Abstract
The microbial degradation of xylan is a key biological process. Hardwood 4-O-methyl-d-glucuronoxylans are extensively decorated with 4-O-methyl-d-glucuronic acid, which is cleaved from the polysaccharides by α-glucuronidases. In this report we describe the primary structures of the α-glucuronidase from Cellvibrio mixtus (C. mixtus GlcA67A) and the α-glucuronidase from Pseudomonas cellulosa (P. cellulosa GlcA67A) and characterize P. cellulosa GlcA67A. The primary structures of C. mixtus GlcA67A and P. cellulosa GlcA67A, which are 76% identical, exhibit similarities with α-glucuronidases in glycoside hydrolase family 67. The membrane-associated pseudomonad α-glucuronidase released 4-O-methyl-d-glucuronic acid from 4-O-methyl-d-glucuronoxylooligosaccharides but not from 4-O-methyl-d-glucuronoxylan. We propose that the role of the glucuronidase, in combination with cell-associated xylanases, is to hydrolyze decorated xylooligosaccharides, generated by extracellular hemicellulases, to xylose and 4-O-methyl-d-glucuronic acid, enabling the pseudomonad to preferentially utilize the sugars derived from these polymers.
Xylan, a β-1,4-linked polymer of xylose, is the most abundant hemicellulose in the majority of plant tissues. The backbone of this polysaccharide is decorated with acetate and sugar moieties that include 4-O-methyl-d-glucuronic acid, linked α-1,2 to the xylan chain. The uronic acid is released from the decorated xylose polymer by α-glucuronidases (4). The primary structures of the eight α-glucuronidases known exhibit similarities and have been placed in glycoside hydrolase family 67 (GH67) (14, 21; http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). The target substrate for the GH67 enzymes is variable. Several bacterial α-glucuronidases appear to cleave only substituted xylooligosaccharides (4, 26), although the enzyme from at least one microbial eukaryote releases 4-O-methyl-d-glucuronic acid from decorated xylan (17).
One of the most extensively characterized xylan-degrading prokaryotes is Pseudomonas cellulosa (2, 7-9, 13, 16, 19). Until now no gene encoding an α-glucuronidase has been isolated from this organism. In this study we characterized a P. cellulosa α-glucuronidase gene (agu67A) and its encoded product (α-glucuronidase 67A [P. cellulosa GlcA67A]), and here we report the sequence of a homologous gene encoding a GH67 α-glucuronidase from Cellvibrio mixtus. The data show that P. cellulosa GlcA67A is membrane bound and hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. The sequences of P. cellulosa and C. mixtus 16S ribosomal DNA indicate that these organisms belong to the same genus, which is not Pseudomonas. The role that P. cellulosa GlcA67A plays in hemicellulose degradation is discussed below.
Characterization of Pseudomonas and Cellvibrio α-glucuronidase genes.
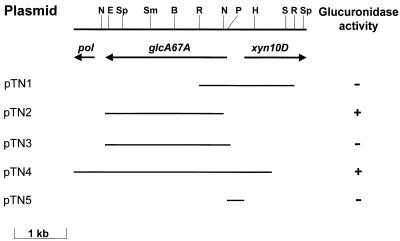
Previously, P. cellulosa and C. mixtus were shown to contain similar xylanase genes, identified as xyn10D (9). In this study the region upstream of xyn10D was isolated from gene libraries of P. cellulosa and C. mixtus (constructed in λZAPII) by using xyn10D as the probe. Approximately 40,000 recombinants from the previously constructed libraries (19) were plated on NZYM top agar at a density of 3 plaques per cm2 and were subjected to plaque hybridization by using the 5′ regions of xyn10D from C. mixtus and P. cellulosa as probes (9). By using custom-made primers, the complete sequence of the cloned region was determined with an ABI Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit and an Applied Biosystems 377A sequencing system. The data revealed a homologous gene, designated agu67A, in both the Pseudomonas and Cellvibrio cloned DNA, which was transcribed in the strand opposite xyn10D and comprised 2,196 and 2,040 bp, respectively. The proteins encoded by P. cellulosa and C. mixtus agu67A, which were designated P. cellulosa GlcA67A and C. mixtus GlcA67A, respectively, had Mrs of 83,027 and 79,838. A physical map of the Pseudomonas agu67A locus is shown in Fig. 1. Upstream (10 bp) of the assigned translational start codon of C. mixtus agu67A is the motif GGAGGA, which strongly resembles a prokaryotic ribosome binding site (12). The initiation codon for P. cellulosa agu67A is less apparent. Two possible candidates are at positions 1 and 64 in the open reading frame; they are both preceded approximately 7 bp by sequences that resemble weak Shine-Dalgarno sequences. The ATG at position 1 was designated the initiation codon as the sequence downstream of this trinucleotide encodes an amino acid sequence that exhibits similarity to a prokaryotic signal peptide (see below), consistent with the membrane location of the pseudomonad enzyme. Southern hybridization performed with the agu67A genes as probes showed that single copies of the α-glucuronidase genes were present in the two bacterial genomes (data not shown).
