for pBLOCK in Fig. 1. The thin arrow inside the pBLOCK plasmid indicates the target region in IROK for IRL attack.
Class II: typical chromosomal transposition events.
A classical transposition event was observed in 11 cases with a 3-bp duplication at the sites of insertion (Fig. 3, class II).
FIG. 3.
Insertion sites of the transposable module into the chromosome of Y. pestis. Superscript italic letters indicate the following. a, the three classes have been defined according to the insertion mechanisms and sites (see Results and Discussion). b, nucleotides duplicated after transposition are indicated in bold. W stands for A or T. c, thick arrows indicate the module orientation on the Y. pestis chromosome. Only the nine external base pairs of IRL and IRR are written, and Y. pestis IR residues which correspond to changes in the original sequence of IROK are double-underlined. d, the sequence of the transposable module in the donor plasmids is written in both orientations. e, the coordinates have been determined referring to Y. pestis CO-92 biovar Orientalis, sequenced at the Sanger Centre. The progenitor of clone I.a is the hypothetical chromosomal structure resulting from the initial transposition event (see Results and Discussion).
For seven clones (class IIA, Fig. 3), the transposable module had moved into five distinct regions. It inserted into IS1541 (19) in clones IIA.a and IIA.b. The insertion site (upstream of IRR) and the duplication (TAT) are identical, but the module orientations and the chromosomal locations (coordinates 4,064,971 and 3,417,535, respectively) are different. An insertion into a palindromic region which precedes another copy of IS1541 was observed in clone IIA.g. In clones IIA.c to IIA.e, independent transposition events occurred at the same site (position 2,398,820) and with the same module orientation. In clone IIA.f, the transposable module inserted between thiE and thiF homologs.
For four clones (class IIB, Fig. 3), transposition has occurred next to a noncharacterized Y. pestis resident IS. Screening of the GenBank database with its nucleotide sequence gave the best score with IS150 (55.5% identity). The distal sequences of its inverted repeats are almost identical to IS1397: Y. pestis IRL shares 19 nucleotides with IS1397 IRs, and Y. pestis IRR shares 21 nucleotides with IS1397 IRL and 23 with IS1397 IRR (Fig. 4). OrfA and OrfB Y. pestis IS proteins share 38.9 and 49.5% identity, respectively, with the corresponding proteins of IS1397. The transposable module has inserted three or five nucleotides downstream from the IR in three (IIB.a to IIB.c) or one (IIB.d) clones, respectively. Although this Y. pestis IS is present in 10 copies (2 of them are truncated) in the Y. pestis genome, the four insertions were all located next to the same copy.
FIG. 4.
Sequence comparisons between IS1397 and Y. pestis IS IRs. IS1397 IRL and IRR are compared with homologous sequences of Y. pestis IRs. Variable nucleotides between the four sequences are indicated in bold.
Interestingly, two sites were found either three or four times independently: clones IIA.c to IIA.e at position 2,398,820 and clones IIB.a to IIB.d at position 4,137,899. The exact site of transposition in clone IIB.d is shifted by 2 bp compared with the three others. Hot spots for insertion of IS1397 have been found in all tested species (6, 31). The finding of a similar phenomenon in Y. pestis confirms that some chromosomal regions are more attractive than others for transposition, independently of the presence of PUs.
Moreover, IS1397 seems to target sequences which share similarities with the E. coli PU consensus in class IIA clones. Indeed, in five regions (represented by a total of seven independent events [Fig. 3, class II]), transposition events took place 11 to 13 bp from the sequence 5′GCCGG3′ (5′GCCGGA3′ in 4 examples) (Table 1), which is part of the sequence of an E. coli PU stem. This sequence is, however, not found in class IIB clones (next to a Y. pestis IS). Similarly, for E. coli (6) and for species containing PUs which are different from those of E. coli, such as K. pneumoniae and K. oxytoca, a few transposition events were found outside PUs but in the vicinity of similar sequences (31). A comparison of all these non-PU insertion sites (Table 1) shows that the 5′GCCGG3′ sequence is present in 11 out of 16 insertion sites, and in these 11 cases, the homology can be further extended (underlined in Table 1). However, this similitude between Y. pestis target consensus and E. coli PUs is limited to the first part of the PU stem, and no palindromic structure was found in this case. This observation suggests that the transposase binds directly the target sequence and that a secondary DNA structure is not required for IS1397 transposition.