FIG. 1.
Physical map of agu67A locus. The positions of the cleavage sites for the following restriction enzymes are indicated: BamHI (B), EcoRI (R), EcoRV (E), HindIII (H), NcoII (N), PstI (P), SalI (S), SmaI (Sm), and SphI (Sp). The arrows show the extents and orientations of the genes encoding GlcA67A (agu67A), Xyn10D (xyn10D), and truncated DNA polymerase II (pol). Plasmid pTN4 was derived from recombinant phage, and pTN3, which encodes full-length GlcA67A, was a PCR product derived from pTN4. Plasmid pTN2, which encodes mature GlcA67A (residues 22 to 732), was derived from pTN3 by cloning the 2.1-kb NcoI-XhoI fragment into the expression vector pET32c. In pTN3 the α-glucuronidase gene was cloned into the pET expression vector pET28a, while pTN5 was derived by cloning a PCR product into the promoter probe vector pRG960SD. Glucuronidase activity refers to the presence of functional soluble enzyme activity in Escherichia coli strains carrying the plasmids.
Sequence and translation of P. cellulosa agu67A and C. mixtus agu67A.
The P. cellulosa and C. mixtus agu67A genes and their encoded proteins showed 71 and 76% sequence identity, respectively. Comparison of P. cellulosa GlcA67A and C. mixtus GlcA67A with protein databases revealed similarity to proteins in GH67, and these enzymes exhibited the highest homology (51%) with the α-glucuronidase from Caulobacter crescentus. P. cellulosa GlcA67A contained a predicted signal peptide comprising a basic hydrophilic N terminus followed by a stretch of small hydrophobic residues capable of forming an α-helix. A possible cleavage site is located between Ala-22 and Gln-23. In contrast, C. mixtus GlcA67A lacked an N-terminal signal peptide. These two GlcA67As do not appear to be modular enzymes; there was no evidence of the serine-rich linker sequences or noncatalytic carbohydrate binding modules that are prevalent in the cellulases and xylanases of these bacteria (8, 12, 13, 16, 19).
The similarity of the primary structures of all known fungal and prokaryotic α-glucuronidases, including P. cellulosa GlcA67A and C. mixtus GlcA67A, suggests that these enzymes evolved from a common ancestral protein, which is in contrast to the findings for other plant cell wall-degrading enzymes. The findings for other plant cell wall-degrading enzymes could be a consequence of the complexity of the substrates, which require a range of different enzymes with varied specificities and modes of action for complete hydrolysis. In contrast, the target substrate for α-glucuronidase is relatively simple, an α-1,2-glycosidic bond between 4-O-methyl-d-glucuronic acid and xylose. Alternatively, α-glucuronidases may have evolved relatively recently, which would be consistent with the lack of 4-O-methyl-d-glucuronoxylan in ancient plant species, and thus there has been insufficient time for the appearance of multiple α-glucuronidase families through convergent or divergent evolutionary mechanisms.
16S rRNA sequences of P. cellulosa and C. mixtus.
The primary structures of plant cell wall hydrolases from P. cellulosa and C. mixtus are very similar (9, 13, 19), suggesting that there is an evolutionary link between these two prokaryotes. To investigate this possible relationship, the 16S rRNA genes of the two organisms were amplified by PCR by using primers which bind to the 5′ and 3′ ends of eubacterial 16S ribosomal DNA (15), and the genes were sequenced. The data showed that the two sequences exhibited 95% sequence identity, indicating that the two bacteria could be classified in the same genus but are different species. Neither of the bacteria could be classified as P. cellulosa as the most closely related pseudomonad 16S rRNA sequence exhibited only 91% similarity with the P. cellulosa gene. Based on these data, we propose that P. cellulosa should be reclassified as Cellvibrio cellulosa.
Biochemical properties of P. cellulosa GlcA67A.
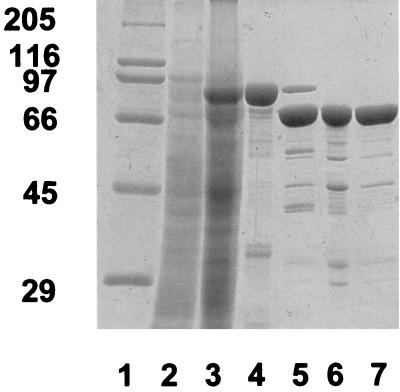
Attempts to express full-length P. cellulosa agu67A (pTN3) failed to produce functional α-glucuronidase as the encoded protein formed inclusion bodies that could not be resolubilized. Plasmid pTN2, a recombinant of pET32C (Novagen) that encoded mature P. cellulosa GlcA67A (residues 22 to 732) fused to the C terminus of thioredoxin, directed the synthesis of a functional α-glucuronidase. The fusion protein, containing an N-terminal His tag, was purified by metal ion affinity chromatography. P. cellulosa GlcA67A released from the fusion protein by the proteinase enterokinase was then further purified by ion exchange and gel filtration (Fig. 2); details of the purification method used were described by Nurizzo et al. (20).
FIG. 2.
Purification of GlcA67A. GlcA67A was purified as described in the text. The various fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis by using a 10% (wt/vol) polyacrylamide gel which contained the following samples: high-molecular-weight standards (lane 1); cell extract of E. coli containing pTN2, cultured in the absence (lane 2) and in the presence (lane 3) of isopropyl-β-d-thiogalactopyranoside (IPTG) (2); and protein purified by metal ion affinity chromatography (lane 4) and after enterokinase treatment (lane 5), anion-exchange chromatography (lane 6), and size exclusion chromatography (lane 7).
The activity of purified P. cellulosa GlcA67A against 4-O-methyl-d-glucuronoxylooligosaccharides was determined by high-performance liquid chromatography. Briefly, substrates were incubated with enzyme in 50 mM sodium phosphate-12 mM citrate buffer (pH 6.5) (PC buffer) for various times at 37°C. At regular intervals aliquots were removed, boiled for 10 min to stop the reaction, and subjected to high-performance liquid chromatography as described previously by using a Carbopac PA-100 column (18). The enzyme released 4-O-methyl-d-glucuronic acid from aldobiouronic acid, aldotriouronic acid, aldotetraouronic acid, and aldopentaouronic acid with specific activities against these substrates (at a concentration of 2 mM) of 58.2 ± 6.9, 61.3 ± 4.1, 41.4 ±3.5, and 84.5 ± 11.7 U/mg of protein, respectively. To evaluate the activity of P. cellulosa GlcA67A against polysaccharides, the colorimetric assay of Khandke et al. was used (17). The reactions were carried out in 100 mM sodium maleate buffer (pH 6.3) at 37°C, aliquots were removed, and the presence of 4-O-methyl-d-glucuronic acid was determined (17). Although the enzyme (10 U) did not release 4-O-methyl-d-glucuronic acid from 4-O-methyl-d-glucuronoxylan, the glycoside hydrolase did produce 20 ± 3 μmol of the uronic acid in 25 min when the substrate had been pretreated with 100 U of P. cellulosa xylanase 10A for 3 h. These data indicate that P. cellulosa GlcA67A does not contain an active site that comprises an open cleft, which is typical of endo-acting glycoside hydrolases (5). Furthermore, as P. cellulosa GlcA67A exhibits similar activities against substrates containing one, two, and three xylose residues, the enzyme is likely to interact only with the aglycone sugar that participates in the target glycosidic bond. The importance of the aglycone moiety is supported by the results of a previous study, which showed that the Clostridium stercorarium and Thermoanaerobacterium saccarolyticum α-glucuronidases did not hydrolyze 4-nitrophenyl-α-d-glucuronide but cleaved 4-O-methyl-d-glucuronoxylooligosaccharides (4, 24). Furthermore, all the substrates evaluated in this study contain the uronic acid decoration at the nonreducing end of the xylose polymer (as indicatred