TABLE 1.
Comparison of non-PU insertion sites of the transposable module in several enterobacteria
| Species | No. of clones | Flanking sequence upstream of the insertion sitea | Duplication | No. of residues identical to 17-bp consensus |
|---|---|---|---|---|
| ATAC | ||||
| E. coli PU consensus | AT GCCGAT GC GCG | NNN | 17 | |
| TGGT | ||||
| Y. pestis | 2 | ATA GCCGGAG GT TTTTC A | TAT | 10 |
| Y. pestis | 3 | TTT GCCGGAT AC AGCCG A | TAG | 12 |
| Y. pestis | 1 | TAC GCCGGAT TA CCCAA | CTG | 9 |
| Y. pestis | 1 | AAT GCCGGTC TG GGGT | AAC | 10 |
| Y. pestisb | 3 | TTT GGGGTTC AG TGCA | CGG | 6 |
| Y. pestisb | 1 | TTT GGGGTTC AG TGCACG | GCC | 7 |
| K. pneumoniae | 1 | TGG GCCGGAT GG GCCGA | CAG | 10 |
| K. pneumoniae | 1 | CCT GCCGGAT GA CACGT | TTA | 13 |
| K. pneumoniae | 1 | CAG GCCGGAT GG AACGG | GGT | 12 |
| K. pneumoniae | 1 | ACG GCCTTCG GG GGCGA | AGTG | 10 |
| K. pneumoniae | 1 | ACT TCCACCA GC AGCGG | AAC | 9 |
| K. oxytoca | 1 | AGT GCCTGAT GG GCGGC | ATG | 13 |
| K. oxytoca | 1 | ACC GCCGGAT CG CGG | CGT | 9 |
| K. oxytoca | 1 | GAC GCCGGAT AC CTGC | GCG | 9 |
| E. coli | 1 | CTT GCCGGAT AT TCA | CAG | 9 |
| E. coli | 1 | CAA GCCGGAT CG TCCGG | GAAC | 10 |
Flanking sequences left to insertion sites are aligned under the E. coli Y PU consensus 5′ end. Identities with the E. coli Y PU consensus are underlined, and identities with the insertion consensus that we have defined (5′ GCCGG3′) are twice underlined. Duplicated nucleotides are indicated in bold.
Insertion in these clones occurred at the same location on the chromosome, but the exact site of transposition in the last clone is shifted by 2 bp compared with the three others.
Class III: nontypical transposition events.
Nine transposition events (Fig. 3) occurred next to the same Y. pestis IS as in class IIB, but they were different for the following reasons. (i) No target duplication was observed. (ii) Three nucleotides, either ATT or AAT, were found between the end of the target site (Y. pestis IR) and IROK. These nucleotides are flanking IS1397 IRs in pBLOCK or pBLOCK+. (iii) Changes in the end of the IS1397 IR, which is distal to Y. pestis IR, were observed: 5′TGCAGTTCA3′ (IS1397 IRL) in clone III.d or 5′TTCAGTTCA3′ (IS1397 IRR) in clones III.a to III.c and III.e to III.h were changed into 5′TTCAGTGCA3′ (change in italics). In all cases, the original sequence was converted into the sequence found at the target site. In clone III.i, 5′TTCAGTTCA3′ (IS1397 IRR) was unchanged, while the sequence of Y. pestis IS at this position is 5′TTCACTTCA3′.