in the Megazyme catalogue). It is possible, therefore, that P. cellulosa GlcA67A cleaves only 4-O-methyl-d-glucuronic acid linked to the nonreducing end of xylose polymers. This view is supported by the crystal structure of GlcA67A, which shows that the enzyme can only accommodate 4-O-methyl-d-glucuronic acid attached to a xylose moiety in which the 4′ hydroxyl is not linked to another sugar (20). Furthermore, Biely et al. showed that the Aspergillus tubingensis α-glucuronidase displayed absolute specificity for 4-O-methyl-d-glucuronic acid groups linked to the nonreducing ends of xylooligosaccharides (3). The pH optimum of P. cellulosa GlcA67A was 6.3, and the enzyme displayed 20 and 60% of the maximal activity at pH 5 and 8, respectively. The enzyme was rapidly inactivated at temperatures above 55°C and was susceptible to proteinase attack, as shown by the observation that Glc67A was inactivated by trypsin at a rate similar to the inactivation rate for the intracellular enzyme malate dehydrogenase. Gel filtration showed that the enzyme had an Mr of 150,000, as judged by gel filtration (data not shown), indicating that it had a dimeric structure.
Expression and location of GlcA67A in P. cellulosa.
P. cellulosa was cultured on Luria broth supplemented with various saccharides at 37°C with aeration, and the level and cellular location of the α-glucuronidase activity were determined. The data (Table 1) showed that the enzyme was expressed only on media containing xylan, while glucose repressed the synthesis of the glycoside hydrolase. RNA dot blots showed that the appearance of agu67A mRNA and α-glucuronidase activity coincided (data not shown). To probe the expression pattern of P. cellulosa agu67A further, the 456-bp 5′ nontranslated region of the gene was cloned into the reporter probe vector pRG9600SD (25) immediately upstream of uidA (encoding β-glucuronidase) to generate pTN5. Wild-type P. cellulosa did not display β-glucuronidase activity, while the bacterium containing pTN5 expressed β-glucuronidase activity when xylan was added to the culture medium (Table 1) but not in the presence of glucuronic acid. Glucose, but not glucuronic acid, repressed uidA expression when the prokaryote was grown on media containing xylan. The pattern of GlcA67A expression is similar to the patterns of expression of α-glucuronidases from other prokaryotes and microbial eukaryotes (6, 22) and is consistent with the assembly of a repertoire of degradative enzymes in response to the presence of a complex substrate, such as xylan.
TABLE 1.
Influence of growth conditions on the expression of glucuronidase by P. cellulosa
| Carbohydrate added to Luria-Bertani growth mediuma | Enzyme activityb
|
|
|---|---|---|
| α-Glucuronidasec | β-Glucuronidased | |
| None | NDe | ND |
| 4-O-Methyl-d-glucuronoxylan | 45 ± 5 | 32 ± 4 |
| Oat spelt xylan | 75 ± 6 | 60 ± 6 |
| Wheat arabinoxylan | 32 ± 3 | 33 ± 7 |
| Rye arabinoxylan | 51 ± 6 | 41 ± 7 |
| Oat spelt xylan and glucose | ND | ND |
| Glucuronic acid | ND | ND |
| β-Glucan | ND | ND |
Carbohydrates were each added to a final concentration of 0.2%.
α-Glucuronidase and β-glucuronidase activities, expressed as nanomoles of product per minute per milligram of total bacterial protein, were measured when the cultures reached the mid-log phase. The substrates employed were aldobiouronic acid (2 mM) to measure α-glucuronidase activity and 4-methylumbelliferyl-β-glucuronide (1 mM) to measure β-glucuronidase activity, as described elsewhere (7).
α-Glucuronidase activity was determined in cultures of wild-type P. cellulosa.
β-Glucuronidase activity was determined in cultures of P. cellulosa harboring pTN5.
ND, no activity detected. The sensitivities of the assays were such that 100 pmol of product/min/mg of protein could be detected with the α-glucuronidase assay and 1 pmol of product/min/mg protein could be detect with the β-glucuronidase assay.
To investigate the cellular location of GlcA67A, P. cellulosa cultured on xylan was fractionated. Briefly, cells from a 400-ml stationary-phase culture were pelleted, resuspended in 10 ml of PC buffer, and sonicated at 4°C by using a Braun Labsonic U sonicator set at 2-min 0.5 s cycles and a power setting of 45. The cell membranes were then pelleted by centrifugation at 100,000 × g at 4°C for 1 h, and the supernatant, consisting of the cytoplasm and the periplasm and referred to as the cell extract, was retained for further use. The purities of the different fractions were assessed by measuring the activities of the marker enzymes malate dehydrogenase (cell extract) and NADH oxidoreductase (membrane fraction and purified inner membrane fraction) as described previously (9). The data (Table 2) confirmed the generally successful separation of the cellular compartments, although the cytoplasm-periplasm fraction contained some NADH oxidase (membrane marker). The segregation of GlcA67A was very similar to the segregation of NADH oxidase, indicating that the former enzyme is located in the membrane in the pseudomonad. Attempts to determine whether GlcA67A was in the inner or outer membrane of P. cellulosa were unsuccessful as lauryl sarcosine (which solubilizes the inner membrane) inactivated the α-glucuronidase. Although it is unclear whether the enzyme is located on the inner or outer membrane, it is interesting that the α-glucuronidase activity displayed by whole cells of P. cellulosa is similar to that displayed by the membrane fraction (data not shown), indicating that the enzyme is situated on the outer membrane. It should be noted, however, that GlcA67A is susceptible to trypsin inactivation, while all other extracellular P. cellulosa hydrolases characterized to date are resistant to proteolytic attack (10, 11, 23). Thus, although the α-glucuronidase is cell associated and is secreted out of the cytoplasm, the precise cellular location of the enzyme remains uncertain. The mechanism by which P. cellulosa GlcA67A is membrane associated is unclear. It is possible that the enzyme contains an uncleaved signal peptide, in which case the predicted cleavage site (Ala-22-Gln-23) would not be recognized by the signal peptidase. If this is correct, then the sequence from Gln-23 to Asp-29, which is not present in C. mixtus Glc67A, may function as a small linker between the enzyme and the membrane.
TABLE 2.
Cellular localization of α-glucuronidasea
| Fraction | % of:
|
||
|---|---|---|---|
| Malate dehydrogenase | NADH oxidoreductase | α-glucuronidase | |
| Cell extract | 93 ± 5 | 28 ± 2 | 32 ± 4 |
| Culture medium | 0 | 0 | 0 |
| Total membrane | 7 ± 5 | 72 ± 2 | 68 ± 4 |
Wild-type P. cellulosa was grown to the late exponential phase on medium containing xylan and was fractionated into the different cellular components as described in Materials and Methods. The enzymes malate dehydrogenase and NADH oxidoreductase were markers for the cell extract (combined cytoplasm and periplasm) and membrane fractions, respectively. A value of 0% indicates that no catalytic acitivity was detected above the sensitivity of the assay, which was 100 pmol of product/min/mg of protein.