What was observed for class II clones corresponds to the hypothetical classical pathway for IS1397 transposition, which consists of the excision of the transposable module encompassing 3 bp flanking either IRR or IRL, followed by a joining. The formation of such circles as transposition intermediates is now well documented in a number of cases (14, 29). After removal of the three extra base pairs, the IS is integrated into its target, with a 3-bp duplication.
Instead, in the class III clones, 3 bp, 5′ATT3′ or 5′AAT3′, which are originally flanking, respectively, the 5′ and 3′ ends of the transposable module on pBLOCK (Fig. 1), were inserted between the Y. pestis resident IS and the transposable module. The trinucleotide 5′ATT3′ sequence is found in six clones, either next to IRL (clones III.c and III.f to III.i) or to IRR (clone III.d), keeping the same configuration as on pBLOCK (i.e., 5′ to the transposable module). However, three clones (clones III.a, III.b, and III.e) displayed the nucleotides 5′AAT3′ at the 5′ end of the transposable module (whereas it is found at its 3′ end on pBLOCK).
Moreover, in the class III clones, not only were the three flanking nucleotides kept, but also the end of the distal IR (actually IRR in eight cases and IRL in one case) was substituted for the sequence found in the Y. pestis target IR. One possible mechanism for the final structure is depicted in Fig. 5. The first step of transposition would be as previously described, leading to the formation of a circular intermediate. A 3-bp shift of the cutting site would lead to a deletion of the last 3 bp of either IS1397 IRs (TCA in both cases), and three extra base pairs flanking either IR (AAT or ATT, AWT in the figure) would be kept instead. This could explain the presence of the 3 bp which are flanking the module. The further steps of transposition would be as described previously: the 3-bp duplication of the insertion site (the three last base pairs of Y. pestis IR) would reconstitute a complete but modified IS1397 IR. In clone III.i, IS1397 IRs were unchanged. However, in this particular example, Y. pestis IR ends with CTTCA whereas IS1397 IRR ends with GTTCA. This means that at the most, the four last nucleotides could have been exchanged with no apparent change in the end product.
FIG. 5.
Hypothetical pathways for the formation of class III (see Fig. 3) clones. Top, the donor plasmid (either pBLOCK or pBLOCK+) is cut 3 bp after IRR (1) or before IRL (2). After recircularization of the module (middle part), the three last base pairs (AWT, which stands for ATT or AAT) of IRL (left) or IRR (right) are removed, instead of the three extra bases, and the module is integrated into the target with a 3-bp duplication (lower part).
Our hypothesis is strengthened by the examination of clone I.a, which harbored a circle where the transposable module has an intact IRR linked to a modified IRL (Y. pestis type) by the trinucleotide AAG. It can be assumed that this circle originates from a molecule different from the initial plasmid (pBLOCK), presumably from a chromosomal insertion site with a structure similar to clone III.d (i.e., IROK containing a modified IRL inserted next to a Y. pestis IR). It is interesting that one of the Y. pestis IRs (at nucleotide 4,417,515) is flanked by AAG, so that the circle observed in clone I.a could come out from the excision of IS1397 previously integrated at this site.
The analysis of the associations between IS1397 IRs and the flanking 3 bp showed that six clones displayed the AAT-IR parental structure (found on the donor plasmid) and three clones had the ATT-IR recombinant structure (Fig. 5). This shows that IS1397 transposase did not strongly discriminate between IRL and IRR for excision and entrapping of the flanking 3 bp (Fig. 5, top). Contrastingly, eight clones had the structure Y. pestis IR- A(A/T)T-IRL-modified IRR and only one (clone III.d) was Y. pestis IR-ATT-IRR-modified IRL, indicating that the transposition intermediates were more often opened inside IRR than IRL (Fig. 5, bottom). It should be noted that the terminal region of Y. pestis IRs (which are involved in catalysis [7]) is more similar to the IS1397 IRR than IRL (Fig. 4), which could be related to the prevalence of the first category (Y. pestis IR-A(A/T)T-IRL-modified IRR). Clone III.d is actually peculiar, since IS1397 IRL exhibited a structure (two changes in the last 8 bp) which is not explained by the model above.