The observation that GlcA67A is cell associated suggests a mechanism by which the bacterium can preferentially utilize the nutrients derived from decorated xylans. Thus, extracellular xylanases digest 4-O-methyl-d-glucuronoxylan into 4-O-methyl-d-glucuronoxylooligosaccharides that are not further metabolized by β-xylosidases, enzymes that are expressed by numerous microorganisms. It appears that the xylanases, at least those from P. cellulosa, can accommodate 4-O-methyl-d-glucuronic acid at the +1 and −3 subsites but not at the −1 or −2 subsite (Gilbert, unpublished data). Thus, hydrolysis of 4-O-methyl-d-glucuronoxylan by the Pseudomonas enzymes catalyzes formation of aldotetraouronic acid in which 4-O-methyl-d-glucuronic acid is linked to the nonreducing terminus of β-1,4-linked xylotriose, preventing attack by β-xylosidases. These oligosaccharides are hydrolyzed on the P. cellulosa membrane by a combination of P. cellulosa GlcA67A and the xylanases Xyn10C and Xyn10D (7), releasing xylose and 4-O-methyl-d-glucuronic acid, which are available to the pseudomonad. This view is consistent with the substrate specificity of P. cellulosa GlcA67A for oligosaccharides rather than polysaccharides. Support for these proposals is provided by the observation that the major 4-O-methyl-d-glucuronoxylooligosaccharide produced by xylanases in the culture medium is aldotetraouronic acid. The location of the uronic acid at the nonreducing end was confirmed by the observation that the oligosaccharide was not cleaved by a β-xylosidase. Furthermore, when aldotetraouronic acid was incubated with whole cells, the major products generated in the extracellular buffer were 4-O-methyl-d-glucuronic acid, xylose, and xylobiose. It is interesting that the primary arabinofuranosidase (Abf51A) of P. cellulosa is also located on the outer membrane of the bacterium (1) and thus may play a role similar to that of GlcA67A. Because Abf51A cleaves the arabinose moieties from arabinoxylooligosaccharides at the surface of the bacterium, both the furanose sugar and the xylose liberated by the action of cell-associated xylanases on the resultant linear xylooligosaccharide can preferentially be utilized by the pseudomonad. Thus, a picture of how P. cellulosa hydrolyzes decorated xylans is beginning to emerge. The polysaccharides are initially cleaved by xylanases into substituted xylooligosaccharides; enzymes associated with the bacterial surface then remove the side chains of these oligomers, and the liberated sugars, together with the xylooligosaccharides, are preferentially metabolized by the bacterium. We further suggest that the proposed role for P. cellulosa GlcA67A can be extended to other prokaryotic α-glucuronidases which have an intracellular location and appear to act on 4-O-methyl-d-glucuronoxylooligosaccharides but not on 4-O-methyl-d-glucuronoxylan (21, 26).
Nucleotide sequence accession numbers.
The nucleotide sequences of the P. cellulosa agu67A and C. mixtus agu67A genes have been deposited in the GenBank database under accession numbers AY065638 and AY065639, respectively. The GenBank accession numbers for the sequences of P. cellulosa and C. mixtus 16S rRNA are AF452103 and AF448513, respectively
REFERENCES
- 1.Beylot, M. H., K. Emami, V. A. McKie, H. J. Gilbert, and G. Pell. 2001. Pseudomonas cellulosa expresses a single membrane-bound glycoside hydrolase family 51 arabinofuranosidase. Biochem. J. 358:599-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beylot, M. H., V. A. McKie, A. G. Voragen, C. H. Doeswijk-Voragen, and H. J. Gilbert. 2001. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem. J. 358:607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biely, P., R. P. de Vries, M. Vrsanska, and J. Visser. 2000. Inverting character of alpha-glucuronidase A from Aspergillus tubingensis. Biochim. Biophys. Acta 1474:360-364. [DOI] [PubMed] [Google Scholar]
- 4.Bronnenmeier, K., H. Meissner, S. Stocker, and W. L. Staudenbauer. 1995. α-d-Glucuronidases from the xylanolytic thermophiles Clostridium stercorarium and Thermoanaerobacterium saccharolyticum. Microbiology 141:2033-2040. [DOI] [PubMed] [Google Scholar]
- 5.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 6.de Vries, R. P., C. H. Poulsen, S. Madrid, and J. Visser. 1998. aguA, the gene encoding an extracellular alpha-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J. Bacteriol. 180:243-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emami, K., T. Nagy, C. M. G. A. Fontes, L. M. A. Ferreira, and H. J. Gilbert. Evidence for temporal regulation of Pseudomonas cellulosa xylanases belonging to glycoside hydrolase family 11. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 8.Ferreira, L. M., T. M. Wood, G. Williamson, C. Faulds, G. P. Hazlewood, G. W. Black, and H. J. Gilbert. 1993. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem. J. 294:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontes, C. M., H. J. Gilbert, G. P. Hazlewood, J. H. Clarke, J. A. Prates, V. A. McKie, T. Nagy, T. H. Fernandes, and L. M. Ferreira. 2000. A novel Cellvibrio mixtus family 10 xylanase that is both intracellular and expressed under non-inducing conditions. Microbiology 146:1959-1967. [DOI] [PubMed] [Google Scholar]
- 10.Fontes, C. M., J. Hall, B. H. Hirst, G. P. Hazlewood, and H. J. Gilbert. 1995. The resistance of cellulases and xylanases to proteolytic inactivation. Appl. Microbiol. Biotechnol. 43:52-57. [DOI] [PubMed] [Google Scholar]
- 11.Hall, J., S. Ali, M. A. Surani, G. P. Hazlewood, A. J. Clark, J. P. Simons, B. H. Hirst, and H. J. Gilbert. 1993. Manipulation of the repertoire of digestive enzymes secreted into the gastrointestinal tract of transgenic mice. Bio/Technology 11:376-379. [DOI] [PubMed] [Google Scholar]
- 12.Hall, J., and H. J. Gilbert. 1988. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol. Gen. Genet. 213:112-117. [DOI] [PubMed] [Google Scholar]
- 13.Hall, J., G. P. Hazlewood, N. S. Huskisson, A. J. Durrant, and H. J. Gilbert. 1989. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol. Microbiol. 3:1211-1219. [DOI] [PubMed] [Google Scholar]
- 14.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphry, D. R., A. George, G. W. Black, and S. P. Cummings. 2001. Flavobacterium frigidarium sp. nov., an aerobic, psychrophilic, xylanolytic and laminarinolytic bacterium from Antarctica. Int. J. Syst. E vol. Microbiol. 51:1235-1243. [DOI] [PubMed] [Google Scholar]
- 16.Kellett, L. E., D. M. Poole, L. M. Ferreira, A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandke, K. M., P. J. Vithayathil, and S. K. Murthy. 1989. Purification and characterization of an α-d-glucuronidase from a thermophilic fungus, Thermoascus aurantiacus. Arch. Biochem. Biophys. 274:511-517. [DOI] [PubMed] [Google Scholar]
- 18.McKie, V. A., G. W. Black, S. J. Millward-Sadler, G. P. Hazlewood, J. I. Laurie, and H. J. Gilbert. 1997. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo- mode of action. Biochem. J. 323:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millward-Sadler, S. J., K. Davidson, G. P. Hazlewood, G. W. Black, H. J. Gilbert, and J. H. Clarke. 1995. Novel cellulose-binding domains, NodB homologues and conserved modular architecture in xylanases from the aerobic soil bacteria Pseudomonas fluorescens subsp. cellulosa and Cellvibrio mixtus. Biochem. J. 312:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nurizzo, D., T. Nagy, H. J. Gilbert, and G. J. Davies. 2002. The structural basis for catalysis and specificity of the Pseudomonas cellulosa α-glucuronidase, GlcA67A. Structure 10:547-556. [DOI] [PubMed]
- 21.Ruile, P., C. Winterhalter, and W. Liebl. 1997. Isolation and analysis of a gene encoding α-glucuronidase, an enzyme with a novel primary structure involved in the breakdown of xylan. Mol. Microbiol. 23:267-279. [DOI] [PubMed] [Google Scholar]
- 22.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spurway, T. D., C. Morland, A. Cooper, I. Sumner, G. P. Hazlewood, A. G. O'Donnell, R. W. Pickersgill, and H. J. Gilbert. 1997. Calcium protects a mesophilic xylanase from proteinase inactivation and thermal unfolding. J. Biol. Chem. 272:17523-17530. [DOI] [PubMed] [Google Scholar]
- 24.Uchida, H., T. Nanri, Y. Kawabata, and K. Murakami. 1992. Purification and characterisation of intracellular α-glucuronidase from Aspergillus niger 5-16. Biosci. Biotechnol. Biochem. 56:1608-1615. [Google Scholar]
- 25.Van den Eede, G., R. Deblaere, K. Goethals, M. Van Montagu, and M. Holsters. 1992. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol. Plant-Microbe Interact. 5:228-234. [DOI] [PubMed] [Google Scholar]
- 26.Zaide, G., D. Shallom, S. Shulami, G. Zolotnitsky, G. Golan, T. Baasov, G. Shoham, and Y. Shoham. 2001. Biochemical characterization and identification of catalytic residues in α-glucuronidase from Bacillus stearothermophilus T-6. Eur. J. Biochem. 268:3006-3016. [DOI] [PubMed] [Google Scholar]