In order to know whether class III nontypical transposition events, all occurring next to the Y. pestis IS, were due to the activity of Y. pestis endogenous transposase(s) rather than to IS1397 transposase, we tested whether transposition of IROK cloned into a transposase-free plasmid could be selected, using SacB toxicity. We first tested for E. coli whether sucrose toxicity was sufficient to counter-select pBLOCKS. Since we did not know if the transposase production under uninduced conditions (i.e., without IPTG) was sufficient to promote transposition, we also induced transposase production under sublethal conditions, i.e., 10−6 M IPTG. Twelve independent clones transformed with pBLOCKS were streaked on plates containing Sac and KAN or Sac, KAN, and 10−6 M IPTG. In each case, 152 colonies were subsequently checked for plasmid loss on plates containing either CHL or 10−3 M IPTG. We obtained 146 independent Kmr IPTGR Cms clones after limited transposase induction (IPTG, 10−6 M) and 129 independent Kmr IPTGR Cms clones without transposase induction. Ten of each were examined for the presence of IROK on the chromosome by Southern blot hybridization and displayed one or two bands ranging from 2.5 to 10 kb (data not shown). We sequenced the new flanking regions of the transposable module for six candidates (three induced and three not induced). In all cases, an insertion into the loop of a PU was observed (data not shown), confirming that selection of transposition events can be selected by sucrose toxicity. This result shows that the uninduced level of OrfAB production is sufficient to promote transposition. In a next step, pIROKCS was used to test whether transposition of IROK could occur into E. coli and Y. pestis chromosomes without IS1397 transposase. Twelve independent clones containing pBLOCKS were streaked on plates containing Sac and KAN. In the cases of both E. coli and Y. pestis, all clones obtained had kept the plasmid since they were all CHL resistant. This indicates that IS1397 transposase is indispensable for IROK transposition in both E. coli and Y. pestis and that class III transposition events are not due to an endogenous Y. pestis transposase activity alone. We can hypothesize that class III transposition events could result from co-action of an endogenous Y. pestis transposase and IS1397 transposase. Further experiments outside the scope of this paper would be necessary to confirm this hypothesis.
Conclusion.
Our previous results demonstrated that IS1397 transposes preferentially into PUs in PU-containing species (6, 31), with a preference for E. coli consensus PUs. In the present study, we show that for Y. pestis, a species which does not contain PUs, transposition can take place. Circles and nontypical transposition events that we observed suggest that IS1397 has adopted the two-step transposition mechanism (23) already described for other IS3 family members (25). We demonstrate here our previous hypothesis: IS1397 transposition can take place via direct recognition of a consensus (5′GCCGG3) target sequence which shares sequence similarities with PUs and which is found 11 to 13 bp upstream from the insertion site. This suggests that transposition of IS1397 does not necessarily involve PU-associated structures. Indeed the results with Y. pestis show that no PU specific host factor is strictly required for IS1397 transposition. However, IS1397 transposition depends on factors other than sequence recognition. First, for all species so far examined, hot spots of insertion were observed, which indicates a possible role of local chromosomal structures. Second, for E. coli, PUs are almost exclusive targets for IS1397 rather than other sites containing the consensus 5′GCCGG3′ sequence, which are, however, abundantly represented throughout the chromosome outside PUs. This indicates that the consensus sequence is sufficient to enable IS1397 transposition but that the specificity of insertion into PUs could be due to PU structure and/or specifically bound factors.
Acknowledgments
We thank M. Chandler for helpful discussion.
This work was supported in part by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires from Ministère Français de l'Education Nationale de la Recherche et de la Technologie.
Footnotes
Dedicated to M. Hofnung, who died on 28 June 2001 during the writing of this paper.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bachellier, S., J.-M. Clément, and M. Hofnung. 1999. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 150:627-639. [DOI] [PubMed] [Google Scholar]
- 3.Bachellier, S., J.-M. Clément, M. Hofnung, and E. Gilson. 1997. Bacterial interspersed mosaic elements (BIME) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics 145:551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachellier, S., W. Saurin, D. Perrin, M. Hofnung, and E. Gilson. 1994. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs). Mol. Microbiol. 12:61-70. [DOI] [PubMed] [Google Scholar]
- 5.Boccard, F., and P. Prentki. 1993. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 12:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clément, J.-M., C. Wilde, S. Bachellier, P. Lambert, and M. Hofnung. 1999. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J. Bacteriol. 181:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbyshire, K. M., L. Hwang, and N. D. Grindley. 1987. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc. Natl. Acad. Sci. USA 84:8049-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espéli, O., and F. Boccard. 1997. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol. Microbiol. 26:767-777. [DOI] [PubMed] [Google Scholar]
- 9.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 10.Gilson, E., J.-M. Clément, D. Brutlag, and M. Hofnung. 1984. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 3:1417-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilson, E., D. Perrin, and M. Hofnung. 1990. DNA polymerase I and a protein complex bind specifically to E. coli palindromic units highly repetitive DNA: implications for bacterial chromosome organization.Nucleic Acids Res. 18:3941-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haren, L., B. Ton-Huang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 14.Kiss, J., and F. Olasz. 1999. Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Mol. Microbiol. 34:37-52. [DOI] [PubMed] [Google Scholar]
- 15.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon, J., and M. Chandler. 1998. Insertion Sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez, E., B. Bartolome, and F. De La Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ-alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 18.Metcalf, W., W. Jiang, and B. L. Wanner. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Odaert, M., A. Devalckenaere, P. Trieu-Cuot, and M. Simonet. 1998. Molecular characterization of IS1541 insertions in the genome of Yersinia pestis. J. Bacteriol. 180:178-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oppenheim, A. B., K. E. Rudd, I. Mendelson, and D. Teff. 1993. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol. Microbiol. 10:113-122. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polard, P., M. F. Prere, O. Fayet, and M. Chandler. 1992. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 11:5079-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau, P., C. Normand, C. Loot, C. Turlan, R. Alazard, G. Duval-Valentin, and M. Chandler. 2002. Transposition of IS911, p. 367. In N. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 24.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sekine, Y., K. Aihara, and E. Ohtsubo. 1999. Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol. 294:21-34. [DOI] [PubMed] [Google Scholar]
- 26.Shafferman, A., R. Kolter, D. Stalker, and D. R. Helinski. 1982. Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J. Mol. Biol. 161:57-76. [DOI] [PubMed] [Google Scholar]
- 27.Stalker, D. M., R. Kolter, and D. R. Helinski. 1982. Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J. Mol. Biol. 161:33-43. [DOI] [PubMed] [Google Scholar]
- 28.Stern, M. J., E. Prossnitz, and G. Ferro-Luzzi Ames. 1988. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol. 2:141-152. [DOI] [PubMed] [Google Scholar]
- 29.Ton-Hoang, B., L. Polard, C. Haren, C. Turlan, and M. Chandler. 1999. IS911 transposon circles give rise to linear forms that undergo integration in vitro. Mol. Microbiol. 32:617-627. [DOI] [PubMed] [Google Scholar]
- 30.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilde, C., S. Bachellier, M. Hofnung, and J.-M. Clément. 2001. Transposition of IS1397 in the family Enterobacteriaceae and first characterization of ISKpn1, a new insertion sequence associated with Klebsiella pneumoniae palindromic units. J. Bacteriol. 183:4395-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, Y., and G. Ferro-Luzzi Ames. 1988. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc. Natl. Acad. Sci. USA 85:8850-8854. [DOI] [PMC free article] [PubMed] [Google Scholar]